Abstract
Phosphotriesterases catalyze the hydrolytic detoxification of phosphotriester pesticides and chemical warfare nerve agents with various efficiencies. The directed evolution of phosphotriesterases to enhance the breakdown of poor substrates is desirable for the purposes of bioremediation. A limiting factor in the identification of phosphotriesterase mutants with increased activity is the ability to effectively screen large mutant libraries. To this end, we have investigated the possibility of coupling phosphotriesterase activity to cell growth by using methyl paraoxon as the sole phosphorus source. The catabolism of paraoxon to phosphate would occur via the stepwise enzymatic hydrolysis of paraoxon to dimethyl phosphate, methyl phosphate, and then phosphate. The Escherichia coli strain DH10B expressing the phosphotriesterase from Agrobacterium radiobacter P230 (OpdA) is unable to grow when paraoxon is used as the sole phosphorus source. Enterobacter aerogenes is an organism capable of growing when dimethyl phosphate is the sole phosphorus source. The enzyme responsible for hydrolyzing dimethyl phosphate has been previously characterized as a nonspecific phosphohydrolase. We isolated and characterized the genes encoding the phosphohydrolase operon. The operon was identified from a shotgun clone that enabled E. coli to grow when dimethyl phosphate is the sole phosphorus source. E. coli coexpressing the phosphohydrolase and OpdA grew when paraoxon was the sole phosphorus source. By constructing a short degradative pathway, we have enabled E. coli to use phosphotriesters as a sole source of phosphorus.
Organophosphates (OPs) have been used as pesticides and chemical warfare agents during the last 60 years. OPs are considered nonpersistent compounds, breaking down in the environment over time to form relatively nontoxic compounds due to the action of light, water, and soilborne organisms. The bioremediation of sites contaminated with OPs is of concern in circumstances where there is a high risk of contact with humans. Escherichia coli, Moraxella sp., and Pseudomonas putida expressing recombinant bacterial phosphotriesterases (PTEs) have been used to detoxify solutions of phosphotriester pesticides (24, 34, 47) and could potentially be used for the bioremediation of contaminated soil and water. PTEs have been characterized with high activity towards particular OPs (e.g., paraoxon), but relatively low activities towards other OPs (e.g., demeton) (12, 20, 29). For the bioremediation of sites contaminated with poor PTE substrates, it is desirable to evolve PTEs with enhanced activity towards such substrates.
Several groups have undertaken the directed evolution of PTEs for higher activity towards poor substrates (7, 8, 49). The major difficulty in generating such an enzyme is effectively screening large mutant libraries for mutants with higher activity. To this end, we are investigating the possibility of coupling PTE activity with cell growth that uses a phosphotriester as the sole phosphorus source. The enzyme expression levels would be adjusted such that phosphotriesterase activity is growth rate limiting. Under these conditions, mutants with higher activity would enable their host to grow faster than their neighboring cells and would appear as larger colonies. Potential positive colonies could then be further screened by more traditional techniques. Such a process would potentially be more specific than using fluorigenic analogues and allow the screening of PTE mutants for substrates without a colored product.
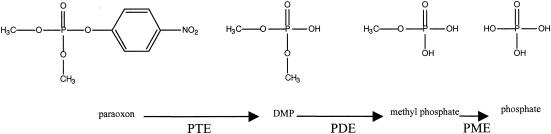
The proposed catabolic pathway for the breakdown of a typical phosphotriester, dimethyl paraoxon, to phosphate is outlined in Fig. 1. The breakdown of paraoxon involves three hydrolytic steps. PTEs catalyze the hydrolysis of paraoxon to dimethyl phosphate (DMP). Phosphodiesterases catalyze the hydrolysis of DMP to methyl phosphate. Phosphomonoesterases like alkaline phosphatase catalyze the hydrolysis of methyl phosphate to phosphate.
FIG. 1.
Proposed pathway for the mineralization of paraoxon. PTE, phosphotriesterase; PDE, phosphodiesterase, PME, phosphomonoesterase.
For the screening of PTE mutants, use of an easily transformed and well-defined host like E. coli is desirable. However, it has been reported that E. coli is unable to use a typical PTE product, DMP, as a sole source of phosphorus when it is added to the media (48). The reason(s) why E. coli cannot grow on DMP is unknown, although there are at least two possible explanations. The first explanation could be the lack of a phosphodiesterase with sufficient activity for DMP, a particularly stable compound. The second explanation could be an inability to transport DMP into the cytoplasm. The ability of E. coli expressing a PTE like the organophosphorus hydrolase from Agrobacterium radiobacter P230 (OpdA) (20) to grow on paraoxon would be evidence that E. coli has a phosphodiesterase capable of hydrolyzing DMP. If not, then growth would require the coexpression of OpdA and a phosphodiesterase from another source.
A range of organisms has been found with the ability to use dialkyl phosphates as phosphorus sources (10). Two such organisms are Enterobacter aerogenes and Delftia acidovorans. E. aerogenes was reported to be able to use DMP as a sole phosphorus source (48). The phosphohydrolase responsible for the hydrolysis of DMP was isolated and characterized. The phosphohydrolase was described as a cytoplasmic hexamer of 29-kDa subunits with broad specificity for phosphodiesters and phosphomonoesters (17). The activities are greatest under acidic conditions and require the presence of Zn2+, Cd2+, Co2+, Mn2+, or Ni2+ (16). The amino acid composition was determined, but the primary structure, the nucleotide sequence, and the operon of which the phosphohydrolase is a component, remained unknown.
D. acidovorans was reported to be able to grow with diethylthiophosphate as a sole phosphorus source (10). The gene encoding the enzyme responsible for the hydrolysis of diethylthiophosphate or diethyl phosphate was isolated and the protein was characterized (GenBank accession number AF548455). The pdeAgene encodes a 30-kDa protein, which is active as a trimer and displays moderate but broad activity towards phosphodiesters and phosphonomonoesters. In contrast with the phosphodiesterase activity of the phosphohydrolase from E. aerogenes, the phosphodiesterase activity of PdeA is greater under alkaline conditions and activated with the addition of Mg2+. The pdeA gene is located on a DNA fragment bearing the genes for a probable glycerol-3-phosphate (G3P) binding protein and the transposon insertion sequence IS1071. The use of PdeA as part of a system for the complete mineralization of parathion in P. putida has been investigated (44).
