Abstract
The Arabidopsis mutants eto1 (ethylene overproducer) and eto3 produce elevated levels of ethylene as etiolated seedlings. Ethylene production in these seedlings peaks at 60 to 96 h, and then declines back to almost wild-type levels. Ethylene overproduction in eto1 and eto3 is limited mainly to etiolated seedlings; light-grown seedlings and various adult tissues produce close to wild-type amounts of ethylene. Several compounds that induce ethylene biosynthesis in wild-type, etiolated seedlings through distinct 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) isoforms were found to act synergistically with eto1 and eto3, as did the ethylene-insensitive mutation etr1 (ethylene resistant), which blocks feedback inhibition of biosynthesis. ACS activity, the rate-limiting step of ethylene biosynthesis, was highly elevated in both eto1 and eto3 mutant seedlings, even though RNA gel-blot analysis demonstrated that the steady-state level of ACS mRNA was not increased, including that of a novel Arabidopsis ACS gene that was identified. Measurements of the conversion of ACC to ethylene by intact seedlings indicated that the mutations did not affect conjugation of ACC or the activity of ACC oxidase, the final step of ethylene biosynthesis. Taken together, these data suggest that the eto1 and eto3 mutations elevate ethylene biosynthesis by affecting the posttranscriptional regulation of ACS.
The gaseous hormone ethylene has been shown to influence numerous plant growth and developmental processes, including germination, root-hair initiation, leaf and flower senescence and abscission, fruit ripening, nodulation, and the response to a wide variety of stresses (Mattoo and Suttle, 1991; Abeles et al., 1992). Much progress has been made in elucidating the mechanisms of ethylene perception and signal transduction (Ecker, 1995; Kieber, 1997a, 1997b), as well as the ethylene-biosynthetic pathway (Kende, 1993). However, to fully understand the mechanism of ethylene action, it is important to delineate how its biosynthesis is regulated. We have chosen 3-d-old etiolated Arabidopsis seedlings as a model system to unravel this circuitry. Here we describe the characterization of two mutants that affect the regulation of ethylene biosynthesis in etiolated seedlings.
Almost all plant tissues have the ability to make ethylene, although in most cases the amount made is very low. Ethylene production increases dramatically during a number of developmental events such as germination, leaf and flower senescence and abscission, and fruit ripening (Yang and Hoffman, 1984; Mattoo and Suttle, 1991; Abeles et al., 1992). A diverse group of factors modify the level of ethylene biosynthesis, and a major effect of most of these is to increase the steady-state level of ACS mRNA (see Olson et al., 1991, 1995; Rottmann et al., 1991; Botella et al., 1993, 1995), although there is accumulating evidence to suggest that this enzyme is also posttranslationally regulated (Nakajima et al., 1990; Spanu et al., 1994; Oetiker et al., 1997; Vogel et al., 1998b).
The ethylene-biosynthetic pathway (for review, see Yang and Hoffman, 1984; Kende, 1993) starts with the conversion of Met to AdoMet by the enzyme Met adenosyltransferase. ACS, which converts AdoMet to ACC (Adams and Yang, 1979), is the first committed and generally rate-limiting step in ethylene biosynthesis. ACS is encoded by a small gene family comprising at least three to six members in the species that have been closely examined. Distinct subsets of ACS genes are expressed in response to various developmental, environmental, and hormonal factors.
In Arabidopsis six ACS genes have been identified (ACS1–ACS6), two of which are nonfunctional (Liang et al., 1992, 1995; Van der Straeten et al., 1992; Vahala et al., 1998). Inhibition of protein synthesis by cycloheximide treatment induces expression of the functional genes, suggesting that they are under negative control (Liang et al., 1992). Wounding, auxin, LiCl, and anaerobiosis differentially induce these genes (Liang et al., 1992, 1996; Van der Straeten et al., 1992). ACS2 expression is higher in young, developing leaves and flowers compared with more mature tissues from these organs, and its expression is also correlated with the initial stages of lateral root formation (Rodrigues-Pousada et al., 1993). ACS5 is the major isoform involved in the production of ethylene in response to low doses of cytokinin in etiolated seedlings, and this regulation is primarily via a posttranscriptional mechanism (Vogel et al., 1998b). ACS4 transcription is induced by auxin, and several auxin-responsive elements have been identified upstream of the ACS4 coding region (Abel et al., 1995). The steady-state level of ACS6 transcript is increased by treatment with ozone (Vahala et al., 1998). The ACS3 gene is most likely a pseudogene and ACS1 encodes a nonfunctional ACS (Liang et al., 1995).
The final step of ethylene biosynthesis, the conversion of ACC to ethylene, is catalyzed by the enzyme ACC oxidase, which can also play an important role in regulating ethylene biosynthesis, especially under conditions of high ethylene production (Nadeau et al., 1993; Kim and Yang, 1994; Tang et al., 1994; Barry et al., 1996; Lasserre, 1996; Mekhedov and Kende, 1996). ACC oxidase, like ACS, appears to be encoded by a gene family whose members are differentially regulated in a number of plants. There are several ACO genes in Arabidopsis (Newman et al., 1994), at least one of which is induced by ethylene itself (Gomez-Lim et al., 1993).
ACC can be conjugated to an inactive form, malonyl-ACC, by the enzyme ACC malonyltransferase (Amrhein et al., 1981; Hoffman et al., 1982; Kionka and Amrhein, 1984). A second ACC conjugate, 1-(γ-l-glutamylamino)cyclopropane-1-carboxylic acid, has also been identified (Martin et al., 1995), although recent evidence suggests that this is a much less abundant conjugate (Peiser and Yang, 1998). There is some evidence that the level of ACC conjugation is regulated, which may contribute to control of ethylene production (Jiao et al., 1986).
