Abstract
Background
Crohn's disease (CD) has the highest prevalence among individuals of Ashkenazi Jewish (AJ) descent compared to non-Jewish Caucasian populations (NJ). We evaluated a set of well-established CD-susceptibility variants to determine if they can explain the increased CD risk in the AJ population.
Methods
We recruited 369 AJ CD patients and 503 AJ controls, genotyped 22 single nucleotide polymorphisms (SNPs) at or near 10 CD-associated genes, NOD2, IL23R, IRGM, ATG16L1, PTGER4, NKX2-3, IL12B, PTPN2, TNFSF15 and STAT3, and assessed their association with CD status. We generated genetic scores based on the risk allele count alone and the risk allele count weighed by the effect size, and evaluated their predictive value.
Results
Three NOD2 SNPs, two IL23R SNPs, and one SNP each at IRGM and PTGER4 were independently associated with CD risk. Carriage of 7 or more copies of these risk alleles or the weighted genetic risk score of 7 or greater correctly classified 92% (allelic count score) and 83% (weighted score) of the controls; however, only 29% and 47% of the cases were identified as having the disease, respectively. This cutoff was associated with a >4-fold increased disease risk (p < 10e-16).
Conclusions
CD-associated genetic risks were similar to those reported in NJ population and are unlikely to explain the excess prevalence of the disease in AJ individuals. These results support the existence of novel, yet unidentified, genetic variants unique to this population. Understanding of ethnic and racial differences in disease susceptibility may help unravel the pathogenesis of CD leading to new personalized diagnostic and therapeutic approaches.
Keywords: Crohn's Disease, Ashkenazi Jewish, genetic risk score
Background
Crohn's disease (CD) is one of the forms of inflammatory bowel disease (IBD) resulting from defects in the regulation of mucosal immune responses to enteric bacteria in genetically susceptible individuals (reviewed in [1,2]). Familial aggregation [3,4] and higher concordance rates in monozygotic than dizygotic twins [5,6] have provided robust evidence for the involvement of genetic factors in the disease etiology. Also, an important epidemiological feature of CD is that it occurs at significantly different frequencies in different ethnic, demographic and racial groups [7], and has the highest prevalence among individuals of Ashkenazi Jewish (AJ) descent [8,9]. Although the underlying mechanism(s) responsible for ethnic differences remain unclear, it has been hypothesized that the substantially increased risk in AJ versus NJ CD can be explained by the higher frequency and/or greater magnitude of the effect of susceptibility gene variants.
The today largest meta-analysis of six genome-wide association studies (GWAS) have reported 71 confirmed genetic variants predisposing to CD [10], some whose pathogenesis is reasonably well defined (reviewed in [11]).
The strongest and most consistently replicated genetic CD-risk factors are three variants within the nucleotide-binding oligomerization domain containing 2 gene (NOD2, also known as CARD15), namely, p.G908R, p.R702W, and p.L1007fs [12-14]. These three variants have been shown to increase the odds of developing CD by two- to four-fold [15]. There are additional variants of the NOD2 gene that also have been implicated to increase CD risk [16-18]. Another well-validated gene is the interleukin 23 receptor (IL23R), a regulatory cytokine involved in the initiation of innate and adaptive immune responses. In the IL23R gene, a rare non-synonymous variant, p.R381Q, appears to be protective against developing CD in both AJ and NJ populations [16]. Also, several common variants at the autophagy-inducing genes, ATG16L1, autophagy-related 16-like 1, and IRGM, immunity-related guanosine triphosphatase, have been reported through GWAS to be associated with CD in Caucasian populations [17,19-21]. In addition, a locus on chromosome 5p13.1 in the vicinity of the prostaglandin E receptor 4 (subtype EP4) gene, PTGER4, also emerged as having CD susceptibility [18,19].
Modest variation in CD risk has been attributed to additional genes, NKX2-3, a member of the NKX family of homeodomain-containing transcription factors involved in the intestinal inflammatory response [19-21]; IL12B, a cytokine which is a member of the IL12/IL23 pathway playing a key role in chronic intestinal inflammation [19,20]; PTPN2, a protein tyrosine phosphatase that serves as a key negative regulator of inflammatory responses [19-21]; TNFSF15, a member of the tumor necrosis factor superfamily previously reported to be associated with CD in European and Japanese populations [19,22], and STAT3, a signal transducer and activator of transcription with a central role in Th17 differentiation and IL10 signaling [19,23].
Despite consistent replication of the association of these gene variants with CD in various cohorts of European descent, a number of unanswered questions remain regarding the actual risk associated with carrying multiple variants, and the degree to which these variants may explain the excess risk of CD observed in the AJ population. Therefore, this study was designed to determine allele frequencies and to test the joint contribution of the 22 independently replicated susceptibility loci to CD risk in a clinically well-defined AJ population, and to evaluate if combinations of these alleles could help predict CD status.
Materials and methods
Study Population
A total of 369 unrelated individuals of AJ descent with CD were recruited from the New York metropolitan area and from Israel. All patients were seen by gastroenterologists, their diagnoses were based on clinical, endoscopic, radiological, and/or histopathological findings using established criteria [24], and each patient provided a blood sample. A total of 503 AJ individuals referred for prenatal carrier testing for Jewish genetic diseases at the Mount Sinai Medical Center served as controls [25]. No phenotypic information, including the CD status, was available on controls. Self-reported ethnicity was assigned on the basis of having four grandparents of AJ origin in both cases and controls. The study was approved by the relevant institutional review board(s), and informed consent was obtained from each participant.