The aim of this study was to examine the possibility of creating a catabolic pathway in E. coli that would enable growth with paraoxon as the sole phosphorus source. To this end, we found that E. coli expressing the phosphotriesterase OpdA was unable to use paraoxon as the sole phosphorus source. Consequently, we isolated and expressed in E. coli a fragment of E. aerogenes genomic DNA that permitted E. coli to grow using DMP as the sole phosphorus source. Coexpression of this E. aerogenes fragment with OpdA from separate plasmids enabled E. coli to use paraoxon as the sole phosphorus source. The previously described phosphohydrolase was found to be part of an operon homologous to the G3P uptake operon of E. coli (ugp). The phosphohydrolase displayed good activity towards a typical phospholipid metabolite, glycerophosphorylethanolamine (GPE), but an amino acid sequence comparison to the sequence of E. coli glycerophosphodiester phosphodiesterase UgpQ showed no similarity. Consequently, we have named the phosphohydrolase GpdQ or glycerophosphodiester phosphodiesterase (EC 3.1.4.46).
MATERIALS AND METHODS
Strains, plasmids, and chemicals.
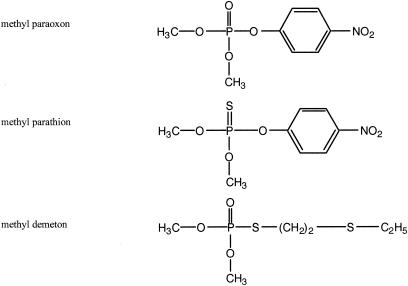
The strains used in this study and their characteristics are listed in Table 1. A field strain of E. aerogenes (number 251) was obtained as a gift from David Gordon (18). The E. coli strain AN1459 (45) was used for the overexpression of GpdQ. The E. coli strain DH10B (Gibco BRL) and pBluescript SK− (Stratagene) were used to screen E. aerogenes genomic DNA fragments for GpdQ activity. Plasmid pSYM5 was constructed to express OpdA in a GpdQ-containing strain, DH10B. The opdA gene was amplified by PCR from pH 2 (20) and then cloned between the NcoI and BamHI sites of pTrcHisB (Invitrogen) to produce pSYM3. The expression cassette (containing opdA) and lacIq were isolated on a 3.9-kb SphI/AviII fragment. The NcoI site in the medium-copy-number plasmid pACYC184 (6) was removed by site-directed mutagenesis. The 3.9-kb fragment was ligated between the SphI and EcoRV sites of pACYC184 to produce pSYM4. The −35 region of the trc promoter was mutated to that of the lac promoter (by site-directed mutagenesis) to produce pSYM5. Modified MOPS (morpholinepropanesulfonic acid) minimal medium plates containing various phosphorus sources were supplemented with the 20 amino acids (the final concentration was 10% of the suggested amount) and the recommended concentrations of vitamins (35). Methanol (1%) was included in all minimal medium plates to solubilize the phosphotriesters. Bacto agar (Difco) used in these plates was rinsed with distilled water three times before use. In all other cases, strains were grown in Luria-Bertani (LB) medium. Strains were grown at 37°C. Strains containing pBluescript and/or pACYC184 (and derivatives) were grown in the presence of ampicillin (100 μg/ml) and/or chloramphenicol (34 μg/ml), as required. Dimethyl paraoxon (paraoxon) was purchased from Chem Service. Dimethyl demeton (demeton) and dimethyl parathion (parathion) were purchased from Riedel-de Haen. The structures of the phosphotriesters used in this study are depicted in Fig. 2. DMP was purchased from Acros Organics. Bis(p-nitrophenyl) phosphate (bpNPP) and p-nitrophenylphosphate (pNPP) were purchased from Sigma. dl-α-Cephalin was purchased from Pfaltz and Bauer. Phospholipase B (Sigma) was used to hydrolyze dl-α-cephalin to GPE (23). The concentration of GPE was estimated by calculating the total phosphate concentration after total acid hydrolysis, performed according to Kates (23). Phosphate concentration was estimated by the phosphate detection assay of Cogan et al. (9). The purity of the substrates was >95%, with the exception of GPE, which contained ∼10% inorganic phosphate. Other enzymes and biochemicals were purchased from Roche or Sigma.
TABLE 1.
Bacterial strains and plasmids used in this study
| Plasmid or strain | Characteristic(s) | Reference or origin |
|---|---|---|
| Plasmids | ||
| pBluescript | High-copy-number plasmid | Stratagene |
| pACYC184 | Medium-copy-number plasmid | 6 |
| pSYM1 | pBluescript containing 5.6-kb fragment from E. aerogenes | This study |
| pSYM2 | pSYM1 with 0.5-kb deletion between BamHI and XhoI sites in E. aerogenes fragment | This study |
| pSYM3 | pTrcHisB opdA | This study |
| pSYM4 | Expression cassette from pSYM3 cloned into pACYC184 | This study |
| pSYM5 | pSYM4 with trp promoter mutated to lac promoter | This study |
| Strains | ||
| Enterobacter aerogenes | Field strain of Enterobacter aerogenes #251 | 18 |
| DH10B | Escherichia coli strain | Gibco BRL |
| GpdQ− | DH10B with pBluescript | This study |
| GpdQ+ | DH10B with pSYM1 | This study |
| GpdQ+ UgpB− | DH10B with pSYM2 | This study |
| GpdQ− OpdA+ | DH10B with pBluescript and pSYM5 | This study |
| GpdQ+ OpdA− | DH10B with pSYM1 with pACYC184 | This study |
| GpdQ+ OpdA+ | DH10B with pSYM1 with pSYM5 | This study |
FIG. 2.
Structural presentation of the phosphotriester pesticides used in this study.
Isolation of the E. aerogenes ugp operon.