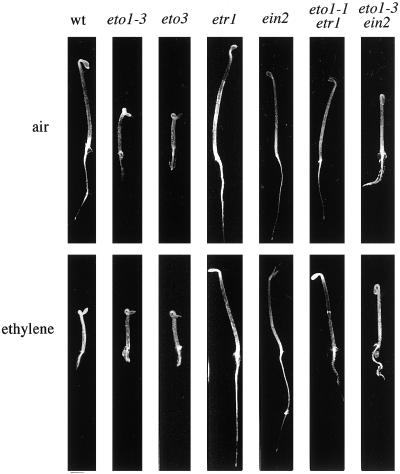
Treatment of etiolated seedlings with ethylene results in a morphology known as the triple response. In Arabidopsis the triple response consists of shortening and radial swelling of the hypocotyl, inhibition of root elongation, and exaggeration of the curvature of the apical hook (see Fig. 1). This response has been used to identify mutants disrupted in ethylene perception and signaling (Bleecker et al., 1988; Guzman and Ecker, 1990; Kieber et al., 1993), as well as mutants affected in the regulation of ethylene biosynthesis. Mutants in the latter class fall into two categories: (a) those that fail to induce ethylene in response to a particular inducer (cytokinin-insensitive mutants, Cin; Vogel et al., 1998a, 1998b); and (b) those that overproduce ethylene (ethylene overproducer, Eto; Guzman and Ecker, 1990; Kieber et al., 1993). Three Eto loci have been identified: eto1 is inherited as a recessive mutation, and eto2 and eto3 are dominant. eto2 was recently found to be the result of a disruption of the carboxy-terminal 11 amino acids of ACS5 (Vogel et al., 1998b). Here we describe the physiological characterization of the eto1 and eto3 mutants. This analysis suggests that these mutations are affected in the posttranscriptional regulation of ACS.
Figure 1.
Phenotypes of 3-d-old etiolated wild-type (wt), single-mutant, and double-mutant Arabidopsis seedlings (as indicated) grown in air (top panels) or 10 μL L−1 ethylene (bottom panels). Representative seedlings were picked and photographed.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
The Columbia ecotype of Arabidopsis was used in this study. Seeds were surface-sterilized as described previously (Vogel et al., 1998b), resuspended in a suitable volume of top agar (0.8% low-melt agarose), and spread onto Murashige and Skoog agar (Murashige and Skoog salts [GIBCO-BRL], 2% Suc, and 0.8% agar, pH 5.7). Seeds were cold treated for 4 d (4°C), exposed to light for 2 h, and then moved to a dark incubator at 23°C. The time at which the vials were moved to 23°C was designated as time 0. Adult plants were grown in potting soil (Metro Mix 250, Grace-Sierra, Boca Raton, FL) under continuous illumination at 23°C. The eto1-1 and eto3 mutants and the eto1-1/etr1-3 double mutant were identified previously (Guzman and Ecker, 1990; Kieber et al., 1993; Roman et al., 1995). The eto1-3 allele was isolated from an x-ray-mutagenized population (ecotype Columbia) (Kieber et al., 1993). This allele was used for all of the experiments, with the exception of the eto1/etr1 double-mutant analysis, which used the eto1-1 allele. The eto1-3/ein2 double mutant was obtained by crossing an eto1-3 homozygote to a ein2-1 homozygote. The F1 population was allowed to self, and seedlings that were phenotypically Eto− were then selected in the F2 line. Tall progeny from these individuals were then selected and self-set seed was collected. A line that retested as being ethylene insensitive and that overproduced ethylene was identified and used in further experiments.
Ethylene Measurement
Hormones and CuSO4 were added to about 20 2-d-old etiolated seedlings that were grown in 22-mL GC vials containing 3 mL of Murashige and Skoog agar by pipetting 200 μL of solution on top of the seedlings. An equal volume of water plus solvent was added to the control treatments. These vials were then flushed with hydrocarbon-free air and capped, and the accumulated ethylene measured 24 to 48 h later, as described previously (Vogel et al., 1998b). Ethylene production was normalized to the number of seedlings in each vial and the time between capping and sampling. All observations are from at least three replicates, and each experiment was repeated at least once with comparable results. To measure ethylene from adult plants, tissues were detached, weighed, and then placed in 22-mL vials containing 3 mL of Murashige and Skoog agar. The vials were flushed with hydrocarbon-free air, sealed, and incubated in the light for the indicated times. The amount of ethylene produced by light-grown seedlings was determined by putting capped vials in a lighted growth chamber for 72 h.
ACS Assays
ACS was assayed from 3-d-old etiolated seedlings as described previously (Peck and Kende, 1995), with some modifications. Sterilized seeds were plated on filter paper on Murashige and Skoog agar (10,000 seeds per 150-mm plate), cold incubated (4°C) for 4 d, and then moved to a dark chamber for 3 d at 23°C. Ten grams of tissue was added to 15 mL of buffer A (250 mm phosphate buffer, pH 8.0, 10 μm pyridoxal phosphate, 1 mm EDTA, 2 mm PMSF, and 5 mm DTT) and the sample was homogenized on ice for 4 min (maximum speed with a PowerGen 700 homogenizer [Fisher Scientific]). The sample was centrifuged at 15,000g for 15 min and the supernatant was respun at 15,000g for 15 min. One milliliter of the supernatant was placed into a 22-mL GC tube and 100 μL of 5 mm AdoMet was added. This was incubated for 1 h at 22°C. The ACC formed was converted to ethylene by addition of 100 μL of 20 mm HgCl, followed by 100 μL of a 1:1 mixture of saturated NaOH:bleach (Lizada and Yang, 1979). The tubes were capped immediately after addition of the NaOH/bleach and incubated on ice for 10 min. Five milliliters of headspace was removed with a syringe and injected into a new vial, and the ethylene was measured as described previously (Vogel et al., 1998b). All reactions were done in triplicate and compared with controls, to which AdoMet was not added. Protein concentration was determined using the Bradford assay as described by the manufacturer (Bio-Rad).
RNA-Blot Analysis
Total RNA was prepared as described previously (Ausubel et al., 1994) and poly(A+) RNA was isolated using Oligotex-dT resin, as described by the manufacturer (Qiagen, Chatsworth, CA). Five micrograms of poly(A+) mRNA was separated on an agarose gel, blotted to a nylon membrane, and hybridized to radiolabeled probes as described previously (Ausubel et al., 1994). Fragments corresponding to each ACS gene and β-tubulin were obtained by amplifying each from Arabidopsis genomic DNA using PCR with oligonucleotide primers specific for each gene, or in the case of ACS6, the insert from the expressed sequence tag clone FAI88 (see below) was used. The signals were quantified with a phosphor imager and normalized to the level of the β-tubulin control. This analysis was repeated once with comparable results.
Isolation of ACS7
We searched the Arabidopsis expressed sequence tag database (Newman et al., 1994) for sequences similar to those of ACS1 to ACS6. Three clones (FAI88, 288D2T7, and 240L12T7), corresponding to an identical gene, were identified that were similar to the previously identified Arabidopsis ACS genes, but encoded a novel ACS isoform. Sequence analysis revealed that these ACS7 cDNA clones were missing the 5′ portion of the coding region (see Results).