SNP Selection and Genotyping
We used previous GWAS and candidate gene studies to select the 22 most replicated CD-related single nucleotide polymorphisms (SNPs) at or near 10 genes (see Additional file 1, Table S1 online). DNA was extracted from peripheral blood using the Purgene procedure (Gentra Systems Inc, Minneapolis, MN). Genotyping was performed using Taqman assays on an Applied Biosystems PRISM 7900HT Sequence Detection System according to the manufacturer's protocol (Life Technologies, Carlsbad, CA). The specific assays used for genotyping are listed in Additional file 1, Table S1 online.
Statistical Analysis
Observed genotype frequencies among the AJ controls were compared with those expected under Hardy-Weinberg equilibrium using a χ2 test. Pairwise linkage disequilibrium (LD) was assessed using Lewontin's D' and r2 as implemented in Haploview [26]. For each variant, univariate logistic regression was used to calculate allelic odds ratios (ORs) of CD for risk allele carriers versus non-carriers. Multivariate logistic regression was applied to estimate the combined effect of the genetic variants on the disease status. In addition, to estimate the predictive value of multiple susceptibility loci on disease status, we constructed two genetic risk profiles based on the SNPs that sustained statistical significance in the multivariate model. First, we assigned each person a score based on the number of risk alleles carried for the SNPs significantly associated with CD risk in the multivariate model. We assigned "0" to common allele homozygote carriers, "1" for heterozygotes, and "2" for rare allele homozygotes. Second, we calculated a score that weighted the number of susceptibility alleles for each SNP by the strength of their association as determined by regression coefficients [27] based on odds ratios obtained in the multivariate model. Namely, we multiplied ORs by 0, 1, or 2 according to the number of risk alleles carried by each person.
We evaluated the number of risk alleles and weighted score as predictors of genetic susceptibility in logistic regression analyses using different cutoff points. A two-tailed P < 0.05 was considered statistically significant. All analyses were performed using SAS/STAT and SAS/Genetics software version 9.1 (SAS Institute, Inc., Cary, North Carolina, USA).
Results
The observed genotype frequencies among the controls did not significantly differ from those expected under Hardy-Weinberg equilibrium for any of the variants under study. The risk allele distribution for CD cases and unaffected controls is shown in Table 1. For the three extensively studied NOD2 variants, each high risk allele was associated with a significantly increased risk of CD, such as 1.9 (95% CI: 1.1-3.2; p = 0.03) for p.R702W, 3.2 (95% CI: 2.2-4.9; p = 6.4*10-9) for p.G908R, and 5.2 (95% CI: 3.3-8.3; p = 2.0*10-12) for p.L1007fs. In univariate analysis, the presence of at least one of the three high-risk NOD2 alleles conferred a 4.1 risk for CD, whereas the presence of two variant copies of the gene, either the same (homozygotes) or different (compound heterozygotes), conferred an 8.7-fold risk (Table 1). Overall, the rate of carriage of any of these risk variants was 43% in AJ CD cases and 15% in AJ controls. Two additional NOD2 risk variants in intron 2, rs17221417, and intron 8, rs2076756, were strongly linked with the coding variants (D' = 0.99, r2 = 0.17 between rs17221417 and p.G908R and D' = 0.97, r2 = 0.15 between rs2076756 and p.L1007fs, see Additional file 1, Table S2 online) and individually showed a 2-fold increased risk of the disease (Table 1).
Table 1.
Univariate Analysis of Susceptibility Loci with Crohn's Disease Status.
| Gene | SNP ID | Amino acid substitution | Allele frequency | OR(95% CI) 1 | P-value | |
|---|---|---|---|---|---|---|
| Controls (n = 503) | Cases (n = 369) | |||||
| NOD2 | rs2066844 | p.R702W | 0.02 | 0.04 | 1.85 (1.08-3.18) | 0.03 |
| rs2066845 | p.G908R | 0.04 | 0.11 | 3.23 (2.15-4.85) | 6.39*10-9 | |
| rs2066847 | p.L1007fs2 | 0.02 | 0.11 | 5.18 (3.26-8.25) | 2.03*10-12 | |
| Carriage3 | 0.14 | 0.34 | 4.13(3.00-5.67) | 1.49*10-19 | ||
| Compound Hetero/Homozygous4 | 0.01 | 0.09 | 8.68 (3.61-20.87) | 1.10*10-8 | ||
| rs17221417 | intron 2 | 0.22 | 0.38 | 2.13 (1.73-2.63) | 1.32*10-12 | |
| rs2076756 | intron 8 | 0.22 | 0.39 | 2.26 (1.83-2.79) | 6.50*10-14 | |
| IL23R | rs11209026 | R381Q | 0.07 | 0.02 | 0.27 (0.16-0.48) | 4.73*10-6 |
| rs7517847 | intron 6 | 0.38 | 0.25 | 0.55 (0.43-0.68) | 6.88*10-8 | |
| rs11805303 | intron 6 | 0.35 | 0.41 | 1.32 (1.09-1.61) | 0.0076 | |
| IRGM | rs13361189 | intergenic | 0.16 | 0.21 | 1.47 (1.15-1.88) | 0.0014 |
| rs11747270 | intergenic | 0.15 | 0.21 | 1.48 (1.16-1.90) | 0.0012 | |
| rs1000113 | intergenic | 0.15 | 0.21 | 1.49 (1.17-1.91) | 0.0011 | |
| PTGER4 | rs1373692 | p.T300A | 0.61 | 0.67 | 1.29 (1.06-1.58) | 0.012 |
| rs1992660 | intergenic, near 5'-UTR | 0.60 | 0.65 | 1.21 (0.99-1.47) | 0.062 | |
| rs4613763 | intergenic | 0.07 | 0.08 | 1.05 (0.74-1.51) | 0.77 | |
| ATG16L1 | rs2241880 | intergenic | 0.59 | 0.63 | 1.18 (0.97-1.43) | 0.10 |
| rs10210302 | intergenic | 0.59 | 0.62 | 1.14 (0.94-1.39) | 0.19 | |
| NKX2-3 | rs11190140 | intergenic, near 5'-UTR | 0.45 | 0.43 | 0.92 (0.76-1.11) | 0.36 |
| IL12B | rs6887695 | intergenic | 0.32 | 0.32 | 1.03 (0.84-1.26) | 0.81 |
| rs10045431 | intergenic | 0.71 | 0.69 | 0.94 (0.77-1.16) | 0.59 | |
| PTPN2 | rs2542151 | intergenic | 0.10 | 0.12 | 1.20 (0.89-1.63) | 0.22 |
| TNFSF15 | rs4263839 | intron 1 | 0.76 | 0.75 | 0.95 (0.77-1.19) | 0.67 |
| STAT3 | rs744166 | intron 1 | 0.61 | 0.62 | 0.99 (0.82-1.21) | 0.93 |
OR (95% CI), Odds ratio (95% confidence interval); 1 Test for allelic trend; 2 fs, Frame shift; 3 Carriage of at least one of the above three NOD2/CARD15 risk alleles, and 4 Homozygous carriage of at least one of the above risk alleles or heterozygous carriage of more than one of the above risk alleles.