E. aerogenes genomic DNA was isolated with the QIAGEN DNeasy tissue kit and then partially digested with the Sau3AI restriction enzyme. Fragments (2 to 10 kb) were ligated into the dephosphorylated BamHI site of pBluescript. DH10B competent cells were transformed with the ligation mix. The transformation mixes were rinsed of media three times with distilled water to remove contaminating sources of phosphorus and then plated on minimal medium plates containing 0.1 mM DMP as the sole phosphorus source. The plasmid from a single colony that grew significantly larger than the other colonies was isolated. The plasmid was found to confer the ability to use DMP as the sole phosphorus source when it was used to retransform DH10B. GpdQ+ cells grew at similar rates with 0.1 mM DMP or 0.1 mM OpdA-hydrolyzed paraoxon as phosphorus sources. Restriction digest analysis showed that the plasmid contained a 5.6-kb fragment. The fragment was sequenced and the plasmid was named pSYM1. A truncated form of pSYM1 was made by removal of a 0.5-kb BamHI/XhoI fragment, thereby producing an operon with an inactive form of the UgpB homologue, open reading frame 5 (ORF5) (pSYM2).
Growth assays.
The E. coli strains described in Table 1 were tested for three qualities: (i) their ability to grow using paraoxon, parathion, or demeton as sole phosphorus sources; (ii) their ability to grow on DMP in the presence of paraoxon, parathion, or demeton; and (iii) their ability to grow on demeton in the presence of paraoxon. GpdQ−/+ cells were transformed with either pACYC184 or pSYM5. The transformation mixes were rinsed with distilled water three times to remove contaminating sources of phosphorus and then plated at a cell density yielding ∼1,000 colonies per 10-cm-diameter minimal medium plate. Three sets of plates were made. The first contained the various phosphotriesters as phosphorus sources (0.1 mM). The second contained 0.1 mM phosphotriester plus 0.1 mM DMP, and a third contained 0.1 mM demeton and 0.1 mM paraoxon. Colony growth was monitored every 24 h for up to 4 days.
Sequence and structural analysis of the E. aerogenes ugp operon.
The nucleotide and conceptual translation sequences of the E. aerogenes fragment were analyzed by using the BLAST program against the nonredundant protein database at the National Center for Biotechnology Information (1). Sequence motifs of GpdQ were identified by using the MotifScan program against the Prosite database of protein families and domains (14). The multiple alignment of sequence motifs was accomplished by using selected sequences from the Pfam multiple sequence alignment of the metallophosphoesterase domain (42). Sequence structure homology was identified by using the FUGUE program (41) to search the HOMSTRAD database (33). Secondary structure prediction was made with the JPred secondary structure prediction server (11).
Overexpression and purification of recombinant GpdQ.
The putative gpdQ gene was amplified by PCR and then cloned between the NdeI and EcoRI sites of the constitutive-expression plasmid pCY76 (49). Apr transformants were selected in AN1459. LB broth (10 ml) was inoculated with single colonies and incubated at 30°C overnight. LB broth (two samples of 500 ml) was inoculated with the overnight culture and incubated at 30°C for 15 h. All of the following steps were done at 4°C. Cells were harvested by centrifugation and then resuspended in 20 mM Tris · Cl, pH 7.6. Cells were lyzed with a French press operated at 14,000 lb/in2. The cell debris was pelleted by centrifugation. The soluble fraction was loaded onto a DEAE-Fractogel (Merck) column at 1 ml/min. A linear NaCl gradient (0 to 1 M) was applied over 10 column volumes to elute bound proteins. The eluted fractions were assayed for phosphodiesterase activity by the reaction of 5 μl of eluate in 100 μl of 1 mM bpNPP in 100 mM Tris · Cl, pH 8.0, over 30 min at 30°C. The majority of the phosphodiesterase activity eluted at ∼400 mM NaCl. The active protein was precipitated with 2 M ammonium sulfate and then pelleted by centrifugation. The pellet was resuspended with 20 mM Tris · Cl, pH 7.6, and then dialyzed against 20 mM Tris · Cl-1.5 M ammonium sulfate, pH 7.6, overnight. The soluble fraction was loaded onto a phenyl Sepharose column (Amersham Biosciences) at 1 ml/min. A linear ammonium sulfate (1.5 to 0 M) gradient was applied over 10 column volumes to elute bound proteins. The eluted fractions were assayed for phosphodiesterase activity as above. The most active fractions were eluted with ∼300 mM ammonium sulfate. The ammonium sulfate was removed from selected fractions by ultrafiltration with a 100-kDa-cutoff Centriplus device (Amicon). The yield was ∼30 to 35 mg of protein per liter of culture. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the concentrated protein showed a single band migrating at ∼31 kDa.
In vitro assays of purified GpdQ.
The activity of GpdQ towards bpNPP in polyacrylamide gels under partially denaturing conditions was detected by incubating the gel containing the run sample in ∼20 ml of 100 mM HEPES (pH 8.0)-1 mM bpNPP for 10 min (19).
Values over a range of substrate concentrations did not fit the Michaelis-Menten equation well; therefore, the specific activity of GpdQ towards the following substrates (each at 1 mM) was determined: pNPP, bpNPP, DMP, GPE, demeton, and paraoxon. Assays were conducted in two buffers: 20 mM sodium acetate (pH 5.0) and 20 mM HEPES (pH 8.0). The hydrolysis of DMP and GPE was assayed by means of the production of inorganic phosphate by calf intestinal alkaline phosphatase used as a means of quantifying the amount of phosphomonoester produced (17). After treatment of the substrate with GpdQ, the pH was raised to 9.0, and calf intestinal alkaline phosphatase was used to hydrolyze the resulting phosphomonoesters. The quantification of inorganic phosphate was accomplished according to the malachite green-ammonium molybdate method of Cogan et al. (9). Standard curves were run in parallel with potassium phosphate. Activities toward pNPP, bpNPP, and paraoxon were determined by transferring reaction mixtures every 1 min over a 10-min period into 200 mM Tris · Cl, pH 8.0, and then measuring the concentration of p-nitrophenolate spectrophotometrically at 410 nm (ɛ410 = 16,200 M−1 cm−1) (3). Activity towards demeton was determined by transferring reaction mixtures every 1 min over a 10-min period into 200 mM Tris · Cl, pH 8.0, 2 mM 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB), and 1% methanol and then measuring the concentration of 3-carboxy-4-nitrothiophenolate at 410 nm (ɛ410 = 13,600 M−1 cm−1) (13). Methanol (1%) was included in the phosphotriesterase assays. All reactions proceeded at room temperature.
Proteolysis of GpdQ.
