Abstract
Genetically constructed microbial biosensors for measuring organic pollutants are mostly applied in aqueous samples. Unfortunately, the detection limit of most biosensors is insufficient to detect pollutants at low but environmentally relevant concentrations. However, organic pollutants with low levels of water solubility often have significant gas-water partitioning coefficients, which in principle makes it possible to measure such compounds in the gas rather than the aqueous phase. Here we describe the first use of a microbial biosensor for measuring organic pollutants directly in the gas phase. For this purpose, we reconstructed a bioluminescent Pseudomonas putida naphthalene biosensor strain to carry the NAH7 plasmid and a chromosomally inserted gene fusion between the sal promoter and the luxAB genes. Specific calibration studies were performed with suspended and filter-immobilized biosensor cells, in aqueous solution and in the gas phase. Gas phase measurements with filter-immobilized biosensor cells in closed flasks, with a naphthalene-contaminated aqueous phase, showed that the biosensor cells can measure naphthalene effectively. The biosensor cells on the filter responded with increasing light output proportional to the naphthalene concentration added to the water phase, even though only a small proportion of the naphthalene was present in the gas phase. In fact, the biosensor cells could concentrate a larger proportion of naphthalene through the gas phase than in the aqueous suspension, probably due to faster transport of naphthalene to the cells in the gas phase. This led to a 10-fold lower detectable aqueous naphthalene concentration (50 nM instead of 0.5 μM). Thus, the use of bacterial biosensors for measuring organic pollutants in the gas phase is a valid method for increasing the sensitivity of these valuable biological devices.
Increasingly, genetically modified bacteria are being developed and tested for use as measuring devices for pollutants in the environment (reviewed recently in references 5, 8, and 19). Such bacterial biosensors either constitutively express a signal, such as bioluminescence, with any signal decrease related to exposure of the cells to pollutants (28), or induce expression of an analytically useful signal specifically and only in the presence of one or a limited set of structurally related compounds (1, 14, 20, 26, 32). For a number of reasons, it is useful to have microorganisms themselves monitor the immediate environment rather than using analytical chemistry. First, bacterial biosensors can be used for quick screening purposes, either for detecting the presence of toxic compounds in aqueous samples or for detecting a predefined set of target compounds for which the biosensors had been developed. Second, having microorganisms themselves “measure” the concentrations of pollutants is thought to be more representative for estimation of the bioavailable fraction and could solve some of the difficulties of extrapolating bioavailable concentrations from differential chemical extraction techniques. Hence, microorganisms which are able to sense and “report” pollutant concentrations (or fluxes) can also be used to study the factors governing pollutant bioavailability to microorganisms in the environment.
Biosensor bacteria (or whole-cell living bioreporters, as they are sometimes called) (2) are usually constructed by means of genetic techniques by coupling the DNA for a promoterless reporter gene which codes for an easily measurable protein to the operator or promoter DNA from a regulatory circuit. In addition, the sensor cell should contain the cognate sensor component (e.g., transcriptional activator or repressor) capable of mediating expression from the same regulatory circuit. In this way, the sensor component will regulate transcription of the reporter gene from the target promoter whenever the target chemical effector molecule is present inside the cell. The level of reporter gene expression is reflected in the activity of the reporter protein, which can be measured continuously or after a previously determined calibration period. Target compound concentrations or fluxes in unknown samples can then be determined from the activity of the reporter protein and interpolated on the calibration curve (32). Since other chemicals present in the sample may inhibit the physiological response of the bioreporter organisms (12), it is often useful to assess any inhibition in unknown samples by spiking them with known concentrations of chemicals (32) or by using a second, constitutively expressed reporter protein (34). Because bioluminescence is a rare trait, it is often applied as a reporter signal in bacterial biosensors. Bioluminescence is mediated by luciferase enzymes, which can have a prokaryotic or eukaryotic origin (reviewed in reference 33). Prokaryotic luciferases, such as those encoded by the luxAB genes, require reduced flavin mononucleotide, oxygen, and a substrate aldehyde for the bioluminescence reaction. Through regeneration of the aldehyde and of the flavin mononucleotide, the light reaction is coupled to general energy metabolic reactions in the cell (12, 33). Firefly luciferase, on the other hand, directly requires ATP for the light-generating reaction (33). Bioluminescence can be quantified very sensitively by using photographic films, luminometers, or liquid scintillation counters (2). Light can be measured noninvasively and sensitively without disrupting the bacterial cell and therefore enables in situ and on-line monitoring (4). The sensitivity even allows the detection of single bioluminescent bacterial cells by using a charge-coupled device or photon-counting (PC) video camera connected to a microscope (29, 31). For remote sampling of light, bioluminescent biosensors have also been immobilized on the tips of fiber-optic cables connected to a photomultiplier (13, 17). Bioluminescent biosensors have even been coupled to a chip that was designed to detect, process, and report low levels of bioluminescence (30). Unfortunately, luciferase activity is energetically costly for the cell, and physiological differences or compounds inhibitory to cells' energy metabolism can reduce light output (7, 12). Compounds interfering with fatty acid metabolism or cell membrane synthesis may even increase nonspecific light output from some biosensors, probably by providing a larger pool of substrates for the light reaction (12).
In most applications, bacterial biosensor cells have been used in aqueous solutions or slurries of soil-water (reviewed in reference 18). This has some obvious advantages, such as ample supply of nutrients to the cells and ease or tradition of sampling. However, it is usually not considered that the target analyte has to be transported from the bulk solute to the cell and that the process of transport in aqueous solutions is not necessarily fast. The consequence of this is that bacterial biosensors for organic compounds usually do not deliver an analytically useful signal at target compound concentrations below 0.5 to 1 μM (18). Here we describe additional possibilities for measuring with biosensor bacteria, using the gas phase rather than the aqueous phase. As a matter of fact, most organic compounds targeted by bacterial biosensors have significant gas phase partitioning (e.g., BTEX, alkanes, naphthalene, trichloroethylene, and chlorophenol) (1, 11, 27, 32). For our model system, we chose a bacterial biosensor for the detection of naphthalene, which is relatively poorly water soluble and hence a good model for the behavior of a large class of hydrophobic compounds with moderate gas phase partition constants (Henry constant, 0.02) (25). Bacterial biosensors for naphthalene have been described and used before (3, 7, 13, 14, 20), but they have not been systematically tested for accurate measurement of naphthalene through the gas phase, although the principle has been applied previously (23). Here we show that bacterial biosensors can be optimally maintained in controlled chemostat cultivation, react very reliably to naphthalene exposure with a proportional bioluminescence signal, and can be applied in gas phase measurements to increase the bioavailable amount of naphthalene to the cells, thereby effectively lowering the detectable naphthalene concentration range.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli DH5α was used as a general host for plasmid cloning. E. coli CC118λpir was used for the propagation of plasmids with an R6K origin of replication, and E. coli HB101(pRK2013) was used as a helper strain, providing transfer functions during conjugation. Pseudomonas putida pPG7 (NAH) was the source for the nahR and nahG genes and is capable of using naphthalene as a sole source of carbon and energy. P. putida pPG7-JAMA21 is a derivative of strain pPG7, containing the nahR-nahG′::luxAB mini-Tn5 construct, and was used for naphthalene biosensor assays (Fig. 1). This strain still contains the NAH plasmid and is therefore capable of using naphthalene as a sole carbon and energy source.