The rare IL23R p.R381Q variant (rs11209026) was more frequently found among AJ controls than among AJ patients (7% versus 2%, respectively) indicating a significant reduction in CD risk for carriers (OR = 0.3, 95% CI: 0.2-0.5; p = 4.7*10-6). Two non-coding IL23R intron 6 variants linked to rs11209026, (rs11805303 with D' = 1.0, r2 = 0.03 and rs7517847 with D' = 0.86, r2 = 0.08) were also individually associated with CD status, with the rs7517847 variant possessing a protective effect and rs11805303 increasing the risk, similar in direction to their effects in NJ CD [16,17] (Table 1).
In addition, a significant association was detected between the IRGM minor alleles at rs13361189, rs11747270, and rs1000113 and CD risk. All of these SNPs have similar allele frequencies, were in strong LD (pairwaise D'>0.99 and r2>0.95) and associated with a 1.5-fold (95% CI: 1.5-1.9; p = 0.001) increased prevalence of CD. Also, carriers of the common tightly linked variants at PTGER4 rs1373692 and rs1992660 (D' = 0.96, r2 = 0.87) had higher odds of developing the disease than non-carriers. A trend toward a higher frequency of the ATG16L1 rs2241880 and rs10210302 polymorphisms (in LD, D' = 0.99, r2 = 0.96) was observed in CD cases compared to controls, but this association did not reach statistical significance. No other variants were detected to be individually associated with CD risk in our AJ cohort.
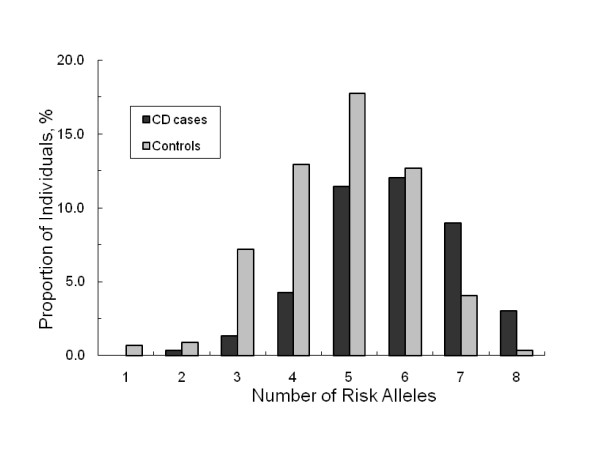
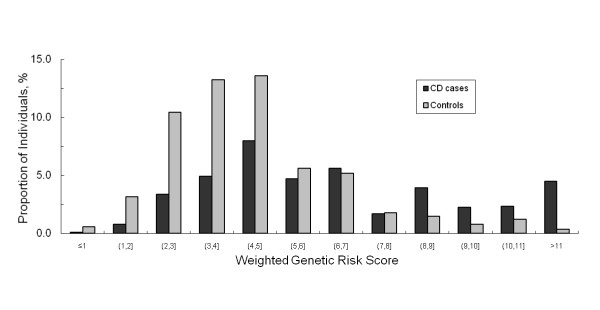
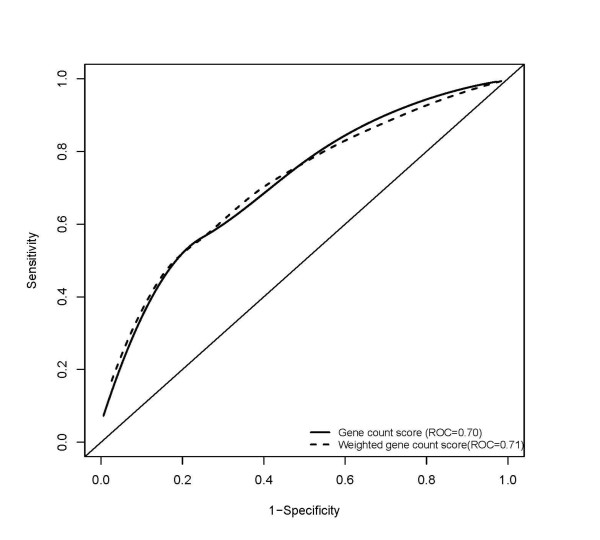
In a multivariate logistic regression model, only three uncommon coding SNPs at NOD2 and one SNP each at IRGM and PTGER4 were independently associated with the increased risk of CD, whereas two SNPs at IL23R showed a protective effect against CD (Table 2). Comparison of the distribution of the total number of these seven loci in cases and controls showed that individuals with CD tended to carry more copies of these susceptibility alleles than unaffected controls (Figure 1). Also, more CD patients than controls had a higher weighted risk score (Figure 2). Using these risk scores to distinguish cases from controls, specificity was high for carriers of ≥7 copies of the seven significant risk alleles and a weighted score of ≥7 (92.2% and 82.5%, respectively) when the highest test accuracy was achieved (65.5% and 67.5%, respectively), implying a relatively low rate of misclassification in the absence of the disease. However, this was at the expense of low sensitivity showing that only 29% and 47% of cases were correctly identified based on the allelic count and weighted genetic risk scores, respectively (Table 3). The area under the receiver operating characteristics (ROC) curve for the two risk models were 70% and 71%, respectively (Figure 3).