GpdQ (16 μg) was incubated with increasing concentrations of trypsin (0, 0.25, 0.50, and 1.0 μg) over a 45-min period at room temperature in 100 mM Tris · Cl, pH 8.0. The activity of the protein was then immediately measured by bpNPP at pH 5 according to the method described above. The protein fragments were analyzed by SDS-PAGE after digestion.
Nucleotide sequence accession number.
The nucleotide sequence of the 5.6-kb fragment containing the E. aerogenes ugp operon was submitted to the GenBank database (accession number AY243367).
RESULTS
Description of E. aerogenes ugp operon.
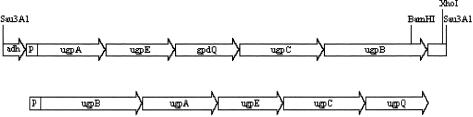
DH10B transformed with a plasmid containing a random fragment of E. aerogenes chromosomal DNA produced a single colony (from 10,000 colonies) that grew on minimal medium plates with DMP as the sole source of phosphorus to a size significantly larger than the rest of the population (∼1 mm compared to pinpricks after 3 days). When the large colony and a few small colonies were streaked on similar plates, the large colony grew to ∼1 mm, whereas the small colonies failed to grow even to their previous size. DH10B pBluescript also produced very small colonies. These results suggest that though there were trace amounts of phosphorus sources in both the rinsed transformation mix and the minimal medium plates, the concentration was low enough to allow a distinction based on colony size between GpdQ− and GpdQ+ cells. It is clear that DH10B lacks the ability to use DMP as a sole phosphorus source under these conditions. Restriction analysis of the presumed GpdQ+ plasmid indicated the presence of a 5.6-kb fragment, whose sequence was determined. BLAST analysis of the nucleotide sequence fragment indicated that the fragment contained at least part of an operon homologous to the G3P uptake (ugp) operon of E. coli. The E. coli ugp operon encodes five proteins: UgpB, a G3P binding protein; UgpA, a permease; UgpC, an ATP-binding protein; UgpE, a permease; and UgpQ, a phosphodiesterase (Fig. 3) (21, 38). ORF1, ORF2, ORF4, and ORF5 from the E. aerogenes fragment are homologous to E. coli ugpA, ugpE, ugpC, and ugpB, respectively. ORF3 encodes a 30.8-kDa protein with an amino acid composition similar to that of the previously reported phosphohydrolase (17). Subsequent cloning and overexpression of ORF3 showed that it has the previously described phosphohydrolase activities. Cloning, overexpression, and purification of the ORF3 protein and in vitro assays confirmed that it has activities similar to those reported by Gerlt and Whitman (17). SDS-PAGE of the purified ORF3 protein showed that it ran as a 31-kDa band when it was previously treated at 90°C in the presence of SDS and as a 95-kDa band when it was not treated. Phosphodiesterase assays conducted on the gel showed a strong yellow band after 5 min in the position of the 95-kDa band detected with Coomassie blue. No activity was associated with the 31-kDa band after 10 min. Therefore, ORF3 most likely encodes GpdQ, and one active form of GpdQ may be a trimer.
FIG. 3.
Diagrammatic comparison of the 5.6-kb fragment isolated from E. aerogenes that enabled E. coli to use DMP as a sole phosphorus source (top) and the 5.2-kb ugp operon from E. coli (bottom). P indicates the promoter region of each operon.
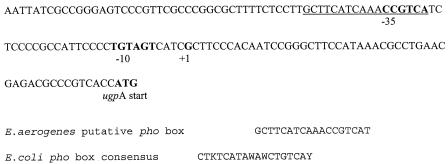
Upstream of the E. aerogenes operon lies a nucleotide sequence (Fig. 4) that is a 78% match to that of the E. coli phosphate box consensus sequence (32). A phosphate box is a region to which the transcription activator PhoB binds, which enhances the binding of RNA polymerase and, consequently, transcription of the following genes under phosphate-limiting conditions (31). GpdQ is expressed in E. aerogenes under phosphate-limiting conditions (48). A comparison by SDS-PAGE of protein expression in GpdQ+ DH10B cells grown under phosphate-limiting (DMP concentration, 1 mM) and excess phosphate (PO4 concentration, 1 mM) conditions showed expression of a 31-kDa protein only under phosphate-limiting conditions. A comparison by SDS-PAGE of GpdQ− and GpdQ+ cells grown under phosphate-limiting conditions (PO4 concentration, 0.05 mM) showed expression of a 31-kDa protein in the GpdQ+ strain but not in the GpdQ− strain. Collectively, these results suggest that this protein is induced from the putative pho promoter of the E. aerogenes ugp operon under phosphate-limiting conditions. There is no evidence of a cyclic AMP (cAMP) receptor protein binding site in the promoter region of the E. aerogenes ugp operon [consensus sequence, TGTGA(N6)TCACA], suggesting that the expression of this operon is not induced under carbon-limited conditions (2).
FIG. 4.
Nucleotide sequence of the E. aerogenes fragment containing the putative pho box (underlined), putative −35 and −10 regions (bold), putative transcription start site (+1, bold) and the putative start codon of ugpA (bold). The alignment of the putative E. aerogenes pho box with the consensus sequence of the pho box from E.coli is shown below the sequence for comparison (where K represents G or T, W represents A or T, and Y represents C or T).
In vitro assays of GpdQ activity.
The results of activity assays of purified GpdQ with various substrates are summarized in Table 2. The phosphohydrolase was previously shown to have primarily phosphodiesterase activity, with weaker phosphomonoesterase activity (17). Our results are consistent with these findings. The substrate with which GpdQ had the greatest specific activity was GPE, a phosphodiester and typical phospholipid metabolite. In addition, we found that GpdQ had activity towards the phosphotriester pesticides demeton and paraoxon. Demeton hydrolysis occurred 2 times faster at pH 5 than at pH 8, whereas paraoxon hydrolysis was 3.5 times higher at pH 8 than at pH 5. The activity towards demeton was approximately 17 times higher than for paraoxon at pH 5. The activity of purified GpdQ was found to increase after proteolytic digestion with trypsin. Treatment of GpdQ with 0.25, 0.50, and 1.0 μg of trypsin resulted in 50, 73, and 90% increases in activity, respectively, over nondigested GpdQ. SDS-PAGE showed the generation of several bands, the most prominent of which migrated at ∼21 and ∼9 kDa, respectively. Most of the 31-kDa band remained undigested even in the sample containing the highest concentration of trypsin, suggesting that digestion was incomplete.