RESULTS
Ethylene Biosynthesis in the eto1 and eto3 Mutants Is Developmentally Regulated
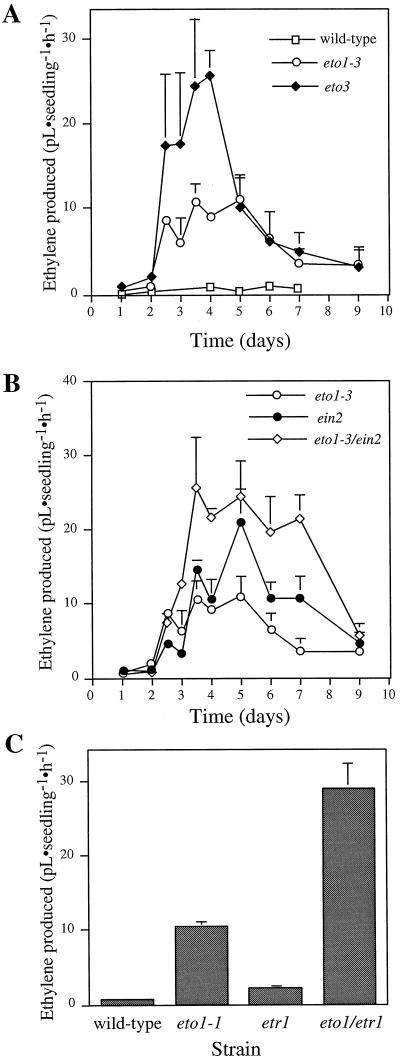
The eto1 and eto3 mutants display a constitutive triple-response phenotype as etiolated seedlings (Fig. 1) caused by an overproduction of ethylene. We analyzed ethylene biosynthesis from wild-type and Eto mutant etiolated seedlings at various intervals during the first several days of growth (Fig. 2). There was almost no detectable ethylene produced by wild-type seedlings during the early stages of germination (<24 h), followed by a low, stable level during the next 6 d. Ethylene biosynthesis in eto1-3 seedlings rose quickly between 48 and 60 h, and remained at a steady, elevated level for the next 60 h, after which it gradually declined to close to wild-type levels. eto3 mutant seedlings displayed a similar pattern, although their peak production was about 2.5-fold higher and the rate of decline of production was increased compared with that observed in eto1-3 seedlings. By d 9, both mutants produced close to wild-type levels of ethylene.
Figure 2.
Ethylene biosynthesis by wild-type, single-mutant, and double-mutant etiolated Arabidopsis seedlings. Seedlings were grown for various times on Murashige and Skoog agar in GC vials and the accumulated ethylene was measured. A, Time course of ethylene production by etiolated wild-type, eto1-3, and eto3 mutant seedlings. B, Time course of ethylene production by etiolated eto1-3, ein2, and eto1-3/ein2 double-mutant seedlings. The data points in A and B represent the ethylene accumulated during the time intervals 0 to 24, 24 to 48, 48 to 60, 60 to 72, 72 to 84, 84 to 96, 120 to 144, 144 to 168, and 168 to 216 h. The second time point in each interval was plotted as the value for the x axis. C, Ethylene accumulated by etiolated wild-type eto1-1, etr1, and eto1-1/etr1 double-mutant seedlings from 72 to 96 h. All values are means (±sd) of three replicates.
To determine if the decline in ethylene biosynthesis was caused by feedback regulation, we examined the time course of ethylene biosynthesis in the double-mutant seedlings of eto1-3 and the mutation ein2 ethylene insensitive (Guzman and Ecker, 1990). The initial rate of increase in ethylene biosynthesis was the same for both eto1-3 and eto1-3/ein2 seedlings (Fig. 2B). However, the double mutant continued to increase its rate of ethylene biosynthesis beyond 60 h, the point at which the rate in eto1-3 seedlings peaked. The peak of biosynthesis in the double mutants was close to what one would predict from an additive interaction, suggesting that these mutations act independently to regulate ethylene biosynthesis. The rate of ethylene biosynthesis in double-mutant seedlings also returned to close to that observed in wild-type seedlings, although the rate of decline was somewhat slower than in the eto1-3 single mutant. This suggests that the decline in biosynthesis is not the result of negative-feedback regulation from the elevated ethylene levels but, rather, may reflect a developmental change in the regulation of ethylene biosynthesis (although feedback regulation may alter the rate at which this occurs).
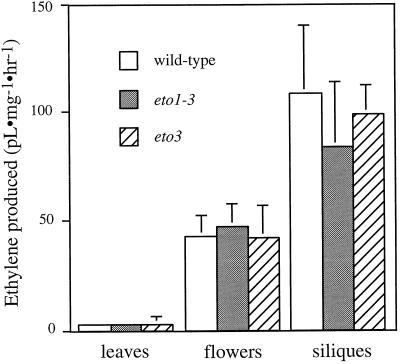
We examined ethylene production from various adult tissues to determine if any were affected by the Eto mutations. Both eto1-3 and eto3 affected ethylene biosynthesis almost exclusively in etiolated seedlings: light-grown seedlings (not shown), adult leaves, flowers, and siliques from eto1-3 and eto3 mutants produced close to wild-type levels of ethylene (Fig. 3). Thus, these mutants are either specific for etiolated seedlings or are involved in regulating ethylene biosynthesis only in the dark.
Figure 3.
Ethylene produced by adult tissues of wild-type, eto1-3, and eto3 mutant plants growing in soil at 23°C under continuous illumination. Ethylene measurements were as described in Methods. Values are means (±sd) of three replicates.
Interaction of the Eto Mutants and Various Inducers of Ethylene Biosynthesis
Ethylene biosynthesis in etiolated Arabidopsis seedlings is strongly induced by a number of plant hormones, as well as by the cupric ion. In some cases the target of these factors has been demonstrated to be distinct ACS genes (Liang et al., 1992; Abel et al., 1995; Vogel et al., 1998b). We examined the interaction of these factors and the eto1-3 and eto3 mutations to begin to address how these signaling pathways are related. The concentration of each inducer was chosen as the concentration that gave the peak of induction in wild-type etiolated seedlings (Woeste et al., 1999).