Table 2.
Genetic effects from multivariate regression model.
| Gene | SNP | 1 Copy of risk allele | 2 Copies of risk allele | Log additive model | |||||
|---|---|---|---|---|---|---|---|---|---|
| Case/Control | OR (95% CI) | P | Case/Control | OR (95% CI) | P | OR (95% CI) | P | ||
| NOD2 | rs2066844 | 28/20 | 2.18 (1.17-4.07) | 0.015 | 2/2 | 2.03 (0.27-15.12) | 0.49 | 2.01 (1.18-3.44) | 0.010 |
| rs2066845 | 67/36 | 3.05 (1.93-4.80) | 1.14*10-6 | 6/0 | ND | - | 3.30 (2.16-5.06) | 3.92*10-8 | |
| rs2066847 | 61/22 | 5.03 (2.93-8.64) | 2.99*10-9 | 11/1 | 36.84 (4.55-298.29) | 0.0007 | 5.24 (3.26-8.44) | 9.18*10-12 | |
| IL23R | rs11209026 | 15/59 | 0.44 (0.23-0.84) | 0.013 | 0/6 | ND | - | 0.43 (0.23-0.80) | 0.0075 |
| rs7517847 | 144/210 | 0.74 (0.54-1.01) | 0.06 | 20/84 | 0.24 (0.13-0.44) | 6.47*10-8 | 0.59 (0.47-0.74) | 9.20*10-6 | |
| IRGM | rs13361189 | 132/139 | 1.70 (1.24-2.35) | 0.0012 | 13/9 | 2.35 (0.93-5.93) | 0.072 | 1.64 (1.24-2.17) | 0.0005 |
| PTGER4 | rs1373692 | 160/238 | 1.18 (0.73-1.90) | 0.50 | 167/188 | 1.75 (1.09-2.83) | 0.021 | 1.37 (1.10-1.70) | 0.0052 |
OR (95% CI), Odds ratio (95% confidence interval); ND, not determined.
Figure 1.
Percentage of participants in each allele count score category among those who developed CD and those who remained CD-free based on 7 SNPs at NOD2, IL23R, IRGM, and PTGER4.
Figure 2.
Percentage of participants in each weighted genetic risk score category among those who developed CD and those who remained CD-free based on 7 SNPs at NOD2, IL23R, IRGM, and PTGER4.
Table 3.
Genetic risk profile using CARD15/NOD2, IL23R, PTGER4, and IRGM susceptibility alleles.
| Risk profile | N (%) | OR (95% CI) | P | Sensitivity | Specificity | PPV | NPV | Accuracy | |
|---|---|---|---|---|---|---|---|---|---|
| CD | Control | ||||||||
| Number of risk alleles | |||||||||
| ≥3 | 366 (99) | 489 (97) | 3.49 (0.99-12.24) | 0.05 | 99.2 | 2.8 | 42.8 | 82.4 | 43.6 |
| ≥4 | 354 (96) | 425 (84) | 4.33 (2.48-7.66) | 5.80*10-8 | 95.9 | 15.5 | 45.4 | 83.9 | 49.5 |
| ≥5 | 316 (86) | 310 (62) | 3.71 (2.64-5.23) | 3.95*10-15 | 85.6 | 38.4 | 50.5 | 78.5 | 58.4 |
| ≥6 | 214 (58) | 152 (30) | 3.19 (2.41-4.22) | 1.94*10-16 | 57.9 | 69.8 | 58.5 | 69.4 | 64.5 |
| ≥7 | 107 (29) | 39 (8) | 4.86 (3.27-7.22) | 9.88*10-17 | 29.0 | 92.2 | 73.3 | 63.9 | 65.5 |
| ≥8 | 27 (7) | 3 (0.6) | 13.16 (3.96-43.72) | 4.29*10-8 | 7.3 | 99.4 | 90.0 | 59.4 | 60.4 |
| Weighted Score | |||||||||
| ≥2 | 368 (>99) | 498 (99) | 3.69 (0.43-31.75) | 0.25 | 99.7 | 1.0 | 42.5 | 83.3 | 42.8 |
| ≥3 | 361 (99) | 470 (93) | 3.17 (1.45-6.94) | 0.003 | 97.8 | 6.6 | 43.4 | 80.5 | 45.2 |
| ≥4 | 331 (90) | 377 (75) | 2.91 (1.97-4.31) | 4.13*10-8 | 89.7 | 25.0 | 46.8 | 76.8 | 52.4 |
| ≥5 | 287 (78) | 259 (51) | 3.30 (2.44-4.46) | 2.42*10-15 | 77.8 | 48.5 | 52.6 | 74.8 | 60.9 |
| ≥6 | 216 (59) | 138 (27) | 3.73 (2.81-4.97) | 1.97*10-20 | 58.5 | 72.6 | 61.0 | 70.5 | 66.6 |
| ≥7 | 174 (47) | 88 (17) | 4.21 (3.09-5.72) | 3.65*10-21 | 47.2 | 82.5 | 66.4 | 68.0 | 67.5 |
| ≥8 | 124 (34) | 42 (8) | 5.56 (3.79-8.15) | 5.14*10-21 | 33.6 | 91.7 | 74.7 | 65.3 | 67.1 |
| ≥9 | 108 (29) | 27 (5) | 7.69 (4.89-12.11) | 3.49*10-23 | 29.3 | 94.6 | 80.0 | 64.6 | 67.0 |
| ≥10 | 73 (20) | 20 (4) | 9.45 (5.15-17.34) | 5.74*10-18 | 19.8 | 96.0 | 78.5 | 62.0 | 63.8 |
| ≥11 | 53 (14) | 13 (3) | 14.2 (6.04-33.40) | 1.76*10-15 | 14.4 | 97.4 | 80.3 | 60.8 | 62.3 |
OR (95% CI), Odds ratio (95% confidence interval); PPV, Positive predictive value; NPV, Negative predictive value.