TABLE 2.
Specific activity of GpdQ towards various substratesa
| Substrate | Specific activity (μmol/min/mol of GpdQ)
|
|
|---|---|---|
| pH 5 | pH 8 | |
| pNPP | 9.7 × 102 | 1.8 × 101 |
| bpNPP | 1.5 × 104 | 1.0 × 104 |
| DMP | 2.4 × 102 | 1.5 × 102 |
| GPE | 5.5 × 104 | 2.6 × 104 |
| Demeton | 2.6 × 103 | 1.2 × 103 |
| Paraoxon | 3.1 × 101 | 1.2 × 102 |
Assays were conducted in 20 mM sodium acetate, pH 5.0, and 20 mM HEPES, pH 8.0, at room temperature.
Growth assays.
The results of growth assays are summarized in Table 3. GpdQ− cells were unable to grow by using DMP or the phosphotriesters as phosphorus sources. In addition to growing on DMP, GpdQ+ cells were able to grow on demeton at almost the same rate as on DMP. However, these cells were unable to grow using paraoxon. The addition of paraoxon inhibited cell growth on DMP or demeton. GpdQ+ cells lacking a functional E. aerogenes G3P binding protein (GpdQ+ UgpB−) grew unhindered on demeton but were unable to grow on DMP. This result indicates that a functional E. aerogenes UgpB is probably required for DMP transport across the inner membrane and that demeton moves into the cytoplasm by other means. GpdQ+ cells expressing phosphotriesterase (GpdQ+ OpdA+) were able to grow using paraoxon as a sole phosphorus source, albeit at a lower rate than with DMP or demeton. OpdA expression permitted growth of GpdQ+ cells on DMP in the presence of paraoxon. GpdQ− OpdA+ cells were unable to grow using DMP or the phosphotriesters, indicating that even when DMP has been generated in the cytoplasm, DH10B cells are still unable to use it as a phosphorus source. None of the tested strains was able to use parathion as a phosphorus source, but the growth of the strains able to grow on DMP was not inhibited by parathion (data not shown).
TABLE 3.
Results of growth assays of the E. coli strains used in this study on various substrates on minimum medium plates
| Strain | Growth by substratea
|
||||||
|---|---|---|---|---|---|---|---|
| DMP | Demeton | Paraoxon | Parathion | DMP + demeton | DMP + paraoxon | Demeton + paraoxon | |
| GpdQ− | − | − | − | − | − | − | − |
| GpdQ+ | ++ | ++ | − | − | ++ | − | − |
| GpdQ+ UgpB− | − | ++ | − | − | ++ | − | − |
| GpdQ− OpdA+ | − | − | − | − | − | − | − |
| GpdQ+ OpdA+ | ++ | ++ | + | − | ++ | + | ND |
−, negligible growth after 4 days; +, 1-mm-diameter colonies after 4 days; ++, 1-mm-diameter colonies after 2 days; ND, not determined.
Sequence and structural analysis of GpdQ.
The amino acid sequence of GpdQ was found to be homologous to the sequences of a broad range of phosphatases. The best match was with a putative Ser/Thr protein phosphatase from Pseudomonas syringae pv. tomato strain DC3000 (34% identity). Of the characterized proteins, the closest relatives are the broad-specificity phosphodiesterase (PdeA) from D. acidovorans (32% identity) and cAMP 3′,5′-phosphodiesterase (CpdA) from E. coli (25% identity).
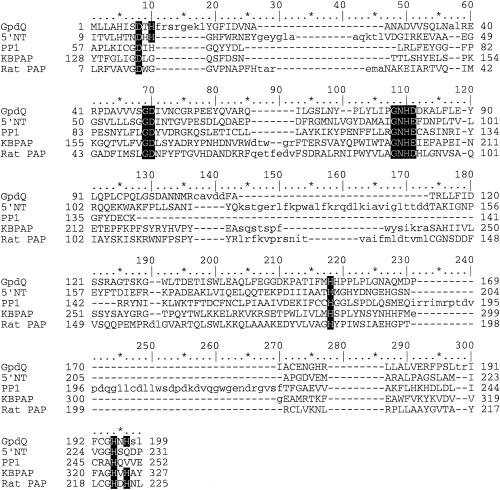
The sequence motif DxH(X)nGDXXD(X)nGNHD/E (28), present in a large group of phosphoesterases, such as Ser/Thr protein phosphatases (30), purple acid phosphatases (PAP) (43, 46), 5′-nucleotidase from E. coli (27), and the MreII nuclease (4), was detected (Fig. 5). Additionally, secondary structure prediction indicated that GpdQ shares a βαβαβ structural motif that is also present in these proteins. This, alongside sequence structure homology, indicates a high likelihood that GpdQ is a member of a diverse metallophosphoesterase family whose members contain binuclear metal centers at their active sites.
FIG. 5.
Multiple alignment of the amino acid sequences of the E. aerogenes GpdQ, 5′ nucleotidase from E. coli (5′ NT), Ser/Thr protein phosphatase 1 from rabbit (PP1), kidney bean PAP (KBPAP), and rat PAP (Rat PAP). Conserved residues involved in metal coordination are shaded.
DISCUSSION
The inability of E. coli expressing OpdA to grow using paraoxon as a sole phosphorus source indicates that one of the reasons why it cannot grow on DMP is its inability to hydrolyze DMP to methyl phosphate under phosphate-limiting conditions. Based on this finding, we sought to isolate and characterize the phosphohydrolase gene from E. aerogenes to test if coexpressing it and OpdA would allow E. coli to grow with paraoxon as the sole phosphorus source.