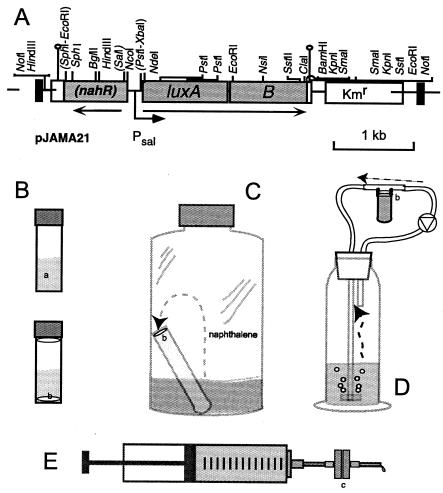
FIG. 1.
Genetic construct for naphthalene detection in P. putida (A) and schemes for the different assay types (B to E). (A) Relevant restriction sites are shown on the genetic map; those removed by cloning are placed within brackets. The black vertical bars indicate the I and O ends of the mini-Tn5 system. Note that the reading frame of nahR was interrupted by removal of the internal SalI site (within brackets). The direction of transcription and the sal promoter are indicated by arrows. (B) Simplest biosensor assay form in a 2-ml glass vial with Teflon-lined cap. Cells were applied either as a suspension in the aqueous phase (a) or immobilized on a filter (b). Filters were placed in solution or in the inside of the cap. (C) Static gas phase detection. Naphthalene evaporates from the aqueous sample and is transported through the gas phase to the filter-immobilized cells (b), which are placed on top of an agar layer in a glass vial. (D) Continuous gas loop device, in which air is recirculated through the system and bubbled through the aqueous sample and along the filter-immobilized cells (b). (E) Syringe advective flow system. Cells are immobilized on a filter which is contained in a metal filter holder (c). The holder is attached to the syringe and a syringe pump slowly pushes the aqueous sample through the filter.
The plasmids used during this study included pUC19 (35), for general subclonings; pGEM-T-Easy (Promega Corp.), for cloning of DNA fragments generated in PCRs; pUC18Not (16), for providing NotI symmetrical restriction sites; and PCK218 (21), serving as the mini-Tn5 vehicle to deliver the nahR-nahG′::luxAB construct to the chromosome of P. putida pPG7.
Construction of the naphthalene-responsive luciferase expression cassette.
A 1.1-kb fragment containing the nahR gene and the sal promoter was recovered from plasmid pCNB4-lacZ (6) by digestion with PstI and treatment with T4 DNA polymerase and, subsequently, digestion with EcoRI and treatment with Klenow DNA polymerase. This fragment was ligated with the linearized luxAB-containing vector, pJAMA8 (32), which was prepared by digestion with SphI and treatment with T4 DNA polymerase and, subsequently, digestion with XbaI and treatment with Klenow DNA polymerase. Transformants in E. coli were checked for their plasmid content, and plasmids with the proper orientation of the sal promoter with respect to the luxAB genes were designated pJAMA18. The reading frame of the nahR gene in pJAMA18 was interrupted by digestion of the unique SalI site, filling of both ends with Klenow polymerase, and religation (yielding pJAMA19). The nahR-Psal-luxAB fragments of both pJAMA18 and pJAMA19 were recovered as 3.5-kb NotI fragments and ligated with the vector part of PCK218 (yielding pJAMA20 and pJAMA21, respectively) (Fig. 1). By triparental mating and transposon insertion, the fragments were inserted in a single copy into the chromosome of P. putida pPG7 (NAH), as previously described (16). Four independent transconjugants of each mating (i.e., with E. coli containing pJAMA20 or pJAMA21) were purified and checked for the intactness of the nahR-Psal-luxAB fragments by PCR and Southern blotting. All transconjugants were subsequently tested for the induction of the luciferase by incubation with naphthalene and were all more or less similar in the capacity to be induced. One of them, the strain P. putida pPG7::pJAMA21, which displayed the highest inducible luciferase expression, was used for the experiments in this work.
Media and growth conditions.
All E. coli strains were cultivated in Luria-Bertani medium (24) (both liquid and solid), supplemented with ampicillin or kanamycin (both at 50 μg/ml) wherever necessary, at 37°C. P. putida strains were grown at 30°C in Luria-Bertani broth, nutrient broth (NB) (Biolife, Milan, Italy), or type 21C mineral medium (MM) (9) plus 2% (vol/vol) Hutner's trace element solution, 0.2% (vol/vol) vitamin solution (9), with succinate or glucose (both at 10 mM) as carbon sources. P. putida was grown on naphthalene on agar plates which were incubated in a gas-tight jar with an open petri dish containing naphthalene crystals. Media for P. putida pPG7-JAMA21 included 50 μg of kanamycin per ml.
Chemostat cultivation.
P. putida pPG7-JAMA21 was continuously cultivated in chemostats for most naphthalene biosensor assays. Chemostats consisted of 500-ml Schott flasks, with special metal lids, which were filled with 100 ml of medium and were operated at a dilution rate of 0.05 h−1 at 30°C, with constant stirring at 200 rpm. Pressurized air was used to aerate the vessel medium constantly. The medium consisted of MM as the base, with 10 mM succinate as a carbon source. The pH of the medium was buffered at 6.8, as no pH control was used in the chemostat flask itself. The strain was inoculated from a −80°C glycerol stock for each new chemostat experiment in NB with 50 μg of kanamycin per ml. During continuous cultivation, no kanamycin was added to the medium. A small sampling port was installed in the metal lid, through which 2-ml portions of the bacterial suspension in the chemostat could be retrieved by suction with a syringe. P. putida cells could generally be kept for approximately 2 weeks under continuous cultivation. The culture was sampled every day to determine its optical density at 600 nm and to check its purity on NB plates without kanamycin. Steady-state values of the optical density at 600 nm fluctuated slightly around 0.9, with an average number of viable cells of 1.3 × 109 per ml. The culture was kept for seven volume changes before naphthalene biosensor assays were performed.