Figure 3.
Receiver operating curve (ROC) for allele count and weighted genetic risk scores based on 7 SNPs at NOD2, IL23R, IRGM, and PTGER4. Diagonal line indicates prediction by chance alone.
The CD risks in the participants with ≥7 copies of the risk alleles or a weighted score of ≥7 compared to those with <7 copies of the risk alleles or a weighted score of <7 were 4.9-fold (95% CI: 3.3-7.2) and 4.2-fold (95% CI: 3.1-5.7), respectively (Table 3).
Discussion
This study was designed to assess the contribution of established risk variants in selected CD-associated genes to CD risk in the AJ population, a group known to have an unusually high prevalence of the disease compared to NJ European or North American Caucasians. We confirmed previous reports of significant association between variants in several immunity-related genes and disease susceptibility in Caucasian populations, but they do not appear to account for the increased CD risk in the AJ population. NOD2, the first gene associated with CD, is a polymorphic gene involved in innate immune responses. To date, over 60 NOD2 variations have been identified, several of which are specific to AJ individuals [28,29], with p.R702W, pG908R, and p.L1007fs accounting for 81% of the genetic variation [30]. These three variants were present in 43% of our CD patients compared to 15% of controls. Rare variants identified by sequencing of the NOD2 promoter and exonic regions in AJ families revealed no association with disease risk [28]. In the present study, carriage of any one of the three NOD2 variants was found to be associated with an over 4-fold increased risk of CD, while carriage of at least two of the SNPs increased the odds of AJ CD almost nine times. This is comparable with the estimates of 2.39 (95% CI: 2.00-2.90) and 17.1 (95% CI: 10.7-27.2)-fold increased risk, respectively, reported in a large meta-analysis of 42 studies conducted predominantly in NJ populations [15]. Compared to a sub-analysis of six studies that analyzed a total of 1,658 individuals of AJ descent, each variant allele in our study conferred a slightly higher risk for p.R702W, p.G908R and p.L1007fs (OR of 1.85 (95% CI: 1.08-3.18), 3.23 (95% CI: 2.15-4.85) and 5.18 (95% CI: 3.26-8.25) versus 1.74 (95% CI: 0.88-3.42), 1.93 (95% CI: 1.25-3.00), and 2.45 (95% CI: 1.51-3.98), respectively). While additional more common non-coding NOD2 variants were individually associated with CD status in univariate analyses, their significance disappeared in the presence of the three established NOD2 risk loci.
Since the original report showing a protective effect on susceptibility to CD of the IL23R non-synonymous coding variant rs11209026 (p.R381Q) [16], numerous reports including GWAS, have underscored the importance of this allele in lowering IBD risk [18,31,32]. In our study, the magnitude of protection from CD associated with the minor allele at IL23R rs11209026 (p.R381Q) was comparable to that reported previously in AJ and NJ cohorts (OR range 0.26-0.41) [16,33] with one exception, in which rs11209026 was not associated with protection from CD in an AJ population in Canada [34]. Notably, we observed higher frequencies of the rs11209026 variant in AJ controls compared to NJ controls suggesting a greater protection in the AJ population. In the present study, two additional non-coding IL23R variants (rs7517847 and rs11805303) were independently associated with CD risk with rs7517847 maintaining its significance in a multivariate model after the adjustment for other susceptibility loci. This marker has been previously implicated in AJ and NJ CD [16,17]. Of note, the largest today GWAS meta-analysis of ulcerative colitis (UC), another form of IBD, has shown that, among the 99 confirmed IBD loci meeting genome-wide significance (P < 5 × 10-8) in UC and/or CD, 28 loci shared association signals between UC and CD, with many common variants identified in the IL23 signaling pathway, specifically IL23R, JAK2, STAT3, IL12B, and PTPN2 [35]. The significance of these findings is underlined by the central role that IL23 plays in the induction of IL-17 by Th17 lymphocytes [36].
In addition to confirming several associations between genetic variants and CD risk, our study, for the first time, replicated IRGM and PTGER4 as CD susceptibility loci in the AJ population. Among several IRGM variants individually associated with CD risk in univariate analyses, only intergenic variant rs13361189 survived the adjustment for other genetic risk factors. That is, in a multivariate model, we detected a 1.64-fold increased CD risk per each copy of the IRGM rs13361189 risk allele. An earlier meta-analysis combining the effects of IRGM rs13361189 across studies of European populations reported a pooled OR of 1.34 per copy of the variant [37]. Detection of this polymorphism in the flanking region of the gene prompted sequencing of the entire gene; however, no non-synonymous variants associated with the disease were identified suggesting that the causal variant is unlikely to change the amino acid sequence of the IRGM protein [21]. Moreover, a 20-kb deletion polymorphism, upstream of IRGM and in perfect linkage disequilibrium with rs13361189, has been reported to have distinct expression patterns modulating the cellular autophagy process in response to intracellular bacteria [38].