The phosphohydrolase gene was selected for its ability to enable E. coli to use DMP as a sole phosphorus source. It was found that the gene was part of an operon homologous to the G3P uptake operon of E. coli (ugp). One function of the ugp operon is to recycle phospholipid metabolites like G3P and glycerophosphoryl diesters (5). A second possible function is to regulate cAMP levels in the cell. Expression of the E. coli ugp operon is induced under both phosphate- and carbon-limiting conditions from separate promoters (22). As the E. aerogenes ugp operon has all of the genes and is induced under phosphate-limiting conditions in a similar way to the E. coli ugp operon, it is likely that the operons have similar functions. The lack of a cAMP receptor protein binding site in the promoter region suggests that the E. aerogenes ugp operon is not expressed under carbon-limiting conditions and has no direct role in regulating cAMP levels, although this idea was not tested further. Although GpdQ displays no sequence similarity to UgpQ, considering the operon in which it is located and the high activity displayed towards a probable natural substrate (GPE), it is likely that the function of the two phosphodiesterases is similar.
While the components of the two operons are similar, the gene arrangement within them differs. In E. coli, the gene order is ugpBACEQ, whereas in E. aerogenes, the order is ugpAEQCB (Fig. 3). In E. coli, the operon is transcribed monocistronically from either of the two promoters (38). We have not investigated the regulation of the E. aerogenes gpdQ operon. The inability of a GpdQ+ UgpB− strain to grow on DMP and its ability to grow on demeton suggest that a functional UgpB is required for growth on DMP. As the function of E. coli UgpB is to bind G3P and glycerophosphoryl diesters to enable their transport into the cytoplasm via the UgpACE complex, it is likely that this lack of growth is due to an inability of the truncated form of UgpB to bind DMP (40). The inhibition of GpdQ+ UgpB+ cell growth on either DMP or demeton by paraoxon and the inhibition of GpdQ+ UgpB− cell growth on demeton by paraoxon are consistent with the notion that paraoxon is a competing substrate for the growth-rate limiting hydrolysis of demeton and/or DMP. These facts also suggest that the site of inhibition is at the point of hydrolysis, not transport.
It is of interest to note that the amino acid sequence of UgpB from E. aerogenes is homologous to that of the putative sugar binding protein isolated on the same fragment as PdeA from D. acidovorans (expect value, 6e − 11). The proximity of a UgpB homologue and a phosphodiesterase in D. acidovorans suggests that PdeA is also part of a ugp-like operon. Unfortunately, it is unknown if these genes are part of a larger operon or if they are expressed under phosphate-limiting conditions.
The results from the in vitro assays of purified GpdQ on various substrates help explain the growth assay results. GpdQ activity towards demeton is evidence that the growth of GpdQ+ cells is due to hydrolysis of demeton to DMP by GpdQ, not by an E. coli enzyme. The low in vitro activity towards paraoxon indicates the most likely point of GpdQ+ growth inhibition on DMP, i.e., a competing substrate. The relatively high specific activity towards demeton suggests that the faster removal of demeton compared to paraoxon allows time for the subsequent hydrolysis of DMP. The reasons why GpdQ has higher activity towards the phosphorothiolate demeton than towards paraoxon remain unknown. The higher activity of GpdQ towards paraoxon at high pH levels in contrast to other tested substrates is intriguing and awaits further examination. Also of interest are the contrasting pH optima of the phosphodiesterase activities of GpdQ and PdeA from D. acidovorans. Considering the amino acid homology between these two enzymes, this difference is interesting and requires further investigation.
The proteolytic activation of GpdQ is reminiscent of that reported for PAP and may be significant considering their structural similarity. Recombinant human PAP displays increased activity after proteolytic cleavage and removal by trypsin of a tripeptide segment adjacent to the active site (15). Size exclusion chromatography (data not shown) indicates that GpdQ remains intact, suggesting limited proteolysis of loop regions in the protein analogous to that seen in PAP. It has been proposed that this cleavage regulates PAP activity by raising kcat and shifting the pH optimum. Considering that GpdQ synthesis is induced under phosphate-limiting conditions, it remains unclear why its activity might be further regulated by proteolysis.
Comparison of the previously determined amino acid composition of GpdQ to a conceptual translation of gpdQ indicates that the two are most likely the same protein (17). Sequence analysis and structure prediction show that GpdQ is a member of the recently identified family of class III 3′,5′-cyclic nucleotide phosphodiesterases (39) and, more broadly, the family of metallophosphoesterases identified by Koonin (28). The conserved amino acids of the phosphoesterase motif are involved in coordination of the binuclear metal center at the active site and metal-bound water nucleophiles in other members of this metallophosphoesterase family (26, 27). Accordingly, it is likely that two metal ions are coordinated at the catalytic center of GpdQ. PdeA is activated by Mg2+, but the effect of other divalent metal ions was not reported (44). Previous characterization of pH suggested that three Zn2+ and three Mn2+ ions were bound per hexamer, although Gerlt and Wan reported difficulties in determining the metal ion concentration with certainty (16). Considering the similarity of GpdQ to binuclear metallophosphatases and the lack of relationship with mononuclear metallophosphatases, it is probable that GpdQ has two metal ions per monomer, possibly one Zn2+ and one Mn2+ per monomer. The predicted secondary and tertiary structure of GpdQ, namely a double-β sandwich surrounded on both sides by α helices and characterized by the βαβαβ motif, is similar to that of other binuclear metallophosphatases such as PAP (43) and the Ser/Thr protein phosphatases (25).
The inhibition of growth of a GpdQ+ strain on DMP by paraoxon suggests that under certain conditions organophosphate pesticides can act as antibiotics. The term antibiotics is used here in a general sense to describe a compound that can inhibit the growth of an organism under particular conditions, rather than in the usual and more specific sense to describe bacterial growth inhibitors in a medical environment. The ability of OpdA to enable growth in the presence of paraoxon raises the possibility that the role of phosphotriesterases in bacteria may be more than simply the generation of phosphate sources from organophosphate pesticides. In effect, phosphotriesterases can act as antibiotic resistance enzymes. While it is true that, in the long term, paraoxon would be broken down to provide a phosphorus source, in the short term, the generation of a phosphorus source from paraoxon is of secondary importance compared to the removal of paraoxon as a growth inhibitor. The establishment of a phosphotriesterase via gene transfer or gene duplication and divergence from GpdQ would provide a growth advantage under such conditions.