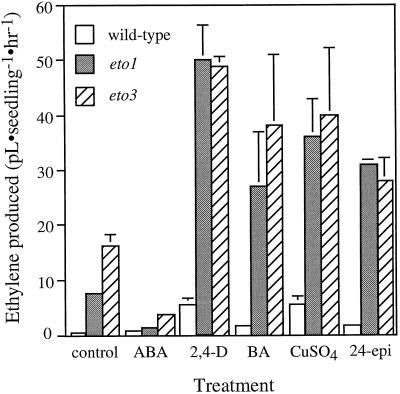
Both eto1-3 and eto3 displayed a synergistic interaction with auxin, cytokinin, cupric ion, and 24-epibrassinolide (Fig. 4). The level of ethylene produced was generally close to multiplicative. For example, the eto1-3 mutation elevated ethylene biosynthesis approximately 15-fold above wild-type levels, 5 μm BA increased ethylene approximately 4-fold in etiolated, wild-type seedlings, and BA-treated eto1-3 seedlings made close to 60-fold more ethylene than untreated, wild-type seedlings. In general, the synergism of these inducers with the eto3 mutation was slightly less than that observed with eto1-3. This synergism indicates that the eto1-3 and eto3 mutations may act interdependently with these factors in regulating ethylene biosynthesis. Interestingly, exogenous ABA strikingly dampened the ethylene overproduction observed in the Eto mutants, although it did not appear to affect ethylene production in wild-type seedlings.
Figure 4.
Ethylene produced by wild-type and mutant etiolated Arabidopsis seedlings in response to treatment with ABA (75 μm), 2,4-D (160 μm), BA (5 μm), CuSO4 (20 mm), and 1 μm 24-epibrassinolide (24-epi). For all treatments except BA, seedlings were grown in GC vials, and 200 μL of the solutions of the indicated concentrations was added at 48 h. The vials were flushed with hydrocarbon-free air, sealed, and returned to the dark at 23°C. The ethylene that accumulated during the next 24 h was measured. For BA treatment, the seedlings were germinated on Murashige and Skoog medium supplemented with 5 μm BA and the ethylene production during the first 72 h of germination was measured. Values are means (±sd) of three replicates.
Interaction with Mutants Affected in Ethylene Biosynthesis
Ethylene-insensitive mutations produce elevated levels of ethylene, most likely because they block the feedback inhibition of ethylene biosynthesis in vegetative Arabidopsis tissue (Guzman and Ecker, 1990). To determine the effect of ethylene feedback on the Eto mutants, we constructed double mutants of eto1 and the ethylene-insensitive mutants ein2 and etr1-3 (ethylene resistant) (Fig. 1). The average length of hypocotyls from the double mutants was not significantly different from that of each ethylene-insensitive parent (not shown), which indicates that the etr1-3 and ein2 mutations are epistatic to eto1. This confirms the expectation that mutations defective in the perception of ethylene act downstream of those affecting ethylene biosynthesis (Roman et al., 1995). However, under the conditions that we used, the hypocotyl lengths of ein2 etiolated seedlings were much more variable than those of either the wild-type or eto1-3/ein2 etiolated seedlings (not shown). It is interesting that the apical hooks of the eto1-3/ein2 double-mutant etiolated seedlings generally appeared to be more closed that those of the ein2 single parents (Fig. 1), which have a hook that is less angled than that of wild-type etiolated seedlings.
When combined with the eto1-1 mutation, the etr1-3 mutation displayed a synergistic interaction in terms of the amount of ethylene produced (Fig. 2C). However, the amount of ethylene produced by the eto1-3/ein2 double mutant appeared to be close to additive relative to the parental seedlings (Fig. 2B). This difference may reflect a branch in the ethylene-response pathway or perhaps subtle differences between the eto1-1 and eto1-3 alleles.
eto1 and eto3 Etiolated Seedlings Have Elevated Levels of ACS Activity
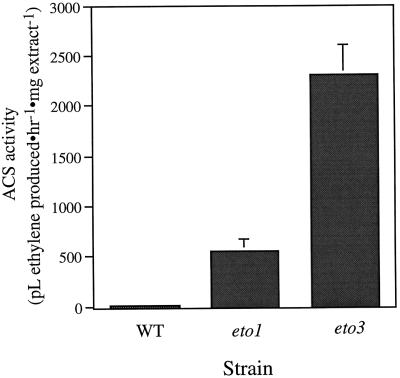
One likely target of these ethylene-overproducing mutants is ACS, the rate-limiting step of ethylene biosynthesis. Previous work demonstrated that ACS is the target of various inducers of ethylene biosynthesis in etiolated Arabidopsis seedlings by both transcriptional and posttranscriptional mechanisms (Liang et al., 1992, 1996; Van der Straeten et al., 1992; Abel et al., 1995; Vogel et al., 1998b). We assayed the level of ACS in crude extracts from wild-type and eto1-3 and eto3 etiolated seedlings (Fig. 5). Both mutants showed high elevated levels of ACS activity compared with wild-type etiolated seedlings, which had barely detectable levels of ACS activity. This indicates that increases in ACS activity may be responsible for the elevated ethylene biosynthesis observed in the mutant seedlings.
Figure 5.
ACS activity in crude extracts from 3-d-old, etiolated wild-type (WT), eto1, and eto3 mutant seedlings. The enzyme was assayed by incubating crude extracts in buffer with or without AdoMet, and then measuring the amount of ACC formed by converting it to ethylene (see Methods). The activity was calculated by subtracting the amount of ethylene produced in the absence of added AdoMet and then normalizing to the protein concentration of each sample. Values are means (±sd) of three replicates.
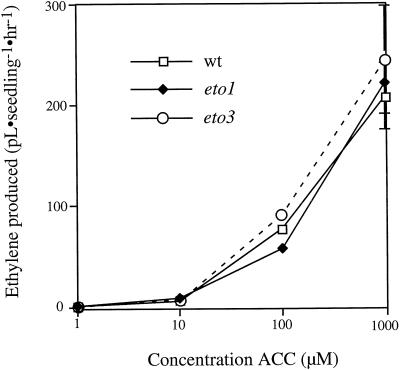
The formation of ACC is generally the rate-limiting step of ethylene biosynthesis, but regulation may also be mediated by changes in ACC oxidase levels or by changes in the amount of ACC that is conjugated. To address this issue, we examined the ability of intact wild-type and mutant seedlings to convert exogenous ACC to ethylene, which should reflect the level of ACC oxidase activity minus any ACC that becomes conjugated (Fig. 6). Both eto1-3 and eto3 mutants were indistinguishable from wild-type seedlings in their conversion of ACC to ethylene over a wide range of exogenous ACC concentrations. Consistent with this, eto1-3 and eto3 seedlings contained wild-type or slightly elevated levels of ACC N-malonyltransferase and γ-glutamyltranspeptidase activity (not shown). These data suggest that these mutants elevate ethylene biosynthesis primarily by increasing ACS activity.