Several GWAS have identified a 250-kb region of chromosome 5p13.1 containing multiple SNPs with strongly suggestive evidence of disease association [10,18-21,39]. Despite the fact that this locus is contained within a 1.25-Mb gene desert, there is consistent evidence that disease-associated alleles correlate with PTGER4 expression levels. A common intergenic variant near PTGER4, rs1373692, has been associated with a 1.59-fold increase in CD risk in a Caucasian population [18]. Our results show that the PTGER4 rs1373692 risk allele frequency was higher in AJ cases than in AJ controls, translating to a 1.37-fold increase in odds of developing the disease per one copy of the risk allele. While the exact mechanism of the PTGER4 variation on the pathophysiology of CD remains unclear, it is likely that variants in this locus may modulate PTGER4 expression levels [18].
Importantly, while similar effect sizes were determined for the PTGER4 and IRGM risk alleles in the AJ cohort as in NJs, considerable differences in frequencies were observed between AJ controls and those reported in the literature for NJ controls. For the variants that remained significant in the multivariate model, IRGM rs13361189 was as twice as common in AJ controls than in NJs (16% versus 8%). Further studies are warranted to understand the cause and consequences of population differences in risk allele distribution.
Although we found a trend toward a slightly higher frequency of the ATG16L1 rs2241880 G allele among AJ CD cases in univariate analyses, this association did not reach statistical significance. A previous study found no association between this variant and the risk of CD in an AJ population [34]. However, a series of recent meta-analyses have concluded that this variant is associated with increased risk of CD in NJ Caucasian populations [37,40,41]. The meta-analysis of 17 studies that included over 30,000 subjects showed a significant association between the ATG16L1 rs2241880 polymorphism and CD risk with an OR of 1.39 (95% CI: 1.27-1.51) and 1.87 (95% CI: 1.69-2.05) for heterozygote and homozygote risk allele carriers, respectively, compared to wild-type homozygotes [40]. Two additional meta-analyses have also shown a strong association between CD and ATG16L1 rs2241880, finding a 1.62 (95% CI: 1.37-1.91) and 1.28 (95% CI: 1.06-1.54)-fold increase in CD risk for single allele carriers [37,41]. Based on our sample size, we had 80% statistical power to detect an OR of 1.32 or higher, assuming a log-additive genetic model. Yet, we cannot rule out a smaller effect of the ATG16L1 rs2241880 variant or involvement of additional SNPs that might confer a stronger effect in individuals of AJ descent.
We evaluated the ability to predict CD risk using the combined information from the seven variants of the four genes that were found to be independently significant in the multivariate model. The genetic risk profile was based on the number of multiple risk alleles carried by each individual and their weighed effects. The highest test prediction accuracy was achieved with the cut-off of equal to or greater than seven for both allelic count score and the weighted genetic risk score. While the specificity of the tests was reasonably high, with 83-92% of controls predicted to not have CD, only about 29%-47% of the cases were identified as having the disease.
Genetic risk scores have previously been applied by other investigators [27,42-44]. A combination of NOD2, IBD5, NOD1, and TNFSF15 genotype and smoking status demonstrated comparable sensitivities and specificities in an earlier study of NJ Caucasian CD cases [27]. Another report claimed to successfully predict a CD genetic risk profile similar to our study using 10 alleles at five genes including NOD2, ATG16L1, IRGM, IL23R and 5p13 (corresponding to the PTGER4 region) in a European NJ population [44]. Even though these studies used different combinations of susceptibility loci in their predictive models, the tradeoff between the sensitivity and specificity was similar. Further work will be needed before predictive models can be translated into direct clinical utility and help clinicians rule out CD in individuals with symptoms of IBD leading to more efficient patient management and resource utilization.
Limitations of our study include the fact that our controls were individuals referred for unrelated genetic testing and provided no information on their CD status and other phenotypic characteristics. Given the low disease prevalence, it is highly unlikely that more than two or three affected individuals would be found in the control group. Nonetheless, even if such misclassification did occur, this would bias our results toward null. In addition, AJ ethnicity was self-reported and not verified using ancestry informative markers. However, a recent study has shown that within self-described Americans of European descent, there is a clear genetic corollary which would permit near perfect inference of AJ ancestry [45]. Moreover, a relatively small sample size may have prevented us from detecting modest effects, such as for the NKX2-3, IL12B, PTPN2, TNFSF15, or STAT3 polymorphisms (OR < 1.3; see Additional file 1, Table S1 online) that were confirmed by the recent and largest CD meta-analysis [10]. However, even though we cannot rule out smaller effects of these variants in individuals of AJ descent, it is unlikely that they are responsible for excess disease prevalence in this population.
Also, differences in linkage disequilibrium patterns between AJ and NJ populations may result in the contribution of different genetic markers in the region to disease susceptibility in individuals of AJ descent. In addition, due to a limited number of studies that reported effect sizes of selective CD-related risk variants in cohorts of AJ descent, we were restricted to using the same dataset to develop and validate the predictive models. For the weighted risk score estimation, we used odds ratios derived using an additive genetic model. This means that the logarithm of the odds ratio was assumed to relate linearly to the number of copies of the higher-risk allele [46]. Thus, the odds ratio reported was that associated with one copy of the higher-risk allele, which likely underestimated the effect of risk of homozygous carriage. This was due to the fact that an accurate estimation of the genotype-based odds ratios was not possible for some of the susceptibility loci. Further studies are warranted to evaluate the performance of our models in independent AJ populations.