The reason(s) why DH10B is unable to use DMP as a sole phosphorus source remain uncertain. Assuming that DH10B has a functional ugp operon, our results suggest at least two reasons. On the one hand, the inability of GpdQ− OpdA+ cells to grow with paraoxon as the sole phosphorus source suggests that E. coli does not express an enzyme capable of hydrolyzing DMP under phosphate-limiting conditions. On the other hand, the inability of GpdQ+ UgpB− cells to grow on DMP lends weight to the argument that E. coli lacks the capacity to take up DMP. It also remains unclear what enzyme in the GpdQ+ OpdA+ strain hydrolyzes methyl phosphate to phosphate. Given the phosphomonoesterase activity of GpdQ, it is possible that GpdQ catalyzes this step in addition to the phosphodiesterase step. However, an E. coli phosphomonoesterase is more likely responsible. Alkaline phosphatase turns over methyl phosphate readily (36). However, as this enzyme is expressed in the periplasm, the issue of methyl phosphate transport from the cytoplasm to the periplasm is raised. Little has been reported about this issue.
Despite the similarity of the E. aerogenes ugp operon to the E. coli ugp operon, its in vivo function remains unclear. The similarity suggests that the functions of the two operons are similar, i.e., the catabolism of phospholipids. However, the high activity of GpdQ towards the phosphorothiolate pesticide demeton clouds the issue. As the amino acid sequence of the previously described phosphohydrolase was not reported, it is uncertain if the two proteins are identical. If the two enzymes are different in sequence and activity, then the demeton activity may be due to evolutionary changes in response to pesticides in the environment. If the two enzymes are the same, then the demeton activity may be a remarkable example of catalytic promiscuity (37).
The growth of strain DH10B coexpressing GpdQ and OpdA when paraoxon is the sole phosphate source shows that E. coli can be used for selection of OpdA mutants with the ability to hydrolyze paraoxon. In addition, the GpdQ+ strain might also be useful for the bioprospecting of new phosphotriesterases from wild-type organisms that produce DMP from phosphotriesters, especially in cases where the other product is not colored or fluorescent. The use of this system for selecting OpdA mutants with higher activity towards poor substrates appears promising and is under further investigation.
Acknowledgments
We thank David Gordon for providing E. aerogenes and Nick Dixon (RSC, ANU) for providing strain AN1459 and plasmid pACYC184. Thanks to Nick Dixon and Geoff Smith (BAMBI, ANU) for their critiques of the text.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, O. G., and P. H. von Hippel. 1988. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J. Mol. Biol. 200:709-723. [DOI] [PubMed] [Google Scholar]
- 3.Bessey, O. A., O. H. Lowry, and M. J. Brock. 1946. A method for the rapid determination of alkaline phosphatase with five cubic millimetres of serum. J. Biol. Chem. 164:321-329. [PubMed] [Google Scholar]
- 4.Bressan, D. A., H. A. Olivares, B. E. Nelms, and J. H. Petrini. 1998. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzoska, P., and W. Boos. 1988. Characteristics of a ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the ugp transport system of Escherichia coli. J. Bacteriol. 170:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen-Goodspeed, M., M. A. Sogorb, F. Wu, and F. M. Raushel. 2001. Enhancement, relaxation, and reversal of the stereoselectivity for phosphotriesterase by rational evolution of active site residues. Biochemistry 40:1332-1339. [DOI] [PubMed] [Google Scholar]
- 8.Cho, C. M., A. Mulchandani, and W. Chen. 2002. Bacterial cell surface display of organophosphorus hydrolase for selective screening of improved hydrolysis of organophosphate nerve agents. Appl. Environ. Microbiol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogan, E. B., G. B. Birrell, and O. H. Griffith. 1999. A robotics-based automated assay for inorganic and organic phosphates. Anal. Biochem. 271:29-35. [DOI] [PubMed] [Google Scholar]
- 10.Cook, A. M., C. G. Daughton, and M. Alexander. 1978. Phosphorus-containing pesticide breakdown products: quantitative utilization as phosphorus sources by bacteria. Appl. Environ. Microbiol. 36:668-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 12.Dumas, D. P., S. R. Caldwell, J. R. Wild, and F. M. Raushel. 1989. Purification and properties of the phosphotriesterase from Pseudomonas diminuta. J. Biol. Chem. 264:19659-19665. [PubMed] [Google Scholar]
- 13.Ellman, G. L. 1959. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82:70-77. [DOI] [PubMed] [Google Scholar]
- 14.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funhoff, E. G., C. H. Klaassen, B. Samyn, J. Van Beeumen, and B. A. Averill. 2001. The highly exposed loop region in mammalian purple acid phosphatase controls the catalytic activity. Chembiochem. 2:355-363. [DOI] [PubMed] [Google Scholar]
- 16.Gerlt, J. A., and W. H. Wan. 1979. Stereochemistry of the hydrolysis of the endo isomer of uridine 2′, 3′-cyclic phosphorothioate catalyzed by the nonspecific phosphohydrolase from Enterobacter aerogenes. Biochemistry 18:4630-4638. [DOI] [PubMed] [Google Scholar]
- 17.Gerlt, J. A., and G. J. Whitman. 1975. Purification and properties of a phosphohydrolase from Enterobacter aerogenes. J. Biol. Chem. 250:5053-5058. [PubMed] [Google Scholar]
- 18.Gordon, D. M., and F. FitzGibbon. 1999. The distribution of enteric bacteria from Australian mammals: host and geographical effects. Microbiology 145:2663-2671. [DOI] [PubMed] [Google Scholar]
- 19.Harcourt, R. L., I. Horne, T. D. Sutherland, B. D. Hammock, R. J. Russell, and J. G. Oakeshott. 2002. Development of a simple and sensitive fluorimetric method for isolation of coumaphos-hydrolysing bacteria. Lett. Appl. Microbiol. 34:263-268. [DOI] [PubMed] [Google Scholar]