Figure 6.
Ethylene produced by etiolated wild-type (wt), eto1, and eto3 Arabidopsis seedlings in response to varying concentrations of supplemented ACC. Seedlings were grown on 3 mL of Murashige and Skoog agar in GC vials. ACC of the indicated concentrations, in a total volume of 200 μL, was added to the seedlings 48 h after moving to 23°C. The vials were then flushed with hydrocarbon-free air and sealed, and the ethylene produced during the next 24 h was measured. Values are means (±sd) of three replicates.
eto1 and eto3 Etiolated Seedlings Have Wild-Type Levels of ACS mRNA
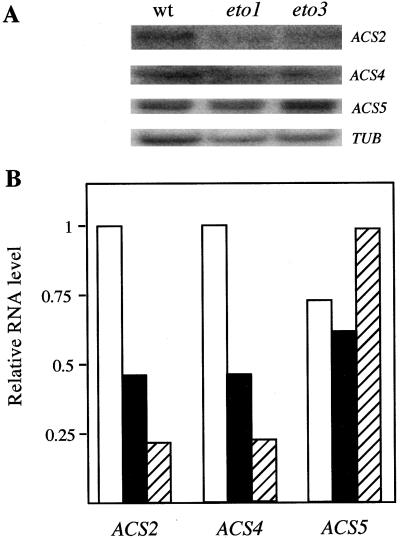
Elevation of ethylene biosynthesis has often been correlated with increases in the steady-state levels of ACS mRNAs, although in a few cases posttranscriptional control has also been demonstrated. To determine if elevated ACS mRNA contributes to the increased ethylene biosynthesis observed in eto1 and eto3 etiolated seedlings, we analyzed ACS mRNA levels by northern blotting (Fig. 7). We analyzed the expression of ACS2, ACS4, ACS5, and ACS6, the three previously identified, active ACS genes in Arabidopsis. In addition, we analyzed a novel ACS gene that we found by searching the Arabidopsis expressed sequence tag database (Newman et al., 1994), which we have named ACS7 (Fig. 8).
Figure 7.
RNA-blot analysis of ACS mRNA levels in etiolated seedlings. Five micrograms of poly(A+) RNA from 3-d-old wild-type (open bars), eto1 (closed bars), and eto3 (hatched bars) seedlings was separated by agarose-gel electrophoresis, blotted to a nylon membrane, and hybridized to an ACS2, ACS4, ACS5, or β-tubulin (TUB) probe. A, The original images of the blots. B, The quantification of the blots in A. The signal from each band was quantified using a phosphor imager. The value for each ACS band was divided by its β-tubulin loading control. The highest level in each set was assigned a value of 1, and the other values are expressed relative to this. The ACS5 blot was from an independent RNA blot and was normalized to its own β-tubulin loading control (not shown).
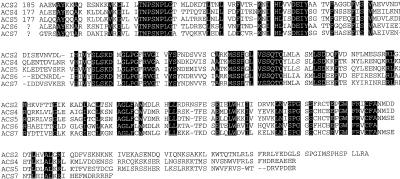
Figure 8.
A comparison of the protein sequences deduced from the cDNA clones for the Arabidopsis ACS genes. Regions of amino acid identity are shaded. The alignment was produced using Clustal software. The sequences for ACS2, ACS4, and ACS5 were from Liang et al. (1992). The sequence for ACS6 was from Vahala et al. (1998). The ACS7 protein sequence was deduced from the DNA sequence of the expressed sequence tag FAI88 (Newman et al., 1994). The amino acid position of the first residue shown for each protein is indicated in the first line.
The ACS7 cDNA clones all appear to lack the 5′ end of the gene, because none was as large as the size predicted from RNA gel-blot analysis (approximately 1.6 kb) and the open reading frame continued to the 5′ end of the longest sequence. However, the 3′ end of the gene is intact and an in-frame stop codon is present, as is a short poly(A+) tail. A comparison of the predicted amino acid sequence of ACS7, derived from the sequence of a non-full-length cDNA expressed sequence tag clone, with the other Arabidopsis ACS genes reveals that this clone lacks the variable carboxy-terminal extension present in other ACS proteins (Fig. 8). This carboxy-terminal domain has been demonstrated in at least two cases to negatively regulate the function of the ACS protein (Li and Mattoo, 1994; Vogel et al., 1998).
Overall, the level of expression of all five ACS genes was very low, in most cases just above the level of detection using 10 μg of poly(A+) mRNA. The steady-state level of mRNA for ACS5 in eto1-3 and eto3 etiolated seedlings was close to the level observed in wild-type seedlings. This was also confirmed by quantitative reverse-transcriptase PCR analysis (not shown). ACS2 and ACS4 steady-state levels were actually lower in the Eto mutants, perhaps reflecting negative feedback from the elevated ethylene levels. There was no detectable ACS6 or ACS7 expression in either wild-type or mutant etiolated seedlings, although treatment with the protein-synthesis inhibitor cycloheximide resulted in high levels of ACS7 expression (not shown), as is the case with the other Arabidopsis ACS genes (Liang et al., 1992). These results indicate that the increased ACS activity is not caused by elevated gene expression, but is likely attributable to a posttranscriptional mechanism.
DISCUSSION
The eto1 and eto3 mutants were isolated as seedlings that displayed a constitutive triple-response phenotype caused by an elevation of ethylene biosynthesis. We demonstrate that this ethylene overproduction is brought about by increased ACS activity, and that this is most likely the result of posttranscriptional regulation. This conclusion is based on measurements of ACS activity and mRNA levels, as well as the interactions with various inducers of ethylene biosynthesis. This is consistent with previous reports of posttranscriptional regulation of ACS in etiolated Arabidopsis seedlings (Vogel et al., 1998b), and suggests that this may be a major mechanism for modulating the level of ethylene biosynthesis in this tissue. Alternatively, it is possible that an additional ACS gene(s) is present in Arabidopsis and that the transcription of this gene is elevated in the Eto mutants. However, if this were the case, one would predict an additive interaction of these mutations with inducers that act through independent ACS genes, which is clearly not the case with either auxin or cytokinin. Thus, the model that is most consistent with the data presented here is that the Eto mutations affect the posttranscriptional regulation of ACS.
One possible mechanism for this posttranscriptional regulation is that ACS protein could be modified to increase its activity or stability, and the Eto mutations affect this modification. Protein phosphorylation has been found to play a role in regulating the function of ACS in tomato (Spanu et al., 1994), and perhaps eto1 and eto3 affect the phosphorylation state of ACS. Alternatively, the translation of ACS mRNA could be enhanced by the eto1 and eto3 mutations, leading to increased levels of ACS enzyme.
Arabidopsis does not display a detectable burst of ethylene in the first 24 h of germination, as is observed in some other plant species (Yang and Hoffman, 1984; Abeles et al., 1992), even though ethylene can stimulate germination in Arabidopsis (Bleecker et al., 1988). In contrast to the wild-type, eto1, eto3, and ein2 mutant seedlings display a sharp increase in ethylene production starting at about 48 h. This increase is then followed by a gradual decline to close to wild-type levels. The constitutive triple-response phenotype of eto1 and eto3 seedlings becomes decreasingly distinct as etiolation continues beyond 4 d (K.E. Woeste and J.J. Kieber, unpublished data), which reflects this diminution of ethylene biosynthesis. One model for this decrease in ethylene biosynthesis in the Eto mutants is that there is a developmental change in the regulation of ethylene biosynthesis. Alternatively, it may reflect an exhaustion of some metabolite required for ethylene biosynthesis, although the observation that eto1/ein2 double mutants decrease ethylene production more slowly suggests that this is not the case, because they presumably have similar metabolic limitations. The hypothesis that developmental changes affect the function of ETO1 and ETO3 is also supported by the observation that mutations in these genes affect only etiolated seedlings. It is possible that the ETO1 and ETO3 gene products are only required in etiolated seedlings, or, alternatively, that they affect the regulation of ethylene in multiple tissues but only in the dark, perhaps playing a role in the circadian regulation of ethylene biosynthesis (Finlayson et al., 1998).
In Arabidopsis vegetative tissue, ethylene biosynthesis appears to be autoinhibitory (Guzman and Ecker, 1990), although the mechanism for this negative-feedback regulation is unknown. Autoinhibition in other systems has been linked to decreased ACS, decreased ACC oxidase, and/or up-regulation of ACC conjugation (Abeles et al., 1992). The ethylene-insensitive mutants etr1 and ein2 block this negative-feedback regulation in etiolated seedlings, leading to an increase in ethylene production (Guzman and Ecker, 1990). The additive interaction of eto1 and ein2 suggests that these mutations act in parallel to regulate ethylene biosynthesis. etr1, unlike ein2, displays a synergistic interaction with eto1, which suggests that eto1 and etr1 act in an interdependent manner to regulate ethylene production. For example, etr1 could elevate ethylene biosynthesis by increased ACS transcription or decreased ACC conjugation, either of which, when coupled with the effect of eto1 on the posttranscriptional regulation of ACS, would lead to a synergistic interaction. The observation that etr1 and ein2 differ in their interaction with eto1 suggests that feedback regulation may occur via multiple mechanisms, and that the feedback pathway may be complex.
We evaluated the effects of a number of inducers of ethylene biosynthesis on eto1 and eto3 mutants to determine how they interact. We found that both cytokinin, which in low doses acts almost exclusively through the ACS5 isoform (Vogel et al., 1998b), and auxin, which acts through ACS4 (Abel et al., 1995), appear to interact with the Eto mutations in a synergistic fashion. In addition, cupric ion and 24-epibrassinolide also act synergistically with eto1 and eto3. This suggests that these factors, like etr1, act interdependently to regulate ethylene biosynthesis. This is readily explained in the case of auxin: auxin elevates ACS4 mRNA levels, and eto1 and eto3 increase ACS gene function posttranscriptionally, which together would lead to a synergistic effect. Likewise, cupric ion has also been shown to elevate ACS mRNA levels in tobacco (Avni et al., 1994), and thus its synergism with the Eto mutants could occur by a similar mechanism. However, the synergism of cytokinin and the Eto mutations is somewhat surprising, because both appear to affect the posttranscriptional regulation of ACS. This interaction suggests that cytokinin and the Eto mutants affect distinct posttranscriptional mechanisms, which could include translational efficiency of ACS mRNA and protein stability or activity.
To evaluate the possibility that the Eto mutations affect the regulation of ACC oxidase, we provided wild-type and mutant plants with an excess of substrate (ACC) and determined that Eto mutants and wild-type plants convert ACC to ethylene at the same rate over a 3-log range of concentrations (Fig. 6). These results suggest that ACC oxidase is not the rate-limiting step of ethylene biosynthesis in etiolated Arabidopsis seedlings, as is also the case in many other plant tissues, and that the Eto mutations do not affect the metabolism of ACC. This, coupled with direct measurements of ACC malonyltransferase and glutamyltranspeptidase activities, which showed the Eto mutants did not decrease ACC conjugation, supports the hypothesis that eto1 and eto3 primarily affect ACS activity.
ABA treatment reduces ethylene production in eto1 and eto3 mutant etiolated seedlings. ABA has also been shown to reduce the induction of ethylene biosynthesis by a number of factors in other plant tissues, including IAA-stimulated and drought-stressed leaves (Wright, 1980; Yoshii and Imaseki, 1981; McKeon et al., 1982; Tan and Thimann, 1989). The reduction of ethylene biosynthesis by ABA has been linked to both decreased ACC oxidase levels and increased ACC conjugation (McKeon et al., 1982; Corbineau et al., 1989; Tan and Thimann, 1989). The difference in the effect of ABA on Eto mutants versus its effect on wild-type etiolated seedlings can be explained if regulation by ABA is only required under conditions of high ethylene production, conditions not normally found in germinating wild-type Arabidopsis seedlings. The Eto mutations do not display any other obvious defects in ABA responses. ABA may act to suppress excess ethylene production downstream of the wild-type gene product of both Eto mutations.
The emerging picture from these and other studies is that posttranscriptional events play an important role in regulating ethylene biosynthesis. The interactions of the Eto mutations with other regulators of ethylene biosynthesis suggest that the pathways regulating this biosynthetic pathway are complex, reflecting the multitude of regulatory inputs that affect ethylene biosynthesis. Cloning of the genes corresponding to the eto1 and eto3 mutations should shed further light on the role that these genes play in regulating ethylene biosynthesis.
ACKNOWLEDGMENT
We thank Melinda Martin for assaying malonyltransferase and glutamyltranspeptidase activities.
Abbreviations:
- ACS
ACC synthase
- AdoMet
S-adenosyl-Met
Footnotes
This work was supported by U.S. Department of Agriculture grant nos. 95-37304-2294 and 97-01425 to J.J.K.
LITERATURE CITED
- Abel S, Nguyen MD, Chow W, Theologis A. ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. J Biol Chem. 1995;270:19093–19099. doi: 10.1074/jbc.270.32.19093. [DOI] [PubMed] [Google Scholar]
- Abeles FB, Morgan PW, Saltveit ME., Jr . Ethylene in Plant Biology. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Adams DO, Yang SF. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc Natl Acad Sci USA. 1979;76:170–174. doi: 10.1073/pnas.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrhein N, Schneebeck D, Skorupka H, Tophof S, Stockigt J. Identification of a major metabolite of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid in higher plants. Naturwissenschaften. 1981;68:619–620. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Avni A, Baily BA, Mattoo AK, Anderson JD. Induction of ethylene biosynthesis in Nicotiana tabacum by a Trichoderma viride xylanase is correlated to the accumulation of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase and ACC oxidase transcripts. Plant Physiol. 1994;106:1049–1055. doi: 10.1104/pp.106.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 1996;9:525–535. doi: 10.1046/j.1365-313x.1996.09040525.x. [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Botella JR, Arteca RN, Frangos JA. A mechanical strain-induced 1-aminocyclopropane-1-carboxylate synthase gene. Proc Natl Acad Sci USA. 1995;92:1595–1598. doi: 10.1073/pnas.92.5.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella JR, Schlagnhaufer CD, Arteca JM, Arteca RN, Phillips AT. Identification of two new members of the 1-aminocyclopropane-1-carboxylate synthase-encoding multigene family in mung bean. Gene. 1993;123:249–253. doi: 10.1016/0378-1119(93)90132-m. [DOI] [PubMed] [Google Scholar]
- Corbineau F, Rudnicki RM, Come D. ACC conversion to ethylene by sunflower seeds in relation to maturation, germination and thermodormancy. Plant Growth Regul. 1989;8:105–115. [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Finlayson SA, Lee IJ, Morgan PW. Phytochrome B and the regulation of circadian ethylene production in sorghum. Plant Physiol. 1998;116:17–25. doi: 10.1104/pp.119.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lim MA, Valdes-Lopez V, Cruz-Hernandez A, Saucedo-Arias LJ. Isolation and characterization of a gene involved in ethylene biosynthesis from Arabidopsis thaliana. Gene. 1993;134:217–221. doi: 10.1016/0378-1119(93)90096-l. [DOI] [PubMed] [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman NE, Yang SF, McKeon T. Identification and metabolism of 1-(malonylamino)cyclopropane-1-carboxylic acid as a major conjugate of 1-aminocyclopropane-1-carboxylic acid, an ethylene precursor in higher plants. Biochem Biophys Res Commun. 1982;104:765–770. doi: 10.1016/0006-291x(82)90703-3. [DOI] [PubMed] [Google Scholar]
- Jiao X-Z, Philosoph-Hadas S, Su L-Y, Yang SF. The conversion of 1-(malonylamino)cyclopropane-1-carboxylic acid to 1-aminocyclopropane-1-carboxylic acid in plant tissues. Plant Physiol. 1986;81:637–641. doi: 10.1104/pp.81.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. Ethylene biosynthesis. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:283–307. [Google Scholar]
- Kieber JJ. The ethylene response pathway in Arabidopsis. Annu Rev Plant Physiol Plant Mol Biol. 1997a;48:277–296. doi: 10.1146/annurev.arplant.48.1.277. [DOI] [PubMed] [Google Scholar]
- Kieber JJ. The ethylene signal transduction pathway in Arabidopsis. J Exp Bot. 1997b;48:211–218. doi: 10.1093/jxb/48.2.211. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenburg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim W, Yang S. Structure and expression of cDNAs encoding 1-aminocyclopropane-1-carboxylate oxidase homologs isolated from excised mung bean hypocotyls. Planta. 1994;194:223–229. [PubMed] [Google Scholar]
- Kionka C, Amrhein N. The enzymatic malonylation of 1-aminocyclopropane-1-carboxylic acid in homogenates of mung bean hypocotyls. Planta. 1984;194:226–235. doi: 10.1007/BF00397444. [DOI] [PubMed] [Google Scholar]
- Lasserre E, Bouquin T, Hernandez JA, Bull J, Pech JC, Balague C. Structure and expression of three genes encoding ACC oxidase homologs from melon (Cucumis melo L.) Mol Gen Genet. 1996;251:81–90. doi: 10.1007/BF02174348. [DOI] [PubMed] [Google Scholar]
- Li N, Mattoo AK. Deletion of the carboxyl-terminal region of 1-aminocyclopropane-1-carboxylic acid synthase, a key protein in the biosynthesis of ethylene, results in catalytically hyperactive monomeric enzyme. J Biol Chem. 1994;269:6908–6917. [PubMed] [Google Scholar]
- Liang X, Abel S, Keller JA, Shen NF, Theologis A. The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1992;89:11046–11050. doi: 10.1073/pnas.89.22.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Oono Y, Shen NF, Köhler C, Li K, Scolnik PA, Theologis A. Characterization of two members (ACS1 and ACS3) of the 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Gene. 1995;167:17–24. doi: 10.1016/0378-1119(95)00694-x. [DOI] [PubMed] [Google Scholar]
- Liang X, Shen NF, Theologis A. Li+-regulated 1-aminocyclopropane-1-carboxylate synthase gene expression in Arabidopsis thaliana. Plant J. 1996;10:1027–1036. doi: 10.1046/j.1365-313x.1996.10061027.x. [DOI] [PubMed] [Google Scholar]
- Lizada MCC, Yang SF. A simple and sensitive assay for 1-aminocyclopropane-1-carboxylic acid. Anal Biochem. 1979;100:140–145. doi: 10.1016/0003-2697(79)90123-4. [DOI] [PubMed] [Google Scholar]
- Martin MN, Cohen JD, Saftner RA. A new 1-aminocyclopropane-1-carboxylic acid-conjugating activity in tomato fruit. Plant Physiol. 1995;109:917–926. doi: 10.1104/pp.109.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AK, Suttle JC. The Plant Hormone Ethylene. Boca Raton, FL: CRC Press; 1991. [Google Scholar]
- McKeon TA, Hoffman NE, Yang SF. The effect of plant-hormone pretreatments on ethylene production and synthesis of 1-aminocyclopropane-1-carboxylic acid in water-stressed wheat leaves. Planta. 1982;155:437–443. doi: 10.1007/BF00394473. [DOI] [PubMed] [Google Scholar]
- Mekhedov SI, Kende H. Submergence enhances expression of a gene encoding 1-aminocyclopropane-1-carboxylate oxidase in deepwater rice. Plant Cell Physiol. 1996;37:531–537. doi: 10.1093/oxfordjournals.pcp.a028976. [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Zhang XS, Nair H, O'Neill SD. Temporal and spatial regulation of 1-aminocyclopropane-1-carboxylate oxidase in the pollination-induced senescence of orchid flowers. Plant Physiol. 1993;103:31–39. doi: 10.1104/pp.103.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima N, Nakagawa N, Imaseki H. Molecular size of wound-induced 1-aminocyclopropane-1-carboxylate synthase from Cucurbita maxima Duch. and change of translatable mRNA of the enzyme after wounding. Plant Cell Physiol. 1990;29:989–998. [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M and others. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetiker JH, Olsen DC, Shiu OY, Yang SF. Differential induction of seven 1-aminocyclopropane-1-carboxylate synthase genes by elicitor in suspension cultures of tomato (Lycopersicon esculentum) Plant Mol Biol. 1997;34:275–286. doi: 10.1023/a:1005800511372. [DOI] [PubMed] [Google Scholar]
- Olson DC, Oetiker JH, Yang SF. Analysis of LE-ACS3, a 1-aminocyclopropane-1-carboxylic acid synthase gene expressed during flooding in the roots of tomato plants. J Biol Chem. 1995;270:14056–14061. doi: 10.1074/jbc.270.23.14056. [DOI] [PubMed] [Google Scholar]
- Olson DC, White JA, Edelman L, Harkins RN, Kende H. Differential expression of two genes for 1-aminocyclopropane-1-carboxylate synthase in tomato fruits. Proc Natl Acad Sci USA. 1991;88:5340–5344. doi: 10.1073/pnas.88.12.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck SC, Kende H. Sequential induction of the ethylene biosynthetic enzymes by indole-3-acetic acid in etiolated peas. Plant Mol Biol. 1995;28:293–301. doi: 10.1007/BF00020248. [DOI] [PubMed] [Google Scholar]
- Peiser G, Yang SF. Evidence for 1-(malonylamino)cyclopropane-1-carboxylic acid being the major conjugate of 1-aminocyclopropane-1-carboxylic acid in tomato fruit. Plant Physiol. 1998;116:1527–1532. doi: 10.1104/pp.116.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Pousada RA, De Rycke R, Dedonder A, van Caeneghem W, Engler G, van Montagu M, Van der Straeten D. The Arabidopsis 1-aminocyclopropane-1-carboxylate synthase gene 1 is expressed during early development. Plant Cell. 1993;5:897–911. doi: 10.1105/tpc.5.8.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann WH, Peter GF, Oeller PW, Keller JA, Shen NF, Nagy BP, Taylor LP, Campbell AD, Theologis A. 1-Aminocyclopropane-1-carboxylate synthase in tomato is encoded by a multigene family whose transcription is induced during fruit and floral senescence. J Mol Biol. 1991;222:937–961. doi: 10.1016/0022-2836(91)90587-v. [DOI] [PubMed] [Google Scholar]
- Spanu P, Grosskopf DG, Felix G, Boller T. The apparent turnover of 1-aminocyclopropane-1-carboxylate synthase in tomato cells is regulated by protein phosphorylation and dephosphorylation. Plant Physiol. 1994;106:529–535. doi: 10.1104/pp.106.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z-Y, Thimann KV. The roles of carbon dioxide and abscisic acid in the production of ethylene. Physiol Plant. 1989;75:13–19. [Google Scholar]
- Tang X, Gomes AMTR, Bhatia A, Woodson WR. Pistil-specific and ethylene-regulated expression of 1-aminocyclopropane-1-carboxylate oxidase gene in petunia flowers. Plant Cell. 1994;6:1227–1239. doi: 10.1105/tpc.6.9.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahala J, Schlagnhaufer CD, Pell EJ. Induction of an ACC synthase cDNA by ozone in light-grown Arabidopsis thaliana leaves. Physiol Plant. 1998;103:45–50. [Google Scholar]
- Van der Straeten D, Rodrigues-Pousada RA, Villarroel R, Hanley S, Goodman HM, van Montagu M. Cloning, genetic mapping, and expression analysis of an Arabidopsis thaliana gene that encodes 1-aminocyclopropane-1-carboxylate synthase. Proc Natl Acad Sci USA. 1992;89:9969–9973. doi: 10.1073/pnas.89.20.9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Schuerman P, Woeste KW, Brandstatter I, Kieber JJ. Isolation and characterization of Arabidopsis mutants defective in induction of ethylene biosynthesis by cytokinin. Genetics. 1998a;149:417–427. doi: 10.1093/genetics/149.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Woeste KW, Theologis A, Kieber JJ. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc Natl Acad Sci USA. 1998b;95:4766–4771. doi: 10.1073/pnas.95.8.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste K, Vogel JP, Kieber JJ (1999) Factors regulating ethylene biosynthesis in etoilated Arabidopsis thaliana seedlings. Physiol Plant (in press)
- Wright STC. The effect of plant growth regulator treatments on the levels of ethylene emanating from excised turgid and wilted wheat leaves. Planta. 1980;148:381–388. doi: 10.1007/BF00388127. [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- Yoshii H, Imaseki H. Biosynthesis of auxin-induced ethylene: effects of indole-3-acetic acid, benzyladenine and abscisic acid on endogenous levels of 1-aminocyclopropane-1-carboxylic acid (ACC) and ACC synthase. Plant Cell Physiol. 1981;22:369–379. [Google Scholar]