Naphthalene biosensor assays.
Generally, naphthalene biosensor solution assays were performed in 2-ml vials with Teflon-lined screw caps (type 27134; Supelco, Bellafonte, Pa.). For calibration series, the vials were filled with 0.97 ml of MM, to which 17 μl of chemostat cell suspension and 10 μl of a naphthalene stock solution (prepared in dimethyl sulfoxide) were added. Final naphthalene concentrations in the aqueous phase (without taking phase equilibria into account) ranged from 0.01 to 10 μM. The vials were incubated at 30°C during the assay, with shaking at 200 rpm. After the assay incubation period (between 30 min and 6 h), samples of 0.2 ml were withdrawn in triplicate from each vial and transferred to a 96-well plate (Microlite TM1; Dynatech Industries, Inc. McLean, Va.). Twenty-five microliters of n-decanal solution was added (to a final concentration of 2 mM) and mixed manually with the cell suspension, and bioluminescence was measured in a Microlumat LB960 luminometer (Berthold AG, Regensdorf, Switzerland) by integrating for 10 s after a 3-min preincubation, as described previously (32).
The effects of glucose or succinate present in the biosensor assay on the performance of P. putida pPG7-JAMA21 for detection of naphthalene were determined by including them at final concentrations of between 10 μM and 10 mM. The following compounds were tested for cross-reactivity with the nahR-nahG′::luxAB sensor: toluene (at 1 and 5 μM), 1- and 2-methylnaphthalene, 1,3-, 1,4-, 1,5-, 2,3-, and 2,6-dimethylnaphthalene (all at a final concentration of 1 μM), and octane (at 0.1, 0.2, and 0.5 μM). All of those compounds were added separately or in combination with 5 μM naphthalene.
To compare naphthalene biosensor assays in solution and in air, we slightly modified the procedure outlined above. A sample of 85 μl of chemostat cell suspension was retrieved, mixed with 5 ml of MM, and immediately filtered through a 25-mm-diameter, 0.45-μm-pore-size nylon membrane disk (Protan BA85; Schleicher and Schuell) by suction to allow for collection of the cells on the filter. Filters were kept moist by placing them with bacteria facing up on an MM agar plate. Three 6-mm-diameter disks were cut out from the main filter with a puncheon. For bioassays in air (Fig. 1C), one disk was either placed on top of a small glass tube with the same diameter which was filled completely with MM agar (cells facing up, to prevent drying of the cells on the filter disk) or directly placed in the lid of the glass tube (cells facing down) (Fig. 1B). For comparison of the biosensor response between the gas and aqueous phases, disks were also placed in aqueous solution. After the required incubation time, the disks were retrieved and placed in a well of a microtiter plate to which 0.2 ml of MM was added, and bioluminescence was measured as described above.
Various settings with biosensor cells were developed in order to test whether the cells could concentrate more naphthalene from solution through advective flow. For a gas advective flow assay, cells were filtered as described above, after which the 6-mm-diameter filter disks were installed in a closed gas loop device. This device consisted of a gas wash flask (ca. 350 ml, with a 100-ml sample volume) through which air was bubbled through a glass frit by a tube pump (type MCP; Ismatec, Glattbrugg, Switzerland) and Tygon R3607 tubing (Ismatec), with small glass filter holders to expose the cells to the passing air (Fig. 1D). After a 2-h exposition time of the cells to the passing air, the filters were recovered and bioluminescence was measured as before. The total gas volume of the device was approximated 250 ml. For a solution advective flow assay, a 50-ml glass syringe was filled with 30 ml of aqueous sample and connected to a stainless steel metal filter holder (25-mm diameter) in which the filter (cellulose acetate; 0.4-μm pore size) (Sartorius) with the cells was placed (Fig. 1E). The sample was then pushed through the filter by means of a syringe pump operating at a speed of 25 or 50 ml/h. After the exposition time, the filter was taken out of the filter holder and placed on the surface of minimal medium agar for the remaining induction period (3 h). Three 6-mm-diameter disks were then punched from the filter, and bioluminescence was measured as described above.
Chemicals.
Naphthalene, purissima, was obtained from Fluka. Substituted naphthalenes were a gift from Matthias Schluepp, BMG Engineering, Schlieren, Switzerland, and were of the highest commercially available grade.
Modeling.
Air-water-biota equilibrium partitioning coefficients were calculated based on a simple model describing the fractions of naphthalene in each of these compartments, using Henry's law to calculate the naphthalene concentration in the gas phase (with the dimensionless Henry constant, H, for naphthalene being 0.02) (25) and the octanol water partitioning coefficient Kow (2,290 for naphthalene) (25) to calculate the naphthalene concentration in the lipid phase of the cells.
RESULTS
General calibration and effect of incubation time.
We reconstructed a bioluminescent naphthalene biosensor of P. putida pPG7 by fusing a DNA fragment containing a dysfunctional nahR gene and the Psal promoter from the NAH7 plasmid to a set of promoterless luxAB genes (Fig. 1) to avoid problems associated with intracellular substrate generation for the luciferase encountered with previously constructed luxCDABE fusions (12). This gene fusion was placed at random in the P. putida pPG7 genome by mini-Tn5 delivery. Several different transconjugants of P. putida were tested for luciferase expression and were all more or less similar in the capacity to be induced by naphthalene (not shown). In these transconjugants, a functional nahR gene was provided in trans from the NAH7 plasmid. Attempts to obtain a functioning naphthalene biosensor by fusing only the Psal promoter to the luxAB genes without the nahR gene in cis were not successful. Although initially a protocol was developed for a naphthalene biosensor assay with the P. putida pPG7-JAMA21 cells grown in batch cultures, similar to a procedure described in the literature (15), we soon began growing the biosensor cells in continuous culture for better reproducibility, ease of measurement, and a faster response time. In its simplest form, the assay consisted of biosensor cells taken from the continuous culture, suspended in 1 ml of sample solution with different naphthalene concentrations, and incubated in closed 2-ml vials.
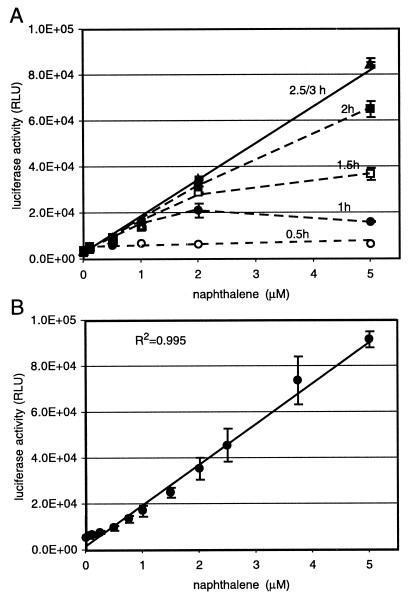
With cells taken directly from the chemostat, the increase in luciferase activity as a function of incubation time and naphthalene concentration showed an interesting pattern (Fig. 2A). Luciferase activity measured after 30 min of incubation time reached a plateau at any naphthalene concentration higher than 0.5 μM. This plateau gradually shifted to higher concentrations at longer incubation times until, at incubation times between 2.5 and 3 h, a nearly complete linear response curve was obtained (Fig. 2B). The probable explanation for this time-dependent behavior lies in the nature of the induction from the Psal promoter. The output from the Psal promoter is dependent on the formation of the inducer salicylate, an intermediate in the metabolism of naphthalene, but not on naphthalene itself. Therefore, the observed plateau may reflect the time needed for the cells to metabolize naphthalene to salicylate. We conclude from this that cells produced the same quantity of salicylate after 30 min of incubation irrespective of the starting concentration, resulting in approximately the same amounts of luciferase enzyme and thus luciferase activity. With longer incubation times, the output from the Psal promoter seemed to continue at higher naphthalene concentrations and luciferase accumulated. It might well be that the output from the Psal promoter stopped at the lower concentrations due to depletion of naphthalene, but because of the stability of the luciferase, the same activity was still measurable after 3 h. In fact, luciferase measurements often showed a slight decrease in light emission between 30 min and 3 h for the concentration range of 0.5 to 1.0 μM (not shown). The lowest naphthalene concentration that was significantly different (P < 0.005; two-tailed paired Student's t test) from the background signal in this type of assay was 0.5 μM (64 μg/liter). This was slightly higher than the 0.35 μM concentration described by Heitzer et al. (15).
FIG. 2.
Calibration of light emission from 2-ml closed vials with biosensor cells suspended in 1 ml of aqueous phase at different (nominal) naphthalene concentrations. The nominal naphthalene concentration indicates the amount added per volume of aqueous phase, without taking partitioning into account. (A) Effect of incubation time on light emission. (B) Calibration curve after 3 h of incubation at 30°C. Bars indicate the average standard deviations calculated for triplicate samples. Light emission is presented as relative light units (RLU), a value produced by the instrument.
Effects of other carbon sources and specificity range of NahR.
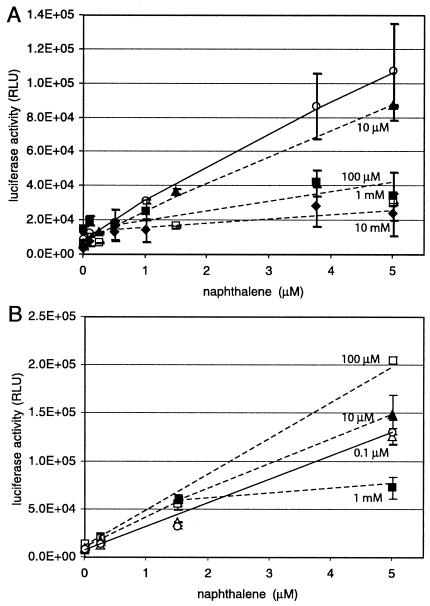
The advantage of using chemostat-grown cells for the biosensor assay is clearly a very fast metabolic response time, since at any moment the carbon concentration in the culture vessel is very low. Therefore, the performance of the biosensor cells is not expected to be inhibited by catabolite-repressive effects exerted by traces of leftover growth substrates. To demonstrate the strong effect of catabolite repression on the biosensor response, we included either succinate or glucose in the assay at different concentrations. Succinate had a very strong effect on the response of the naphthalene biosensor (Fig. 3A). At 10 μM succinate, a 20% inhibition of the signal already occurred, irrespective of the naphthalene concentration, whereas at higher succinate concentrations (100 μM, 1 mM, 10 mM), almost no naphthalene-specific luciferase induction occurred. Only at 1 μM succinate was no inhibition detected. With glucose in the assay, a somewhat opposite effect took place. Glucose at 100 μM stimulated luciferase activity almost twofold, and at 10 μM a 20% increase was still detectable with 5 μM naphthalene. Only with 1 mM glucose in the assay medium was the luciferase activity diminished (Fig. 3B). The stimulating effect of glucose might be due to its providing extra energy for the light-generating reaction, whereas succinate often acts as a catabolite repressor in pseudomonads (22, 36).
FIG. 3.
Effects of different succinate (A) or glucose (B) concentrations on naphthalene-dependent light emission. The system used was suspended cells in a 2-ml vial with 1 ml of aqueous phase. Open circles, no addition; open triangles, 1 μM naphthalene; closed triangles, 10 μM naphthalene; open squares, 100 μM naphthalene; closed squares, 1 mM naphthalene; closed diamonds, 10 mM naphthalene. Solid lines connect the data points for the case without succinate or glucose addition. Dotted lines follow inhibited or stimulated light responses (not drawn for every concentration). Nominal naphthalene concentrations and relative light emission are as described for Fig. 2.
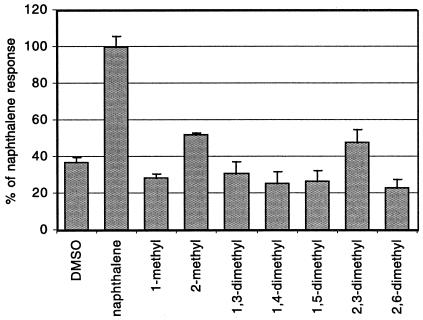
The specificity of the naphthalene sensor was actually quite narrow (Fig. 4). Of various methylated naphthalenes tested, only 2-methyl- and 2,3-dimethylnaphthalene caused an induction of luciferase activity (at 40% of the induction obtained with naphthalene at the same concentration). Toluene (0.1 to 5 μM), octane (0.1 to 1 μM), and phenanthrene (0.5 to 10 μM) did not induce luciferase expression. For salicylate, the light emission was proportional to the salicylate concentration between 1 and 50 μM, although the cells reacted with fivefold lower light emission than naphthalene to the same concentration of salicylate. Nitrate (up to 0.5 mM) and sulfate (up to 0.5 mM) had no effect on naphthalene-inducible luciferase expression.
FIG. 4.