- 20.Horne, I., T. D. Sutherland, R. L. Harcourt, R. J. Russell, and J. G. Oakeshott. 2002. Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolate. Appl. Environ. Microbiol. 68:3371-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasahara, M., K. Makino, M. Amemura, and A. Nakata. 1989. Nucleotide sequence of the ugpQ gene encoding glycerophosphoryl diester phosphodiesterase of Escherichia coli K-12. Nucleic Acids Res. 17:2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasahara, M., K. Makino, M. Amemura, A. Nakata, and H. Shinagawa. 1991. Dual regulation of the ugp operon by phosphate and carbon starvation at two interspaced promoters. J. Bacteriol. 173:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kates, M. 1986. Techniques of lipidology: isolation, analysis and identification of lipids, vol. 3, pt. 2. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 24.Kim, J. W., E. I. Rainina, W. W. Mulbry, C. R. Engler, and J. R. Wild. 2002. Enhanced-rate biodegradation of organophosphate neurotoxins by immobilized nongrowing bacteria. Biotechnol. Prog. 18:429-436. [DOI] [PubMed] [Google Scholar]
- 25.Kissinger, C. R., H. E. Parge, D. R. Knighton, C. T. Lewis, L. A. Pelletier, A. Tempczyk, V. J. Kalish, K. D. Tucker, R. E. Showalter, E. W. Moomaw, et al. 1995. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature 378:641-644. [DOI] [PubMed] [Google Scholar]
- 26.Knofel, T., and N. Strater. 2001. Mechanism of hydrolysis of phosphate esters by the dimetal center of 5′-nucleotidase based on crystal structures. J. Mol. Biol. 309:239-254. [DOI] [PubMed] [Google Scholar]
- 27.Knofel, T., and N. Strater. 1999. X-ray structure of the Escherichia coli periplasmic 5′-nucleotidase containing a dimetal catalytic site. Nat. Struct. Biol. 6:448-453. [DOI] [PubMed] [Google Scholar]
- 28.Koonin, E. V. 1994. Conserved sequence pattern in a wide variety of phosphoesterases. Protein Sci. 3:356-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai, K., N. J. Stolowich, and J. R. Wild. 1995. Characterization of P-S bond hydrolysis in organophosphorothioate pesticides by organophosphorus hydrolase. Arch. Biochem. Biophys. 318:59-64. [DOI] [PubMed] [Google Scholar]
- 30.Lohse, D. L., J. M. Denu, and J. E. Dixon. 1995. Insights derived from the structures of the Ser/Thr phosphatases calcineurin and protein phosphatase 1. Structure 3:987-990. [DOI] [PubMed] [Google Scholar]
- 31.Makino, K., H. Shinagawa, M. Amemura, S. Kimura, A. Nakata, and A. Ishihama. 1988. Regulation of the phosphate regulon of Escherichia coli: activation of pstS transcription by PhoB protein in vitro. J. Mol. Biol. 203:85-95. [DOI] [PubMed] [Google Scholar]
- 32.Makino, K., H. Shinagawa, M. Amemura, and A. Nakata. 1986. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J. Mol. Biol. 190:37-44. [DOI] [PubMed] [Google Scholar]
- 33.Mizuguchi, K., C. M. Deane, T. L. Blundell, and J. P. Overington. 1998. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 7:2469-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulchandani, A., I. Kaneva, and W. Chen. 1999. Detoxification of organophosphate nerve agents by immobilized Escherichia coli with surface-expressed organophosphorus hydrolase. Biotechnol. Bioeng. 63:216-223. [DOI] [PubMed] [Google Scholar]
- 35.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Brien, P. J., and D. Herschlag. 2002. Alkaline phosphatase revisited: hydrolysis of alkyl phosphates. Biochemistry 41:3207-3225. [DOI] [PubMed] [Google Scholar]
- 37.O'Brien, P. J., and D. Herschlag. 1999. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6:R91-R105. [DOI] [PubMed] [Google Scholar]
- 38.Overduin, P., W. Boos, and J. Tommassen. 1988. Nucleotide sequence of the ugp genes of Escherichia coli K-12: homology to the maltose system. Mol. Microbiol. 2:767-775. [DOI] [PubMed] [Google Scholar]
- 39.Richter, W. 2002. 3′, 5′ Cyclic nucleotide phosphodiesterases class III: members, structure, and catalytic mechanism. Proteins 46:278-286. [DOI] [PubMed] [Google Scholar]
- 40.Schweizer, H., and W. Boos. 1984. Characterization of the ugp region containing the genes for the phoB dependent sn-glycerol-3-phosphate transport system of Escherichia coli. Mol. Gen. Genet. 197:161-168. [DOI] [PubMed] [Google Scholar]
- 41.Shi, J., T. L. Blundell, and K. Mizuguchi. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310:243-257. [DOI] [PubMed] [Google Scholar]
- 42.Sonnhammer, E. L., S. R. Eddy, E. Birney, A. Bateman, and R. Durbin. 1998. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 26:320-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strater, N., T. Klabunde, P. Tucker, H. Witzel, and B. Krebs. 1995. Crystal structure of a purple acid phosphatase containing a dinuclear Fe(III)-Zn(II) active site. Science 268:1489-1492. [DOI] [PubMed] [Google Scholar]
- 44.Tehara, S. K., and J. D. Keasling. 2003. Gene cloning, purification, and characterization of a phosphodiesterase from Delftia acidovorans. Appl. Environ. Microbiol. 69:504-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasudevan, S. G., W. L. Armarego, D. C. Shaw, P. E. Lilley, N. E. Dixon, and R. K. Poole. 1991. Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol. Gen. Genet. 226:49-58. [DOI] [PubMed] [Google Scholar]
- 46.Vincent, J. B., and B. A. Averill. 1990. Sequence homology between purple acid phosphatases and phosphoprotein phosphatases. Are phosphoprotein phosphatases metalloproteins containing oxide-bridged dinuclear metal centers? FEBS Lett. 263:265-268. [DOI] [PubMed] [Google Scholar]
- 47.Walker, A. W., and J. D. Keasling. 2002. Metabolic engineering of Pseudomonas putida for the utilization of parathion as a carbon and energy source. Biotechnol. Bioeng. 78:715-721. [DOI] [PubMed] [Google Scholar]
- 48.Wolfenden, R., and G. Spence. 1967. Derepression of phosphomonoesterase and phosphodiesterase activities in Aerobacter aerogenes. Biochim. Biophys. Acta 146:296-298. [DOI] [PubMed] [Google Scholar]
- 49.Yang, H., P. D. Carr, S. Y. McLoughlin, J. W. Liu, I. Horne, X. Qiu, C. M. Jeffries, R. J. Russell, J. G. Oakeshott, and D. L. Ollis. 2003. Evolution of an organophosphate-degrading enzyme: a comparison of natural and directed evolution. Protein Eng. 16:135-145. [DOI] [PubMed] [Google Scholar]