Conclusions
In summary, the present studies determined for the first time the magnitude of individual and combined risks of multiple CD susceptibility loci in the AJ population. We confirmed previously reported associations of the NOD2, IL23R, IRGM and PTGER polymorphisms with CD risk. Genetic effects estimated in the present study were similar to those observed in other Caucasian populations and are, thus, unlikely to explain the excessive prevalence of CD in individuals of AJ descent. Existence of other, yet unidentified, AJ-specific genetic risk factors and environmental triggers remain to be determined. Understanding of population-specific differences in disease susceptibility may help unravel the pathogenesis of CD leading to new personalized diagnostic and therapeutic approaches.
List of Abbreviations
CD: Crohn's Disease; AJ: Ashkenazi Jewish; NJ: non-Jewish; SNP: Single Nucleotide Polymorphism; NOD2: nucleotide-binding oligomerization domain containing 2 gene; IL23R: interleukin 23 receptor gene; IRGM: immunity-related guanosine triphosphatase family, M gene; ATG16L1: autophagy-related 16-like 1 gene; PTGER4: prostaglandin E receptor 4 (subtype EP4) gene; NKX2-3: NKX transcription factor related, locus 3 gene; IL12B: interleukin 12B gene; PTPN2: protein tyrosine phosphatase non-receptor type 2 gene; TNFSF15: tumor necrosis factor (ligand) superfamily, member 15 gene; STAT3: signal transducer and activator of transcription gene;
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
IP conceived the study, participated in its design and statistical data analysis and drafted the manuscript. AAM designed the study, oversaw genotyping, and helped draft the manuscript. LO, participated in the design of the study, oversaw genotyping, and helped draft the manuscript. MTA, DHP, TU, KB, BIK, and LM participated in the design of the study, recruited the patients, and provided critical comments. ME carried out the molecular genetic studies. JH performed the statistical analysis. RJD participated in the design of the study and helped draft the manuscript. The New York Crohn's Disease Working Group recruited the patients. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Supplementary tables.
Contributor Information
Inga Peter, Email: inga.peter@mssm.edu.
Adele A Mitchell, Email: adele.mitchell@mssm.edu.
Laurie Ozelius, Email: laurie.ozelius@mssm.edu.
Monica Erazo, Email: monica.erazo@mssm.edu.
Jianzhong Hu, Email: Jianzhong.Hu@mssm.edu.
Dana Doheny, Email: dana.doheny@mssm.edu.
Maria T Abreu, Email: MAbreu1@med.miami.edu.
Daniel H Present, Email: DanJane1@aol.com.
Thomas Ullman, Email: thomas.ullman@mssm.edu.
Keith Benkov, Email: keith.benkov@mssm.edu.
Burton I Korelitz, Email: bkorelitz@LENOXHILL.NET.
Lloyd Mayer, Email: lloyd.mayer@mssm.edu.
Robert J Desnick, Email: robert.desnick@mssm.edu.
Acknowledgements and Funding
We would like to acknowledge the members of the New York Crohn's Disease Genetics Group: Drs. James Aisenberg, Joel Bauer, Steven Itzkowitz, Barry Jaffin, Arthur Kornbluth, Simon Lichtiger, Peter Rubin, David Sachar, and Anthony Weiss.
This work was funded by the New York Crohn's Disease Foundation to BIK, LM, RJD.
References
- Shanahan F. Crohn's disease. Lancet. 2002;359(9300):62–69. doi: 10.1016/S0140-6736(02)07284-7. [DOI] [PubMed] [Google Scholar]
- Scaldaferri F, Fiocchi C. Inflammatory bowel disease: progress and current concepts of etiopathogenesis. J Dig Dis. 2007;8(4):171–178. doi: 10.1111/j.1751-2980.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- Farmer RG, Michener WM, Mortimer EA. Studies of family history among patients with inflammatory bowel disease. Clin Gastroenterol. 1980;9(2):271–277. [PubMed] [Google Scholar]
- Singer HC, Anderson JG, Frischer H, Kirsner JB. Familial aspects of inflammatory bowel disease. Gastroenterology. 1971;61(4):423–430. [PubMed] [Google Scholar]
- Thompson NP, Driscoll R, Pounder RE, Wakefield AJ. Genetics versus environment in inflammatory bowel disease: results of a British twin study. BMJ. 1996;312(7023):95–96. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tysk C, Lindberg E, Jarnerot G, Floderus-Myrhed B. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut. 1988;29(7):990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata JH, Kantor-Fish S, Frankl H, Godby P, Vadheim CM. Crohn's disease among ethnic groups in a large health maintenance organization. Gastroenterology. 1992;102(6):1940–1948. doi: 10.1016/0016-5085(92)90317-r. [DOI] [PubMed] [Google Scholar]
- Rotter JI, Yang H, Shohat T. In: Genetic diversity among Jews: disease and markers at the DNA level. Bonne-Tamir B, Adam A, editor. Oxford: Oxford University Press; 1992. Genetic complexities of inflammatory bowel disease and its distribution among the Jewish people; pp. 395–411. [Google Scholar]
- Yang H, McElree C, Roth MP, Shanahan F, Targan SR, Rotter JI. Familial empirical risks for inflammatory bowel disease: differences between Jews and non-Jews. Gut. 1993;34(4):517–524. doi: 10.1136/gut.34.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361(21):2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411(6837):603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ. et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357(9272):1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JP. Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am J Gastroenterol. 2004;99(12):2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A. et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3(4):e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40(8):955–962. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D. et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39(7):830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, Cardon L, Takazoe M, Tanaka T, Ichimori T. et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn's disease. Hum Mol Genet. 2005;14(22):3499–3506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O'Malley JT, Kapur R, Levy DE, Kansas GS, Kaplan MH. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178(8):4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2–6. doi: 10.3109/00365528909091339. discussion 16-19. [DOI] [PubMed] [Google Scholar]