Relative response of different methylated naphthalenes on luciferase expression from P. putida pPG7-JAMA21 in a 2-ml closed vial assay with 1 ml of aqueous phase. All compounds were added in 10 μl of dimethyl sulfoxide (DMSO) to a final nominal concentration of 1 μM in the aqueous phase. The induction time was 2.5 h. Light emission with naphthalene was set at 100%. Error bars indicate average standard deviations from the average light emission for triplicate incubations.
Measuring in the gas versus aqueous phase.
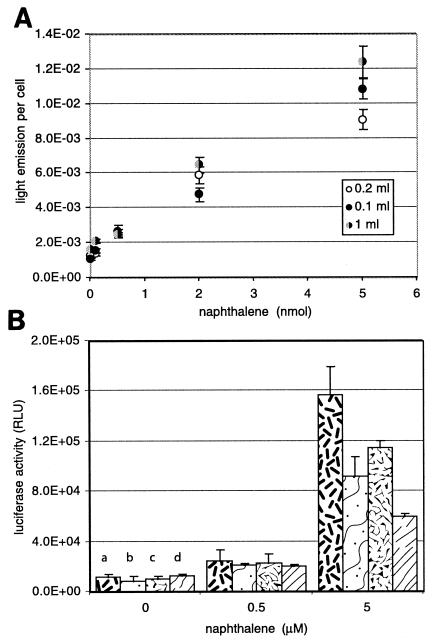
We next considered two other questions that are interesting for biosensor applications: (i) would the biosensor be capable of measuring naphthalene through the gas phase and (ii) would it be possible to use the cells to concentrate naphthalene from a larger sample volume in order to lower the detection limit? To test these questions, the experimental setup for exposing the cells to naphthalene was slightly changed. When the volume of the aqueous phase was reduced from 1 ml to 0.2 or 0.1 ml but the added amount of biosensor cells was kept the same, essentially the same light emission per cell was measured with any naphthalene amount added to the assay (Fig. 5A). As discussed below, this suggests that all naphthalene was depleted from the assay and that no increase of light emission could be expected from changing the aqueous volume/cell number ratio. Next, instead of keeping the biosensor cells in suspension, we filtered them onto a membrane filter which was then incubated with either naphthalene in solution or in the gas phase (Fig. 1B). Three replica filters were cut from each membrane in order to ensure good reproducibility. Nevertheless, measurement fluctuations were higher for biosensor cells on membranes than in suspension. When cells were incubated in 2-ml vials with 1 ml of aqueous solution and different naphthalene concentrations, either on a membrane placed in solution or on a membrane attached to the lid of the vial (gas phase measurement), it was found that membranes exposed to the gas phase exhibited higher luciferase activity than those exposed in solution (Fig. 5B), although the induction ratio (i.e., the ratio between the response with naphthalene and that without it) was significantly higher only at 5 μM naphthalene, not at 0.5 μM. When vials were incubated with cells on a membrane both in solution and in the gas phase, the response was slightly lower than with one membrane only, but still cells exposed in the gas phase displayed higher luciferase activity (and a higher induction ratio with 5 μM naphthalene [11.3, compared to 4.6 in solution]).
FIG. 5.
Biosensor assays in aqueous solution and in air. (A) Effects of different relative volumes of aqueous phase (1.0, 0.2, and 0.1 ml) on light emission per cell. The total amount of cells used for the assay was 2.2 × 107. Light emission per cell was calculated as the luciferase activity, measured in relative light units (RLU), divided by the number of cells used for the luciferase measurement: for 1.0 ml of aqueous phase, 4.4 × 106 cells; for 0.2 and 0.1 ml of aqueous phase, 2.2 × 107 cells. The amount of naphthalene added to the aqueous phase is shown in nanomoles. (B) Effect of measuring with filter-immobilized cells in 2-ml vials with 1 ml of aqueous phase, either in gas phase alone (a), in aqueous phase alone (b), or with two filters simultaneously in gas phase (c) and aqueous phase (d). Error bars represent the average standard deviations for the averages for triplicate filters.
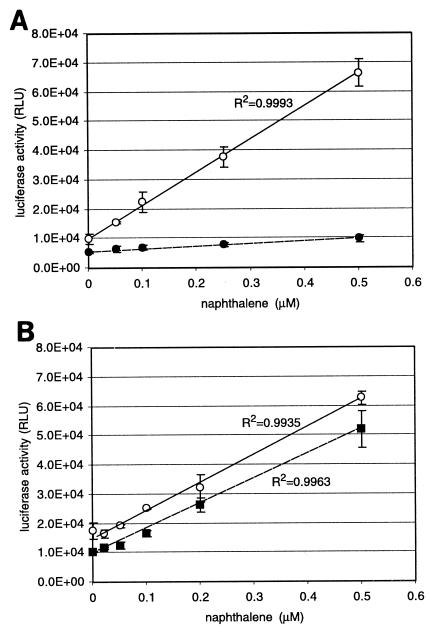
To investigate whether the biosensor cells on filters could also concentrate a larger absolute quantity of naphthalene from a larger sample volume (thereby effectively detecting lower naphthalene concentrations), assays were set up with filter-immobilized biosensor cells exposed to the gas phase of a larger sample volume or attached to a 50-ml syringe, whereby the solution was passed over the filter. Compared to a twofold higher amount of cells incubated in a 1-ml suspension in a 2-ml vial with the same aqueous naphthalene concentrations, the biosensor cells on the membrane in a closed 100-ml flask with 10 ml of aqueous solution outperformed those in solution completely (Fig. 6). In fact, a very nice linear calibration curve was obtained in the range of 0.05 to 0.5 μM naphthalene, with induction factors of 1.5 to 6.5, compared to twofold higher induction at 0.5 μM in the 2-ml vial. Assays in the same flask volume but with twice as much sample volume (20 ml) or assays in a 10-fold larger flask with twice the sample size (20 ml) did not bring any further improvements to either the absolute signal or the induction ratio (Fig. 6B). This showed that cells exposed to the gas phase can take up naphthalene from the gas phase and profit from the larger absolute amount of naphthalene in a larger sample volume (10 ml compared to 1 ml).
FIG. 6.
Effects of measuring with filter-immobilized biosensor cells in the gas phase. (A) Assays in 100-ml closed flasks with 10 ml of aqueous sample and filter-immobilized cells exposed to the gas phase (open circles). For comparison, light emission with suspended cells in a 2-ml closed vial with 1 ml of aqueous phase at the same nominal naphthalene concentration is shown (closed circles). The induction time was 2.5 h. (B) Assays with filter-immobilized biosensor cells exposed to the gas phase in 100-ml flasks with 20 ml of aqueous phase (open circles) or in 1,000-ml flasks with 20 ml of aqueous phase (closed squares). Naphthalene concentration is the nominal concentration added to the aqueous phase. RLU, relative light units.
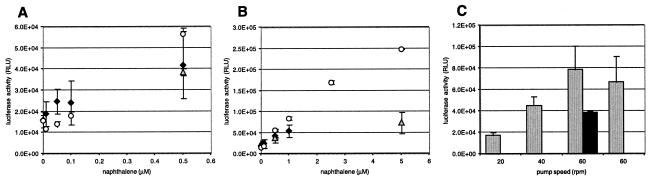
We tried to improve the amount of naphthalene transported to the filter-immobilized biosensor cells by constructing an air-loop device through which air was purged through a sample containing naphthalene and recirculated along the filter, much like a chemical “purge-and-trap” system. Although it could be seen that biosensor cells reacted to naphthalene purged from solution along the filter, the response was far lower than expected for the amount of naphthalene added (Fig. 7B). The response of the cells could be improved by circulating the air at a faster rate (Fig. 7C). Also, when a syringe with 50 ml of sample solution was connected to a metal filter holder with the filter-immobilized sensor bacteria, no improvement of the detection limit was seen compared to that in a regular 2-ml assay with suspended sensor cells (Fig. 7A). Only a slight improvement was visible in the lower concentration range, but there was a clearly higher standard error for replicated measurements (Fig. 7A and B). This indicated that the sensor cells could not concentrate naphthalene from a solution in advective flow.
FIG. 7.
(A) Comparative light emission between a 2-ml closed-vial assay with 1 ml of aqueous phase and suspended biosensor cells (circles), filter-immobilized cells attached to a syringe with 30 ml of aqueous phase flowing through (diamonds), or filter-immobilized cells exposed to the gas phase in the gas-loop device (triangles). See Fig. 1D and E for the schematic setup. (B) Expansion of the same data set for higher naphthalene concentrations. All naphthalene concentrations are given as nominal concentrations in the aqueous phase. (C) Effect of air circulation rate on the biosensor response in the gas-loop device. Light gray, 5 μM nominal naphthalene concentration; black, 0.5 μM nominal naphthalene concentration. Error bars represent the average standard deviations for the average light emission of triplicate filters or vials. RLU, relative light units.
DISCUSSION
For the experimental conditions of 2.2 × 107 cells in a 2-ml assay with 1 ml of aqueous phase, the equilibrium mass fractions between the gas, water, and lipid phases (calculated according to the partitioning model) will equal 0.0195, 0.976, and 0.0049, respectively (Table 1). According to equilibrium partitioning, therefore, only a small fraction of naphthalene (i.e., that present in the lipid phase of the cell membrane) would actually be available for the cells. When changing the partitioning equilibrium by reducing the volume of the aqueous phase, one would expect a stronger reaction of the cells, since with 0.2 and 0.1 ml of aqueous phase, the fractions in the lipid phase would increase to 0.021 and 0.035, respectively. However, when calculating the light output per cell under the conditions of 1, 0.2, and 0.1 ml of aqueous phase, we did not detect any increase (Fig. 5A). We therefore have to assume that equilibrium is not reached in the biosensor assays. The most likely reason for this is that the biosensor bacteria act as a sink for naphthalene, since they are capable of metabolizing it. Furthermore, since cells with the same amount of naphthalene in a larger volume (1 ml compared to 0.2 or 0.1 ml) reacted with the same light emission per cell in the same time frame, we conclude that all of the cells must have detected the same amount of naphthalene, despite the higher dilution and larger diffusion distances in the 1-ml aqueous phase. It seems reasonable to assume that all naphthalene was depleted under those conditions.
TABLE 1.
Phase partitioning at different gas phase volumes
| Vg (ml)a | fgb | flipb | fwb |
|---|---|---|---|
| 1.98 | 0.613 | 0.077 | 0.31 |
| 1.9 | 0.266 | 0.035 | 0.699 |
| 1.8 | 0.149 | 0.021 | 0.83 |
| 1.6 | 0.074 | 0.011 | 0.915 |
| 1.4 | 0.044 | 0.008 | 0.948 |
| 1.2 | 0.029 | 0.006 | 0.965 |
| 1.0 | 0.019 | 0.005 | 0.976 |
Total volume, 2 ml; calculated lipid volume, 2.18 × 10−6 ml.
fg, flip, and fw, fraction in the gas, lipid, and water phases, respectively.
According to the same equilibrium partitioning calculations, only 2% of the naphthalene would be present in the gas phase at a 1:1 volumetric ratio of gas to aqueous phase (Table 1). From the relatively small tendency of naphthalene to partition into the gas phase, one would thus tend to conclude that measurements of naphthalene in the gas phase would be less effective than those in the aqueous phase (which contained 98% of the total amount of naphthalene). Interestingly, however, the cells reacted very efficiently when exposed to naphthalene in the gas phase, which suggests again that no static partitioning equilibrium is formed but that the flux of naphthalene is directed towards the cells. Two effects must play a role here. First, as argued above, since the cells will metabolize naphthalene, they must act as a sink, in which case no partitioning equilibrium exists between naphthalene dissolved in aqueous solution and that in the gas phase (as the lipid phase is constantly depleted). Secondly, since the diffusive transport rate of naphthalene in the gas phase is approximately a factor of 1,000 higher in the gas than in the aqueous phase, naphthalene depleted by the cells will be replenished relatively fast from the gas phase. The two combined effects will result in a net flux of naphthalene from the aqueous phase to the filter-attached cells. The consequence of all these nonequilibrium phenomena is that a larger flux of naphthalene can be maintained toward the cells in the gas phase. Hence, an apparent concentration effect can take place with a larger sample volume and a lower naphthalene concentration, still leading to significant light emission.
Surprisingly, when the efficiency of light production per nanomole of naphthalene and per cell was calculated for the different assays, the suspended cells in the 2-ml assay still reacted with two- to eightfold higher light emission per cell than those immobilized on filters (Table 2). This calculation was performed under the assumption that the total amount of naphthalene added to the system had been transported to the cells. One reason for this apparent contradiction (filter-immobilized cells react more poorly on a per cell basis but can concentrate naphthalene from the gas phase, thereby allowing measurements of lower naphthalene concentrations than in the aqueous phase) might be a geometric effect. Cells immobilized on the surface can only be supplied with naphthalene through one-half of the cell surface. Another reason might have been a technical drawback of exposing cells to the gas phase, unavoidably leading to drying out of the cells, despite our attempts to keep them moist on the surface of an agar layer. If all or a part of the cells were to dry out, this would certainly lead to a physiological loss of cell energy or of activation potential.
TABLE 2.
Calculated efficiencies for light production of the different biosensor settings
| System or assay | Concn of Nahb (μM) | Amt of Nah (nmol) | Amt of Nah/cell | Amt of light produced/cell | Amt of light/nmol of Nah/cell |
|---|---|---|---|---|---|
| 2-ml assaya | 0.05 | 0.05 | 2.2 × 10−9 | 1.5 × 10−3 | 9.7 × 104 |
| 5 | 5 | 2.2 × 10−7 | 2.1 × 10−2 | 8.6 × 104 | |
| Cells on filter in 100-ml flask | 0.05 | 0.5 | 5 × 10−8 | 1.6 × 10−3 | 1.1 × 104 |
| 0.5 | 5 | 5 × 10−7 | 6.7 × 10−3 | 1.1 × 104 | |
| 2-ml assaya | 0.1 | 0.1 | 4.6 × 10−9 | 1.4 × 10−3 | 6.8 × 104 |
| 5 | 5 | 2.2 × 10−7 | 1.2 × 10−2 | 4.6 × 104 | |
| Filter in 2-ml flask and air | 0.5 | 0.5 | 5 × 10−8 | 2.5 × 10−3 | 2.5 × 104 |
| 5 | 5 | 5 × 10−7 | 1.6 × 10−3 | 2.9 × 104 | |
| Syringes, 30 ml | 0.01 | 0.3 | 2.7 × 10−9 | 1.9 × 10−3 | 1 × 105 |
| 0.1 | 3 | 2.7 × 10−8 | 2.4 × 10−3 | 2.9 × 104 | |
| 2-ml assaya | 0.5 | 0.5 | 2.3 × 10−8 | 1.3 × 10−2 | 4.1 × 105 |
| 2.5 | 2.5 | 1.1 × 10−7 | 3.9 × 10−2 | 3.1 × 105 | |
| Gas loop | 0.5 | 25 | 2.3 × 10−7 | 3.8 × 10−3 | 1.7 × 104 |
| 5 | 250 | 2.3 × 10−6 | 7.3 × 10−3 | 3.2 × 103 |
Corresponding assay with suspended cells carried out with the same chemostat.
Nah, naphthalene.
Contrary to the filter-immobilized cells exposed to the gas phase, filtered cells attached to a syringe did not accumulate naphthalene from solution. Apparently, flowing a solution with naphthalene through a layer of cells under the conditions of this sensor assay (25 ml/h) does not increase the effective uptake of naphthalene. The reason for this might be that the final step in naphthalene transport to the cell occurrs via diffusion through a stagnant water layer. Actively forcing the flow to the surface by advection would in this case lead to naphthalene passing by the cells.
The use of genetically modified bacteria to detect and measure organic pollutants in the environment was proven as a concept more than 10 years ago and has been followed up by the construction of quite a number of different biosensor organisms with different specificities (reviewed in references 5, 8, and 19). Unfortunately, most biosensor applications have remained in research laboratories for several different reasons, some of which relate to a lack of standardization, to difficulties in maintaining living microorganisms, and also to poor analytical quality of the assays, poor specificities of detection, and unrealistically high detection limits. In many cases, pollutants will be present in concentrations in the micromolar range or below and cannot be reliably detected with biosensors. Here we have described at least one possible solution for lowering biosensor detection limits, making use of the principle that the cells do not measure compound concentrations per se, but rather compound fluxes. In fact, target compound detection and signal production by biosensor cells are the result of both the specificities of the biological detection system and physicochemical transport phenomena (10). Although in most instances biosensor responses will be calibrated with concentrations of the target compound, it is actually the compound's flux multiplied by the total incubation time period, and therefore the total amount of target compound per cell, which governs the final response. Strategies which can increase the total amount of target compound per cell can thus lead to a lower detection limit. Hence, when viewed from another perspective, the biosensor response not only gives an analytically useful signal for the contamination level in a particular environment, but it also reports the pollutant bioavailability, since those bacteria carrying out in situ biodegradation reactions face the same physicochemical transport phenomena (10). One could therefore envision that the effectiveness of bioremediation strategies to increase pollutant fluxes to the cells can be monitored directly by the sensor bacteria.
We attempted to increase the total amount of naphthalene by transporting it from a larger sample volume to the same number of cells. As it turned out, exposing filtered biosensor cells to the gas phase in a closed system consisting of an aqueous sample and a gas phase led to the required concentrating effect. This partial success in gas phase measurements tempted us to develop a prototype bacterial nose (Fig. 1D). The idea behind this was that in many cases it might actually be interesting to sample the gas phase of a contaminated site, which is usually more accessible than the aqueous or solid phase. For instance, a groundwater contamination with a volatile organic solvent may still be detectable in the gas phase in the unsaturated zone of the soil above (23). Gasoline spills near gasoline stations may easily be detectable in the gas phase. If whole-cell living biosensors are to become a viable future strategy for quick and easy measurements, we should not focus just on aqueous phase detection, but should involve other creative assay types.
Acknowledgments
We thank Stefan Grimberg and Beate Escher for their helpful comments and critical reading of the manuscript.
This work was supported by Swiss National Science Foundation grant SPP-5001-058629.
REFERENCES
- 1.Applegate, B. M., S. R. Kehrmeyer, and G. S. Sayler. 1998. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethylbenzene, and xylene (BTEX) sensing. Appl. Environ. Microbiol. 64:2730-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burlage, R. S. 1997. Emerging technologies: bioreporters, biosensors, and microprobes, p. 115-123. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 3.Burlage, R. S., A. V. Palumbo, A. Heitzer, and G. Sayler. 1994. Bioluminescent reporter bacteria detect contaminants in soil samples. Appl. Biochem. Biotechnol. 46:731-740. [Google Scholar]
- 4.Carmi, O. A., G. S. A. B. Stewart, S. Ulitzur, and J. Kuhn. 1987. Use of bacterial luciferase to establish a promoter probe vehicle capable of nondestructive real-time analysis of gene expression in Bacillus spp. J. Bacteriol. 169:2165-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daunert, S., G. Barrett, J. S. Feliciano, R. S. Shetty, S. Shrestha, and W. Smith-Spencer. 2000. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem. Rev. 100:2705-2738. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo, V., S. Fernandez, M. Herrero, U. Jakubzik, and K. N. Timmis. 1993. Engineering of alkyl-responsive and haloaromatic-responsive gene expression with mini-transposons containing regulated promoters of biodegradative pathways of Pseudomonas. Gene 130:41-46. [DOI] [PubMed] [Google Scholar]
- 7.Dorn, J. G., R. J. Frye, and R. M. Maier. 2003. Effect of temperature, pH, and initial cell number on luxCDABE and nah gene expression during naphthalene and salicylate catabolism in the bioreporter organism Pseudomonas putida RB1353. Appl. Environ. Microbiol. 69:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Souza, S. F. 2001. Microbial biosensors. Biosens. Bioelectr. 16:337-353. [DOI] [PubMed] [Google Scholar]
- 9.Gerhardt, P., R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.). 1981. Manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 10.Harms, H., and T. N. P. Bosma. 1997. Mass transfer limitation of microbial growth and pollutant degradation. J. Ind. Microbiol. 18:97-105. [Google Scholar]
- 11.Hay, A. G., J. F. Rice, B. M. Applegate, N. G. Bright, and G. S. Sayler. 2000. A bioluminescent whole-cell reporter for detection of 2,4-dichlorophenoxyacetic acid and 2,4-dichlorophenol in soil. Appl. Environ. Microbiol. 66:4589-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heitzer, A., B. Applegate, S. Kehrmeyer, H. Pinkart, O. F. Webb, T. J. Phelps, D. C. White, and G. S. Sayler. 1998. Physiological considerations of environmental applications of lux reporter fusions. J. Microbiol. Methods 33:45-57. [Google Scholar]
- 13.Heitzer, A., K. Malachowsky, J. E. Thonnard, P. R. Bienkowski, D. C. White, and G. S. Sayler. 1994. Optical biosensor for environmental on-line monitoring of naphthalene and salicylate bioavailability with an immobilized bioluminescent catabolic reporter bacterium. Appl. Environ. Microbiol. 60:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heitzer, A., O. F. Webb, P. M. DiGrazia, and G. S. Sayler. 1995. A versatile bioluminescent reporter system for organic pollutant bioavailability and biodegradation, p. 191-208. In R. A. Minear, A. M. Ford, L. L. Needham, and N. J. Karch (ed.), Applications of molecular biology in environmental chemistry. Lewis Publishers, New York, N.Y.
- 15.Heitzer, A., O. F. Webb, J. E. Thonnard, and G. S. Sayler. 1992. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl. Environ. Microbiol. 58:1839-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikariyama, Y., S. Nishiguchi, T. Koyama, E. Kobatake, M. Aizawa, M. Tsuda, and T. Nakazawa. 1997. Fiber-optic-based biomonitoring of benzene derivatives by recombinant E. coli bearing luciferase gene-fused TOL-plasmid immobilized on the fiber-optic end. Anal. Chem. 69:2600-2605. [DOI] [PubMed] [Google Scholar]
- 18.Jaspers, M. C. M. 2002. PhD. thesis. Swiss Federal Institute of Technology, Zürich, Switzerland.
- 19.Keane, A., P. Phoenix, S. Ghoshal, and P. C. Lau. 2002. Exposing culprit organic pollutants: a review. J. Microbiol. Methods 49:103-119. [DOI] [PubMed] [Google Scholar]
- 20.King, J. M. H., P. M. DiGrazia, B. Applegate, R. Burlage, J. Sanseverino, P. Dunbar, F. Larimer, and G. S. Sayler. 1990. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science 249:778-781. [DOI] [PubMed] [Google Scholar]
- 21.Kristensen, C. S., L. Eberl, J. M. Sanchez-Romero, M. Givskov, S. Molin, and V. de Lorenzo. 1995. Site-specific deletions of chromosomally located DNA segments with the multimer resolution system of broad-host-range plasmid RP4. J. Bacteriol. 177:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller, C., L. Petruschka, H. Cuypers, G. Burchhardt, and H. Herrmann. 1996. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J. Bacteriol. 178:2030-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ripp, S., D. E. Nivens, Y. Ahn, C. Werner, J. Jarrell IV, J. P. Easter, C. D. Cox, R. S. Burlage, and G. S. Sayler. 2000. Controlled field release of a bioluminescent genetically engineered microorganism for bioremediation process monitoring and control. Environ. Sci. Technol. 34:846-853. [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Schwarzenbach, R. P., P. M. Gschwend, and D. M. Imboden. 1993. Environmental organic chemistry. John Wiley & Sons, Inc., New York, N.Y.
- 26.Selifonova, O., R. Burlage, and T. Barkay. 1993. Bioluminescent sensors for detection of bioavailable Hg(II) in the environment. Appl. Environ. Microbiol. 59:3083-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selifonova, O. V., and R. W. Eaton. 1996. Use of an ipb-lux fusion to study regulation of the isopropylbenzene catabolism operon of Pseudomonas putida RE204 and to detect hydrophobic pollutants in the environment. Appl. Environ. Microbiol. 62:778-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw, L. J., Y. Beaton, L. A. Glover, K. Killham, and A. A. Meharg. 1999. Development and characterization of a lux-modified 2,4-dichlorophenol-degrading Burkholderia sp. RASC. Environ. Microbiol. 1:393-399. [DOI] [PubMed] [Google Scholar]
- 29.Silcock, D. J., R. N. Waterhouse, L. A. Glover, J. I. Prosser, and K. Killham. 1992. Detection of a single genetically modified bacterial cell in soil by using charge-coupled device-enhanced microscopy. Appl. Environ. Microbiol. 58:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson, M. L., G. S. Sayler, B. M. Applegate, S. Ripp, D. E. Nivens, M. J. Paulus, and G. E. Jellison, Jr. 1998. Bioluminescent-bioreporter integrated circuits form novel whole-cell biosensors. Trends Biotechnol. 16:332-338. [Google Scholar]
- 31.Sternberg, C., L. Eberl, L. Kongsbak Poulsen, and S. Molin. 1997. Detection of bioluminescence from individual bacterial cells: a comparison of two different low-light imaging systems. J. Biolumin. Chemilumin. 12:7-13. [DOI] [PubMed] [Google Scholar]
- 32.Sticher, P., M. C. M. Jaspers, K. Stemmler, H. Harms, A. J. B. Zehnder, and J. R. van der Meer. 1997. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl. Environ. Microbiol. 63:4053-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, T., and J. W. Hastings. 1998. Bioluminescence. Annu. Rev. Cell Dev. Biol. 14:197-230. [DOI] [PubMed] [Google Scholar]
- 34.Wood, K. V., and M. G. Gruber. 1996. Transduction in microbial biosensors using multiplexed bioluminescence. Biosens. Bioelectron. 11:207-214. [DOI] [PubMed] [Google Scholar]
- 35.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 36.Yuste, L., I. Canosa, and F. Rojo. 1998. Carbon-source-dependent expression of the PalkB promoter from the Pseudomonas oleovorans alkane degradation pathway. J. Bacteriol. 180:5218-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]