- Scott SA, Edelmann L, Liu L, Luo M, Desnick RJ, Kornreich R. Experience with carrier screening and prenatal diagnosis for 16 Ashkenazi Jewish genetic diseases. Hum Mutat. 2010;31(11):1240–1250. doi: 10.1002/humu.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- McGovern DP, Butler H, Ahmad T, Paolucci M, van Heel DA, Negoro K, Hysi P, Ragoussis J, Travis SP, Cardon LR. et al. TUCAN (CARD8) genetic variants and inflammatory bowel disease. Gastroenterology. 2006;131(4):1190–1196. doi: 10.1053/j.gastro.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Tukel T, Shalata A, Present D, Rachmilewitz D, Mayer L, Grant D, Risch N, Desnick RJ. Crohn disease: frequency and nature of CARD15 mutations in Ashkenazi and Sephardi/Oriental Jewish families. Am J Hum Genet. 2004;74(4):623–636. doi: 10.1086/382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, Taylor KD, Lin YC, Hang T, Wang D, Tang YM, Fischel-Ghodsian N, Targan SR, Rotter JI, Yang H. A novel NOD2/CARD15 haplotype conferring risk for Crohn disease in Ashkenazi Jews. Am J Hum Genet. 2003;72(3):509–518. doi: 10.1086/367848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Zouali H, Cezard JP, Colombel JF, Belaiche J, Almer S, Tysk C, O'Morain C, Gassull M, Binder V. et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70(4):845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugathasan S, Baldassano RN, Bradfield JP, Sleiman PM, Imielinski M, Guthery SL, Cucchiara S, Kim CE, Frackelton EC, Annaiah K. et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40(10):1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W. et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41(2):216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JR, Ahmad T, Geremia A, Beckly J, Cooney R, Hancock L, Pathan S, Guo C, Cardon LR, Jewell DP. Contribution of the novel inflammatory bowel disease gene IL23R to disease susceptibility and phenotype. Inflamm Bowel Dis. 2007;13(9):1063–1068. doi: 10.1002/ibd.20180. [DOI] [PubMed] [Google Scholar]
- Newman WG, Zhang Q, Liu X, Amos CI, Siminovitch KA. Genetic variants in IL-23R and ATG16L1 independently predispose to increased susceptibility to Crohn's disease in a Canadian population. J Clin Gastroenterol. 2009;43(5):444–447. doi: 10.1097/MCG.0b013e318168bdf0. [DOI] [PubMed] [Google Scholar]
- Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A. et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33(2):279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomino-Morales RJ, Oliver J, Gomez-Garcia M, Lopez-Nevot MA, Rodrigo L, Nieto A, Alizadeh BZ, Martin J. Association of ATG16L1 and IRGM genes polymorphisms with inflammatory bowel disease: a meta-analysis approach. Genes Immun. 2009;10(4):356–364. doi: 10.1038/gene.2009.25. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH. et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40(9):1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Hampe J, Rosenstiel P, Becker C, Wagner F, Hasler R, Little RD, Huse K, Ruether A, Balschun T. et al. Systematic association mapping identifies NELL1 as a novel IBD disease gene. PLoS ONE. 2007;2(1):e691. doi: 10.1371/journal.pone.0000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HF, Qiu LX, Chen Y, Zhu WL, Mao C, Zhu LG, Zheng MH, Wang Y, Lei L, Shi J. ATG16L1 T300A polymorphism and Crohn's disease susceptibility: evidence from 13,022 cases and 17,532 controls. Hum Genet. 2009;125(5-6):627–631. doi: 10.1007/s00439-009-0660-7. [DOI] [PubMed] [Google Scholar]
- Marquez A, Nunez C, Martinez A, Mendoza JL, Taxonera C, Fernandez-Arquero M, Diaz-Rubio M, de la Concha EG, Urcelay E. Role of ATG16L1 Thr300Ala polymorphism in inflammatory bowel disease: A Study in the Spanish population and a meta-analysis. Inflamm Bowel Dis. 2009;15(11):1697–1704. doi: 10.1002/ibd.21001. [DOI] [PubMed] [Google Scholar]
- Weedon MN, McCarthy MI, Hitman G, Walker M, Groves CJ, Zeggini E, Rayner NW, Shields B, Owen KR, Hattersley AT. et al. Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. PLoS Med. 2006;3(10):e374. doi: 10.1371/journal.pmed.0030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AF, Jablonski KA, McAteer JB, Saxena R, Pollin TI, Franks PW, Hanson RL, Shuldiner AR, Knowler WC, Altshuler D. et al. Extension of type 2 diabetes genome-wide association scan results in the diabetes prevention program. Diabetes. 2008;57(9):2503–2510. doi: 10.2337/db08-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weersma RK, Stokkers PC, Cleynen I, Wolfkamp SC, Henckaerts L, Schreiber S, Dijkstra G, Franke A, Nolte IM, Rutgeerts P. et al. Confirmation of multiple Crohn's disease susceptibility loci in a large Dutch-Belgian cohort. Am J Gastroenterol. 2009;104(3):630–638. doi: 10.1038/ajg.2008.112. [DOI] [PubMed] [Google Scholar]
- Need AC, Kasperaviciute D, Cirulli ET, Goldstein DB. A genome-wide genetic signature of Jewish ancestry perfectly separates individuals with and without full Jewish ancestry in a large random sample of European Americans. Genome Biol. 2009;10(1):R7. doi: 10.1186/gb-2009-10-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell NP. Statistics for Epidemiology. Chapman & Hall/CRC; 2003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables.