Abstract
Current models suggest that (i) filamentous bacteria are protected against predation by nanoflagellates, (ii) prey size is positively correlated with prey-predator contact probability, and (iii) contact probability is mainly responsible for size-selective predation by interception-feeding flagellates. We used five strains of filamentous bacteria and one bacterivorous nanoflagellate, Ochromonas sp. strain DS, to test these assumptions. The five strains, including one spirochete and four Betaproteobacteria strains, were isolated by the filtration-acclimatization method. All five strains possess flexible cells, but they differ in average cell length, which ranged from 4.5 to 13.7 μm. High-resolution video microscopy was used to measure contact, capture, and ingestion rates, as well as selectivity of the flagellate feeding. Growth and feeding experiments with satiating and nonsatiating food conditions, as well as experiments including alternative well-edible prey, were performed. In contrast to predictions by current models, the flagellate successfully consumed all the tested filamentous strains. The ingestion rate was negatively correlated with bacterial length. On the other hand, the lengths of the filamentous bacteria were not positively correlated to the contact rate and capture rate but were negatively correlated to ingestion efficiency. In experiments including alternative nonfilamentous prey, the flagellates showed negative selection for filamentous bacteria, which was independent of food concentration and is interpreted as a passive selection. Our observations indicate that (i) size alone is not sufficient to define a refuge for filamentous bacteria from nanoflagellate predation and (ii) for the investigated filamentous bacteria, prey-predator contact probability could be more influenced by factors other than the prey size.
Bacteria and bacterivorous protozoans are the basic components of microbial food webs. Among the bacterivorous protists, the bacterivorous nanoflagellates are known as a major factor influencing both the bacterial community structure and the bacterial standing stock (20, 32, 37). The size of bacteria is an important trait that strongly influences the predation of bacteria by protists. Nanoflagellate grazing on bacteria has been shown to be in general size selective, with a preference for medium-sized bacteria (10, 13, 14, 35). Large-sized bacteria, such as filamentous bacteria (15, 16, 18) and microcolony forming bacteria (19), may exceed a species-specific upper ingestion limit of the flagellates, thus providing these bacteria with a refuge from grazing.
The bacterial size equivalent to the upper grazing limit is not well known, not even for otherwise well-investigated flagellate species. Matz et al. have observed an upper ingestion limit of 2.72 μm for Spumella sp. (26). Pernthaler et al. have suggested the division of the natural bacterial communities into several functional sizes that include small cells (<0.4 μm) and “grazing-vulnerable” (0.4 to 1.6 μm), “grazing-suppressed ” (1.6 to 2.4 μm), and “grazing-resistant” bacteria (>2.4 μm) (30). Several size limits for the separation of grazing-sensitive and protected bacteria were used in a variety of publications, and range from 2.4 μm to 10 μm (20). Despite this ambiguity in the upper grazing size limit, a consensus exists among microbial ecologists, who agree that filamentous bacteria are in general protected against predation by nanoflagellates (15, 16, 17, 18, 20, 23, 24, 36, 38, 39). This consensus is supported by a few direct microscopical observations of the interaction of nanoflagellates with filamentous bacteria (2).
Size-selective feeding of nanoflagellates is generally considered a result of predator-prey contact probabilities and uptake limitations of the predator. Theoretical models predict that the contact probabilities in general increase with prey size (11) and, therefore, that the clearance rates increase within a certain size range (10, 12, 13). Such contact probabilities have rarely been demonstrated, however, due to methodological limitations. Live observation techniques provide detailed information on every step (contact, capture, and ingestion) in the feeding process of flagellates (2, 3). So far, the relationship between prey size and contact probabilities has been investigated in only a few experiments with relatively narrow prey size ranges (i.e., 1.30 μm to 2.88 μm) (26).
In this study we tested whether filamentous bacteria are indeed well protected against predation by bacterivorous nanoflagellates. We isolated five strains of filamentous bacteria, which range in mean length from 4.5 to 13.7 μm. Predation by the direct-interception-feeding nanoflagellate Ochromonas sp. strain DS on these filamentous bacteria was investigated in the absence and presence of an alternative well-edible prey (Pseudomonas putida MM1). Video microscopy was used for detailed analysis of each single predation step (contact, capture, and ingestion). Because it was previously demonstrated that food selection by flagellates occurs especially at a high food concentration (5, 25), we performed experiments under both conditions, satiated and nonsatiated prey abundance.
MATERIALS AND METHODS
Microbial strains.
Five strains of filamentous bacteria were isolated by the filtration-acclimatization method (21) from the pelagic zones of three lakes and one pond located in Austria and the People's Republic of China (Table 1; for a brief characterization of the sampling habitats, see reference 21). The phylogenetic position of the isolates was determined by comparative 16S rRNA gene sequence analysis (M. W. Hahn et al., submitted for publication).
TABLE 1.
Characterization of the isolated filamentous bacterial strainsa
| Strain as identified by 16S rRNA gene sequencing | Origin | Length range (μm) | Cell vol range (μm3) | Accession no. of 16S rRNA gene sequence | Closest relative (accession no. of 16S rRNA gene sequence) | Sequence similarity between isolate and closest relative (%) |
|---|---|---|---|---|---|---|
| Hylemonella sp. strain WQH1 | Lake Hallstadt, Austria | 2.0-9.5 | 0.2-1.1 | AJ565430 | H. gracilisc (AF078753) | 96.5 |
| H. gracilis WQT2 | Lake Taihu, P.R. Chinab | 2.5-16.4 | 0.2-1.5 | AJ565424 | H. gracilisc (AF078753) | 99.7 |
| H. gracilis WQM2 | Lake Mondsee, Austria | 4.8-18.4 | 0.4-1.7 | AJ565425 | H. gracilisc (AF078753) | 99.7 |
| H. gracilis WQP1 | Pond 1, Austria | 5.2-21.7 | 0.5-2.1 | AJ565423 | H. gracilisc (AF078753) | 99.7 |
| S. aurantia WQM4 | Lake Mondsee | 4.9-28.3 | 0.4-2.2 | AJ565432 | S. aurantia (M57740) | 99.4 |
The isolates were preliminarily classified based on phylogenetic analysis of their 16S rRNA gene sequences, as well as morphological and other phenotypic traits. Cell length and volume parameters were recorded from early stationary phase in cultures that were used for experiments.
People's Republic of China.
Formerly Aquaspirillum gracile (40).
In our predation experiments the nonfilamentous bacterium P. putida strain MM1, which is a medium-sized prey of good food quality for bacterivorous nanoflagellates (9), was used as alternative prey. The axenically cultured bacterivorous nanoflagellate Ochromonas sp. strain DS was used as a model predator (17).
Long-term predation experiments. (i). Experimental design.
To test the grazing vulnerability of the filamentous bacteria, grazing experiments were conducted separately for each strain. An overview on the experimental design of all predation experiments is presented in Table 2. Five experiments were conducted. Each experiment consisted of triplicate batches containing Ochromonas sp. strain DS and one of the isolates, as well as one negative control for testing the growth of the isolates without flagellate predation. In addition, one positive control for testing the grazing activity of the flagellate on the well-edible prey P. putida MM1 was also set up.
TABLE 2.
Overview of the performed experimentsa
| Type of expt (no. of expts) | Organisms | Measured parameter(s) | Method |
|---|---|---|---|
| Long-term predation expt (5) | Hylemonella sp. strain WQH1 | Flagellate abundance, bacterial abundance, bacterial cell size | Epifluorescence microscopy |
| H. gracilis strain WQT2 | |||
| H. gracilis strain WQM2 | |||
| H. gracilis strain WQP1 | |||
| S. aurantia strain WQM4 | |||
| P. putida MM1b | |||
| Short-term predation exptc (5) | H. gracilis WQT2 + P. putida MM1 (lower concn) | Bacterial abundance | Epifluorescence microscopy |
| S. aurantia WQM4 + P. putida MM1 (lower concn) | |||
| H. gracilis WQT2 + P. putida MM1 (higher concn) | |||
| S. aurantia WQT2 (higher concn) | |||
| P. putida MM1 (higher concn) | |||
| Video microscopy exptc (5) | H. gracilis WQT2 + P. putida MM1 (lower concn) | Contact rate, capture rate, ingestion rate | Video microscopy |
| S. aurantia WQM4 + P. putida MM1 (lower concn) | |||
| H. gracilis WQT2 + P. putida MM1 (higher concn) | |||
| S. aurantia WQT2 (higher concn) | |||
| P. putida MM1 (higher concn) |
Ochromonas sp. DS was used as model predator for all the experiments. The short-term predation experiments and the video microscopy experiments were performed in parallel and used organisms from the same precultures.
Stationary stage, starved and heat killed. It was used as positive control.
P. putida MM1 was at stationary stage, not heat killed.
(ii) Precultures.
P. putida MM1 was cultured in 9-g liter−1 NSY medium, harvested by centrifugation, washed, resuspended in inorganic basal medium, killed with heat (70°C, 2 h), and stored at −20°C until it was utilized in the predation experiments as a reference strain (for the preparation of NSY organic and inorganic basal medium, see reference 21). The isolates Hylemonella sp. strain WQH1, Hylemonella gracilis WQT2, H. gracilis WQM2, and H. gracilis WQP1 were precultured in liquid 9-g liter−1 NSY medium, and Spirochaeta aurantia WQM4 was precultured in 3-g liter−1 NSY medium, until they reached early stationary phase. They were then diluted 10-fold in inorganic basal medium just before the grazing experiments were started. Ochromonas sp. strain DS was precultured axenically with heat-killed P. putida MM1.
(iii) Performance of experiments.
Erlenmeyer flasks containing 100 ml of inorganic basal medium were inoculated with high numbers of bacterial cells (≥13 × 106 cells ml−1) and low numbers of Ochromonas strain DS cells (≤0.3 × 104 cells ml−1). The control for testing the growth of the isolates received no flagellates. All the batches were incubated at 15°C with illumination but without continuous shaking. The bacterial and flagellate populations were monitored for periods of 5 to 15 days depending on the development of the bacterial population. Three-milliliter subsamples were taken at 24-hour intervals during the first 5 days and thereafter at 48-hour intervals, fixed with 2% formaldehyde (final concentration), and analyzed by epifluorescence microscopy within 2 days.
Short-term predation experiments and video microscopy observation. (i) Experimental design.
Five similar batch experiments were performed (Table 2). Each experiment consisted of triplicate batches containing the flagellate Ochromonas sp. strain DS at high abundance and a single strain of tested bacteria or mixtures of one single filamentous bacterium and P. putida MM1 (Table 2), as well as one negative control batch containing bacteria without flagellates.
(ii) Precultures.
H. gracilis WQT2 (intermediate length), S. aurantia WQM4 (long length), and P. putida MM1 (short length) were selected for “classical” grazing efficiency experiments and for video microscopic observations. Precultures of organisms except P. putida MM1 were prepared as described for the long-term predation experiments. P. putida MM1 was cultured in 9-g liter−1 NSY medium, harvested by centrifugation, washed, and resuspended in inorganic basal medium. In contrast to the long-term predation experiments, live P. putida MM1 cells were used in these experiments.
(iii) Performance of short-term experiments.
Erlenmeyer flasks containing 100 ml of inorganic basal medium were inoculated with different numbers of bacterial cells or mixed bacterial cells and high numbers of Ochromonas sp. strain DS (>4.5 × 104 cells ml−1). One control for testing the growth of the bacteria was free of flagellates. The batches were incubated at 15°C with illumination but without continuous shaking. Grazing experiments with a low concentration of bacteria were run for 5 h and sampled in 1-h intervals. Three-milliliter subsamples were fixed with 2% formaldehyde (final concentration) and analyzed by epifluorescence microscopy within 1 day. Experiments with high numbers of bacteria and flagellates were run for 6 h and monitored at 2-h intervals in the same way as mentioned above.
(iv) Performance of video microscopic observations.
Approximately 6 ml of cultures containing a single bacterial strain or mixtures of strains (Table 2) were transferred to an observation petri dish. The flagellates were allowed to attach to the bottom of the petri dish and to adapt to the experimental conditions for 30 min after transfer. The relatively large water volume allowed observation times of up to several hours without significant changes in the culture conditions (e.g., temperature). The feeding behavior was observed with a Zeiss Axiovert 200 microscope equipped with a Plan Neofluar ×100/1.3 oil objective. The microscope was connected with a zoom adapter to a C-2400-77B video camera (Hamamatsu) and an S-VHS recorder (AG 6730; Panasonic). Videotape analysis was carried out by using continuous and single-frame playback mode (2, 3).
In the experiment a single flagellate subpopulation was observed for a total of 25 min as follows: five individual cells were observed subsequently within one petri dish for 5 min each. After each observation period a new flagellate was selected for observation. For each investigated bacterial strain (or combination of bacterial strains), the experiment was repeated in five parallels.
Determination of microbial numbers, size, and cell volume.
Bacterial and flagellate abundance were determined as described previously (17, 31). For the measurement of bacterial cell length and width, images of at least 150 cells per sample were collected with a monochrome charge-coupled device camera (Hitachi Denshi), and sizes were determined with the LUCIA G image analysis systems (Lucia 4.51; Laboratory Imaging, Prague, Czech Republic). Bacterial cell volumes were calculated according to the formula of Andersson et al. (1).
Grazing rate, clearance rate, and ingestion rate. (i) Long-term predation experiments.
The grazing rates (G) (per hour) were estimated from the declining phase at satiating food conditions. At satiating food conditions the per-capita ingestion rates are assumed to be constant, and therefore, we used a linear model (34) that considers losses due to grazing as well as the net growth of bacteria without grazing. In our experiments, no marked growth of bacteria was observed in treatments without grazing due to a lack of substrate supply. The grazing rate was calculated by the equation G = (N0− Nt)/[(N0+ Nt)/2]/Δt, where N0 and Nt referred to the concentrations of bacteria at the beginning and the end of the time interval (Δt) of decline. The clearance rate (C) (nl flagellate−1 h−1) was calculated by the equation C = (G × 1,000,000)/Fm, where Fm (flagellate milliliter−1) was the mean concentration of the flagellate. Fm was calculated for the time of an exponential increase, according to the method of Heinbokel (22): Fm = (Ft− F0)/(ln Ft− ln F0), where F0 and Ft refer to the concentrations of the flagellate at the beginning and at the end of the time interval, respectively. Ingestion rate (I) (bacterial cells flagellate−1 hour−1) was estimated by the equation I = (N0− Nt)/Fm/Δt. Growth rate of Ochromonas sp. strain DS (μ) (hour−1) was calculated by the equation μ = (ln Ft− ln F0)/Δt, where F0 and Ft denote the concentrations of the flagellate at the beginning and at the end of the time interval (Δt) of exponential increase.
(ii) Short-term predation experiments.
For experiments with a low concentration of bacterial cells, exponential decays were found. The grazing rate was calculated according to the formula G = (ln N0 − ln Nt)/Δt while assuming there was no growth of bacteria in the short period. For experiments with a high concentration of bacterial cells, grazing rates, clearance rates, and ingestion rates were calculated in the same way as for the long-term predation experiments while assuming that the abundance of Ochromonas sp. strain DS was stable during the short time of the experiments. A selectivity index for predation of filamentous bacteria (Df) was calculated by the equation Df = Cf/(Cf+ Cm) (8), where Cf and Cm were clearance rates for tested filamentous bacteria and P. putida MM1. Df can range from 0 (uptake of only P. putida MM1) to 1 (uptake of only filamentous bacteria), with a value of 0.5 indicating nonselective feeding.
(iii) Video microscopy experiments.
The definitions of contact, capture, and ingestion were used according to the work of Matz et al. (26). Contact, capture, ingestion, and overall selectivity index were calculated by the formulas CTSI = (CTf/Af)/(CTf/Af + CTm/Am), CPSI = (CPf/CTf)/(CPf/CTf + CPm/CTm), ISI = (If/CPf)/(If/CPf + Im/CPm), and α = (If/Af)/(If/Af + Im/Am), respectively. CTf, CPf, and If were the observed contact, capture, and ingestion rates for filamentous bacteria, CTm, CPm, and Im were the observed contact, capture, and ingestion rates for P. putida MM1, and Af and Am referred to the initial bacterial abundance of filamentous bacteria and P. putida MM1 (8). The selectivity index can again range from 0 to 1.
Statistics.
One-way analysis of variance (ANOVA) was used to compare the growth rate, clearance rate, and ingestion rate for Ochromonas sp. strain DS in the long-term predation experiments. All pairwise multiple comparisons were, therefore, undertaken by Tukey's test. The t test was used to compare the clearance rates and ingestion rates of Ochromonas sp. strain DS on different bacterial strains in the short-term experiments and video microscopy experiments. The differences between selectivity coefficients and the unselectivity (Df and α = 0.5) in the short-term experiments and video microscopy experiments were tested by using a t test as well (33).
RESULTS
Morphological characteristics of the investigated organisms.
All five isolated bacterial strains (Table 1) are filamentous bacteria with a flexible cell shape. Their mean strain-specific lengths ranged from 4.5 to 13.7 μm, and their mean strain-specific cell volume ranged from 0.50 to 1.07 μm3 (Table 3). All five strains possess small cell diameters in a range from 0.31 to 0.38 μm. Large ranges of length and cell volume were observed for each population (Table 1). When growing on NSY agar plates, all five strains demonstrated the ability to penetrate and grow inside the agar. Based on phenotypic traits and results from comparative analysis of the 16S rRNA gene sequences, the isolates were preliminarily classified as Hylemonella spp. and S. aurantia (Table 1).
TABLE 3.
Long-term experiments for examination of predation and growth of the flagellate Ochromonas sp. DS with the filamentous bacteriaa
| Bacterial strain | Length (μm) | Cell vol (μm3) | Initial abundance of bacteria (106 cells ml−1) | Initial abundance of Ochromonas sp. DS (104 cells ml−1) | Final abundance of bacteria in control without grazing (106 cells ml−1) | Ingestion rate (bacteria flagel- late−1 h−1) | Clearance rate (nl flagellate−1 h−1) | Growth rate of Ochromonas sp. strain DS (h−1) |
|---|---|---|---|---|---|---|---|---|
| Hylemonella sp. strain WQH1 | 4.5 ± 1.3 | 0.50 ± 0.14 | 16.5 ± 0.5 | 0.12 ± 0.03 | 14.3b | 22.5 ± 4.6 | 1.85 ± 0.47 | 0.035 ± 0.006 |
| H. gracilis WQT2 | 6.0 ± 2.1 | 0.56 ± 0.19 | 19.0 ± 2.1 | 0.13 ± 0.04 | 18.4c | 16.9 ± 0.6 | 1.43 ± 0.09 | 0.035 ± 0.006 |
| H. gracilis WQM2 | 7.5 ± 2.0 | 0.68 ± 0.18 | 51.6 ± 4.8 | 0.09 ± 0.01 | 68.4d | 13.5 ± 2.5 | 0.69 ± 0.05 | 0.033 ± 0.001 |
| H. gracilis WQP1 | 9.0 ± 3.0 | 0.85 ± 0.27 | 16.2 ± 0.1 | 0.18 ± 0.03 | 15.6e | 13.5 ± 2.6 | 1.19 ± 0.23 | 0.025 ± 0.005 |
| S. aurantia WQM4 | 13.7 ± 3.7 | 1.07 ± 0.28 | 13.0 ± 0.8 | 0.30 ± 0.04 | 13.0f | 6.8 ± 1.6 | 0.69 ± 0.15 | 0.030 ± 0.004 |
| P. putida MM1g | 1.1 ± 0.3 | 0.48 ± 0.03 | 20.4 ± 1.2 | 0.12 ± 0.02 | 27.8 ± 7.7 | 1.88 ± 0.75 | 0.039 ± 0.005 |
Values are means ± standard deviations except where otherwise noted.
Data on day 15.
Data on day 15.
Data on day 12.
Data on day 13.
Data on day 5.
Two predation experiments for P. putida MM1.
In the performed experiments, the average cell diameter of the flagellate Ochromonas sp. strain DS depended on the food conditions and therefore ranged from 4 to 8 μm.
Long-term predation experiments.
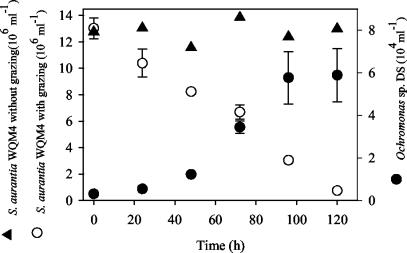
Ochromonas strain DS preyed successfully on all five strains of filamentous bacteria (Table 3; Fig. 1). With predation, the population of the filamentous bacteria decreased in the course of the experiments from initial numbers of more than 13 × 106 cells ml−1 to less than 1 × 106 cells ml−1. In contrast, in the control batches containing no flagellate, the bacteria did not change markedly in numbers (Table 3). In the case of H. gracilis WQM2, an increase of cell numbers was observed towards the end of the experiment. In all experiments an exponential increase of flagellate numbers was observed (Table 3; Fig. 1). The rates (Table 3) determined for growth with the five filamentous strains and P. putida MM1 as the sole sources of food were not significantly different (P = 0.054 [one-way ANOVA]).
FIG. 1.
Predation on S. aurantia WQM4 by Ochromonas strain DS in the long-term predation experiments (also see Table 2). Development of the flagellates and the S. aurantia WQM4 populations with and without predation are shown. Average cell numbers and standard deviations (error bars) for the three parallel preparations are shown except for the control experiment with S. aurantia WQM4 without predation.
Estimated ingestion rates and clearance rates ranged from 6.8 to 27.8 bacteria flagellate−1 h−1 and from 0.69 to 1.88 nl flagellate−1 h−1, respectively (Table 3). The ingestion rate for S. aurantia WQM4 was lower than those for Hylemonella sp. strain WQH1, H. gracilis WQT2, and P. putida MM1 (P = 0.002, P = 0.039, and P < 0.001, respectively [Tukey test after one-way ANOVA]). The clearance rates of the flagellate for H. gracilis WQM2 and S. aurantia WQM4 were lower than those for Hylemonella sp. strain WQH1 (P = 0.011 and P = 0.011 [Tukey test]) and P. putida MM1 (P = 0.02 and P = 0.02 [Tukey test]), while no significant differences were found for the other bacterial strains.
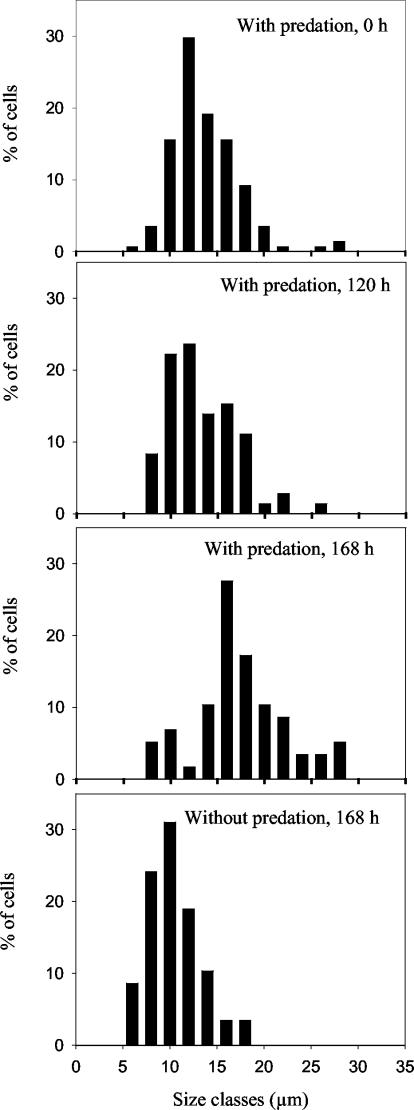
The size distribution of S. aurantia WQM4 (the largest investigated strain) was analyzed over the course of the experiment (Fig. 2). No size shift was observed from the starting point of the experiment to the 120th h, when the abundance of S. aurantia WQM4 was already decreased by the flagellate to 0.74 × 106 cells ml−1 (5.7% of the initial value). Later the size distribution shifted towards larger cells. In the control batch without predation, a shift of the size distribution towards smaller cells was observed.
FIG. 2.
Bacterial size distribution of S. aurantia WQM4 in the long-term predation experiment. The sizes were grouped into 2-μm classes. Cells longer than 27 μm were pooled in one size class (>27 μm).
Short-term predation experiments.
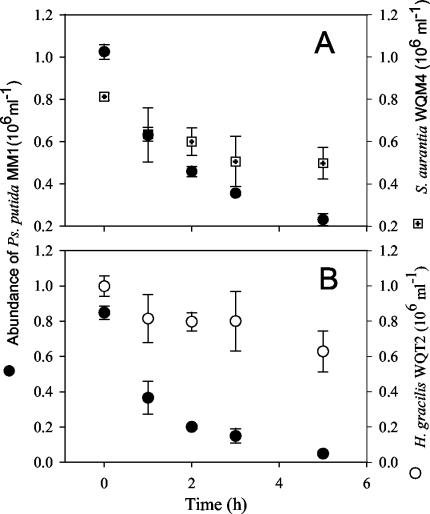
Ochromonas sp. strain DS that was fed with the mixture of P. putida MM1 and S. aurantia WQM4 at a low abundance (Fig. 3A) showed a higher clearance rate (P = 0.002 [t test]) and ingestion rate (P = 0.007 [t test]) for P. putida MM1 than for S. aurantia WQM4. The flagellate selected negatively for S. aurantia WQM4, with a selectivity index of 0.23 ± 0.06 (P < 0.01 [t test]). When the flagellate was fed with a mixture of P. putida MM1 and H. gracilis WQT2 at a low food concentration (Fig. 3B), it also showed a higher clearance rate (P = 0.001 [t test]) and ingestion rate (P = 0.017 [t test]) for P. putida MM1 than for H. gracilis WQT2. The flagellate selected negatively for the filamentous H. gracilis WQT2 as well (Df = 0.11 ± 0.04; P < 0.01 [t test]).
FIG. 3.
Predation on a mixture of P. putida MM1 and S. aurantia WQM4 (A) or of P. putida MM1 and H. gracilis WQT2 (B) by Ochromonas sp. DS at lower food concentrations. Average cell numbers and standard deviations (error bars) for the three parallel preparations are shown.
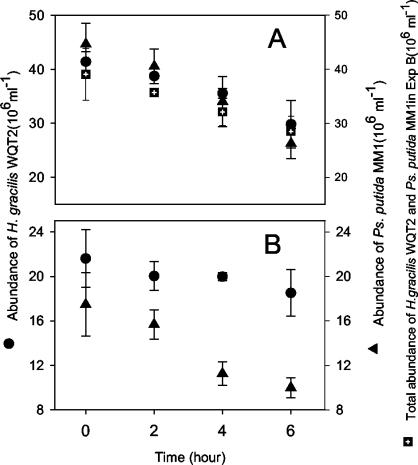
In the experiment with high concentrations of the mixture of P. putida MM1 plus H. gracilis WQT2 (Fig. 4B), the flagellate showed a slightly higher clearance rate and ingestion rate (P = 0.059 [t test]) for P. putida than for H. gracilis WQT2. The selectivity index for H. gracilis WQT2 was 0.25 ± 0.14 (P < 0.1 [t test]). However, no clearance rates and ingestion rates (P = 0.188 [t test]) were found to be largely different when the flagellate was fed with P. putida MM1 and H. gracilis WQT2 separately at high food concentrations (Fig. 4A). Clearance and ingestion rates for predation of a single strain by the flagellate did not differ from total clearance and ingestion rate of feeding by the flagellate on the mixture of P. putida MM1 and H. gracilis WQT2 (Fig. 4A; P = 0.372 [one-way ANOVA]). Selectivity for H. gracilis WQT2 by the flagellate at a low food concentration did not differ significantly (P = 0.171 [t test]) from that observed at satiating food conditions.
FIG. 4.
Predation on either a single population of P. putida MM1 and H. gracilis WQT2 (A) or the mixture of both strains (B) by Ochromonas sp. DS at high food concentrations. For comparison, the total bacterial abundance in the experiment with the mixed bacterial population (B) is also shown in graph A. Average cell numbers and standard deviations (error bars) for the three parallel preparations are shown.
Video microscopic observations.
The direct observation of predation by Ochromonas sp. strain DS showed that the flagellate was able to ingest all of the five strains of filamentous bacteria. We observed that the flagellate did not respond to each contact with the filamentous prey. When filamentous bacteria hit the flagellate cell at low speed, the flagellate frequently failed to react to the contact. Contacts by filamentous bacteria drifting at higher speed were very frequently responded to by the flagellate. The contact and capture rates for the flagellate with P. putida MM1 and the filamentous bacteria were not significantly different in all experiments (Table 4). No positive contact and capture selectivity were observed (Table 4). When the flagellate responded to contacts with filamentous bacteria, it often did not complete the ingestion process successfully. The flagellate built pseudopodia-like structures around the bacterium but could not always manage to enclose the bacterium completely. In these cases, usually an empty vacuole (containing water only) was formed, and the bacterium drifted away or stayed attached to the cell body of the flagellate for a while. A second attempt to ingest such an attached bacterium could be observed up to several minutes after the original contact. Failure to ingest a filamentous bacterium increased with increasing bacterial cell length, but even the longest bacterial filaments were observed being ingested occasionally. Lack of response to contact and failure in ingestion attempts were not frequently observed for the prey P. putida MM1. In all experiments using mixed bacteria as food, the flagellate showed a higher ingestion rate with P. putida MM1 than with filamentous bacteria (Table 4). Furthermore, negative selection for H. gracilis WQT2 and S. aurantia WQM4 were observed. The food concentration had no obvious effect on selectivity for H. gracilis WQT2 (Table 4) (P = 0.958 [t test]). No strong difference in ingestion rates for P. putida and H. gracilis WQT2 were found when the flagellate was fed separately with the two strains at a high food concentration (Table 4).
TABLE 4.
Video microscopic determination of contact rate, capture rate, and ingestion rate of Ochromonas sp. DS with bacterial strainsa
| Parameter | Value for exptl condition(s)b
|
||||
|---|---|---|---|---|---|
| Low abundance of P. pu. MM1 + S. au. WQM4 | Low abundance of P. pu. MM1 + H. gr. WQT2 | High abundance of P. pu. MM1 + H. gr. WQT2 | P. pu. MM1 only | H. gr. WQT2 only | |
| No. of bacteria (106 ml−1) | |||||
| MM1 | 1.03 ± 0.04 | 0.85 ± 0.04 | 17.48 ± 2.85 | 44.66 ± 3.82 | |
| WQM4 | 0.81 ± 0.02 | ||||
| WQT2 | 1.00 ± 0.06 | 21.61 ± 2.56 | 41.38 ± 1.84 | ||
| No. of flagellates (104 ml−1) | 4.59 | 5.52 | 8.97 | 8.97 | 8.97 |
| Contact rate (no. of bacteria flagellate−1 h−1) | |||||
| MM1 | 49.9 ± 7.3 | 69.6 ± 14.3 | 199.2 ± 98.6 | 408.0 ± 175.9 | |
| WQM4 | 50.9 ± 10.4 | ||||
| WQT2 | 57.1 ± 6.9 | 187.2 ± 31.6 | 582.0 ± 47.4 | ||
| t testc | NS | NS | NS | ||
| Capture rate (no. of bacteria flagellate−1 h−1) | |||||
| MM1 | 22.1 ± 5.5 | 28.3 ± 8.0 | 117.6 ± 64.6 | 236.4 ± 81.5 | |
| WQM4 | 14.9 ± 9.5 | ||||
| WQT2 | 24.5 ± 6.2 | 75.6 ± 30.2 | 229.2 ± 51.1 | ||
| t testc | NS | NS | NS | ||
| Ingestion rate (no. of bacteria flagellate−1 h−1) | |||||
| MM1 | 18.7 ± 5.2 | 26.4 ± 8.1 | 40.8 ± 20.1 | 67.2 ± 23.8 | |
| WQM4 | 5.3 ± 2.0 | ||||
| WQT2 | 5.8 ± 3.6 | 7.2 ± 6.6 | 45.6 ± 23.1 | ||
| t testc | * | * | * | ||
| Contact SId | 0.56 ± 0.07 (NS) | 0.42 ± 0.06 (*) | 0.41 ± 0.07 (*) | ||
| Capture SId | 0.37 ± 0.16 (NS) | 0.51 ± 0.08 (NS) | 0.40 ± 0.07 (*) | ||
| Ingestion SId | 0.34 ± 0.12 (*) | 0.19 ± 0.07 (*) | 0.21 ± 0.09 (*) | ||
| Overall SId | 0.27 ± 0.11 (*) | 0.17 ± 0.12 (*) | 0.13 ± 0.13 (*) | ||
The contact rate includes the capture rate and the ingestion rate. The capture rate includes the ingestion rate. Mean lengths of S. aurantia WQM4 and H. gracilis WQT2 were 15.4 and 7.2 μm, with a range from 6.0 to 25.8 μm and 3.0 to 23 μm. P. pu, P. putida; S. au., S. aurantia; H. gr., H. gracilis.
Values are given ± standard deviations.
There was no significant difference between the value for P. putida MM1 only and that for H. gracilis WQT2 only.
SI, selectivity index: Significance is given in parentheses: single asterisk, significant (P < 0.05); NS, not significant.
DISCUSSION
Although many laboratory and field studies have suggested that filamentous bacteria may be resistant to predation by bacterivorous flagellates (15, 16, 17, 18, 23, 24, 36, 38, 39), to date only a few investigations including direct observation of the predation of filamentous bacteria have been performed (2). We isolated several filamentous bacterium strains with flexible cell shapes and investigated the predation of these filamentous bacteria by the nanoflagellate Ochromonas sp. strain DS. Relatives of these bacterial strains have been found in aquatic habitats (6, 7), but not much is known about their ecology.
Are filamentous bacteria generally protected against predation by interception-feeding flagellates?
The results of our experiments challenge the current assumption that filamentous bacteria are protected from grazing by bacterivorous nanoflagellate. Our experiments clearly showed that Ochromonas sp. strain DS is able to prey on all five filamentous strains. Furthermore, the flagellate exhibited exponential increases in cell numbers in all of the experiments (Table 3; Fig. 1).
Previous studies with other filamentous bacteria suggested that these strains formed populations consisting of two differently sized subpopulations that were sensitive and resistant to flagellate predation (17, 18). In contrast, the constancy of the size distribution of S. aurantia WQM4 under intensive predation (Fig. 2), as well as the direct observation of many predation events by video microscopy, indicated that the flagellate is not just preying on the smaller-sized subpopulations of the filamentous strains. The observed shift of the length distribution of S. aurantia WQM4 incubated in the absence of the flagellate toward smaller sizes at the 168th h of grazing could be caused by starvation, while the shift toward a larger cell size under grazing could be a result of enhanced bacterial growth due to the release of organic matter resulting from the grazing of the flagellate. Furthermore, even the smallest cells of some of the investigated strains showed cell lengths above most of the suggested size limits for separation of predation-sensitive and protected bacterial cells (20, 30).
The investigated filamentous bacteria differ from other filamentous bacteria in the flexibility of their cells. This flexibility may have influenced the vulnerability of the investigated bacterial strains. Obviously, further investigations concerning the influence of this trait on the edibility of filamentous bacteria for nanoflagellates need to be conducted.
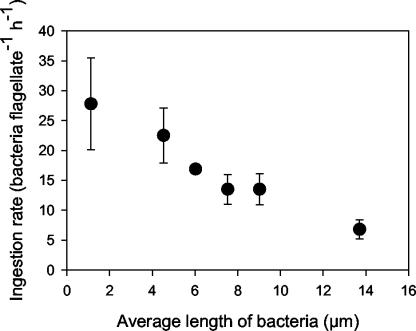
In the long-term grazing experiments, different ingestion rates were found among different bacterial strains (Table 3). The ingestion rates were in general negatively correlated with the average length of the bacteria, including the medium-sized P. putida MM1 (Fig. 5). Despite significant differences in ingestion rates for the different filamentous strains, ingested bacterial cell volume and the growth rate of Ochromonas sp. strain DS were similar for all bacterial strains (Table 3). We assume that this could be a result of satiated food conditions.
FIG. 5.
Flagellate ingestion rates observed in the long-term experiments plotted against the average length of the bacterial strains. The error bars show standard deviations for the three parallel preparations.
We clearly demonstrate that filamentous bacteria are not generally protected against predation by nanoflagellates. To reveal whether this holds true for the majority of filamentous bacteria, more investigations are needed.
Is prey size positively correlated with prey-predator contact probability?
Size-selective feeding of interception-feeding nanoflagellates, such as Ochromonas sp. strain DS, has been explained to be a passive mechanism in which prey size determines the contact rate (11) and subsequently the ingestion rate (12, 13). The geometric model of prey capture by flagellates (11) was modified by Monger and Landry (27) to include the hydrodynamic boundary effects and surface forces (27). Evidence for this model was provided by comparing the size distribution of available prey with the size distribution of ingested prey in the food vacuoles, as well as by determination of ingestion rates for artificial prey with a narrow size range (10, 12, 13, 28, 35). Video microscopic observation allowed us to investigate selectivity during the different steps (3). We found no positive correlations between contact probability and bacterial size (Table 4). It is not clear to what extent these findings can be generalized for all filamentous bacteria or only hold true for the investigated strains.
Is the feeding of interception-feeding flagellates size selective?
Recent studies have shown that overall food selection of nanoflagellates is composed of active (ingestion efficiency and differential digestion) and the above-mentioned passive selection mechanisms (4, 5). Both our classical feeding experiments and the microscopic observations have shown that Ochromonas sp. strain DS prefers P. putida MM1 over S. aurantia WQM4 and H. gracilis WQT2 (Fig. 3 and 4; Table 4). The selectivity did not depend on food concentration as is expected for active food selection mechanisms (5). On the one hand, ingestion of filamentous bacteria was often unsuccessful irrespective of the food concentration. On the other hand, there were no differences in ingestion rates (Table 4) for P. putida MM1 and H. gracilis WQT2 in experiments with single prey. We therefore conclude that food selectivity was due to a passive selection mechanism favoring ingestion of small prey irrespective of the food concentration. Our live observation showed that with filamentous bacteria, only every fourth to fifth captured bacterium was ingested successfully even at nonsatiating food concentrations. In contrast, for P. putida MM1, nearly each captured bacterium was ingested successfully as long as food concentrations were not satiating. This resulted in the observed food concentration-independent selectivity in the presence of alternative prey but comparable predation efficiency when only one strain was present at a satiating food concentration. It can be assumed, however, that the filamentous bacterium strains are a less suitable food when offered at lower, nonsatiating food concentration, since the lower ingestion efficiency may then become important even without the presence of an alternative food source.
It has been suggested that food selection (active food choice) by flagellates is primarily dependent on food concentration (5, 25). We found no influences of food concentration on the selective feeding by the flagellate during ingestion (Table 4). This could be due to the fact that the concentration-dependent feedings observed by other authors were based on the different food quality (beads and bacteria) instead of food size. Selection based on food quality may be an active receptor-mediated selectivity (5, 29). The fact that in our study feeding was selective, independent of food concentration, supports the conclusion that the food size selection is passive rather than active. The responsible mechanism for the investigated filamentous bacteria, however, is the decreasing ingestion efficiency rather than the probability of contact. Even though the flagellate does not actively select small prey, this can be a selective advantage for the investigated filamentous bacteria.
Conclusions.
Current assumptions on the grazing resistance of filamentous bacteria and size-selective grazing do not match our results from experiments with filamentous bacteria. The size of Ochromonas sp. strain DS is a bit larger than that of typical freshwater nanoflagellates; however, these small differences in size cannot explain the observed predation on large filamentous bacteria. More likely, size alone is not sufficient to define a refuge for filamentous bacteria from nanoflagellate predation. More research is needed to reveal the role of flexibility of cells in the interaction between filamentous bacteria and nanoflagellates. We provide evidence that the relevant selective step for the flagellate's feeding on filamentous prey is the efficiency of ingestion and not the probability of contact.
It is unknown whether the results obtained in this study can be generalized for all filamentous bacteria of a comparable size. The investigated filamentous bacteria are not protected from nanoflagellate predation but have a selective advantage due to the decrease in the flagellate's ingestion efficiency with increasing length of the bacteria.
Acknowledgments
We thank Thomas Weisse for valuable comments on a previous version of the manuscript.
This work was partly supported by the Austrian Science Fund (project P15655, awarded to M.W.H.), the National Natural Science Foundation of China (no. 30370278), the Pilot Project of the Knowledge Innovation Program of the Chinese Academy of Sciences (KZCX1-SW-12), and the Austrian Academy of Sciences. Q.L.W was supported by a scholarship from the North-South Dialogue Program, Ministry of Foreign Affairs, Austria.
REFERENCES
- 1.Andersson, A., U. Larsson, and A. Hagstrom. 1986. Size-selective grazing by a microflagellate on pelagic bacteria. Mar. Ecol. Prog. Ser. 33:51-57. [Google Scholar]
- 2.Boenigk, J., and H. Arndt. 2000. Particle handling during interception-feeding by four species of heterotrophic nanoflagellates. J. Eukaryot. Microbiol. 47:350-358. [DOI] [PubMed] [Google Scholar]
- 3.Boenigk, J., and H. Arndt. 2000. Comparative studies on the feeding behavior of the two heterotrophic nanoflagellates: the filter-feeding choanoflagellate Monosiga ovata and the raptorial-feeding kinetoplastid Rhynchomonas nasuta. Aquat. Microb. Ecol. 22:243-249. [Google Scholar]
- 4.Boenigk, J., C. Matz, K. Jürgens, and H. Arndt. 2001. Confusing selective feeding with differential digestion in bacterivorous nanoflagellates. J. Eukaryot. Microbiol. 48:425-432. [DOI] [PubMed] [Google Scholar]
- 5.Boenigk, J., C. Matz, K. Jürgens, and H. Arndt. 2002. Food concentration dependent regulation of food selectivity of interception-feeding bacterivorous nanoflagellates. Aquat. Microb. Ecol. 27:195-202. [Google Scholar]
- 6.Breznak, J. A., and E. Canale-Parola. 1969. Spirochaeta aurantia, a pigmented, facultatively anaerobic spirochete. J. Bacteriol. 97:386-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canale-Parola, E., S. L. Rosenthal, and D. G. Kupfer. 1966. Morphological and physiological characteristics of Spirillum gracile sp. n. Antonie Leeuwenhoek J. Microbiol. Serol. 32:113-124. [DOI] [PubMed] [Google Scholar]
- 8.Chesson, J. 1983. The estimation and analysis of preference and its relationship to foraging models. Ecology 64:1297-1304. [Google Scholar]
- 9.Christoffersen, K., O. Nybroe, K. Jürgens, and M. Hansen. 1997. Measurement of bacterivory by heterotrophic nanoflagellates using immunofluorescence labeling of ingested cells. Aquat. Microb. Ecol. 13:127-134. [Google Scholar]
- 10.Chrzanowski, T. H., and K. Šimek. 1990. Prey-size selection by freshwater flagellated protozoa. Limnol. Oceanogr. 35:1424-1436. [Google Scholar]
- 11.Fenchel, T. 1982. Ecology of heterotrophic microflagellates. I. Some important forms and their functional morphology. Mar. Ecol. Prog. Ser. 8:211-223. [Google Scholar]
- 12.Fenchel, T. 1987. Ecology of protozoa. Science Tech, Inc., Madison, Wis.
- 13.González, J. M., E. B. Sherr, and B. F. Sherr. 1990. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl. Environ. Microbiol. 56:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González, J. M. 1996. Efficient size-selective bacterivory by phagotrophic nanoflagellates in aquatic ecosystems. Mar. Biol. 126:785-789. [Google Scholar]
- 15.Güde, H. 1979. Grazing by protozoa as selection factor for activated sludge bacteria. Microb. Ecol. 5:225-237. [DOI] [PubMed] [Google Scholar]
- 16.Güde, H. 1989. The role of grazing on bacteria in plankton succession, P. 337-369. In U. Sommer (ed.), Plankton ecology. Succession in plankton communities. Springer-Verlag KG, Berlin, Germany.
- 17.Hahn, M. W., and M. G. Höfle. 1998. Grazing pressure by a bacterivorous flagellate reverses the relative abundance of Comamonas acidovorans PX54 and Vibrio strain CB5 in chemostat cocultures. Appl. Environ. Microbiol. 64:1910-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn, M. W., E. R. B. Moore, and M. G. Höfle. 1999. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl. Environ. Microbiol. 65:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, M. W., E. R. B. Moore, and M. G. Höfle. 2000. Role of microcolony formation in the protistan grazing defense of the aquatic bacterium Pseudomonas sp. MWH1. Microb. Ecol. 39:175-185. [DOI] [PubMed] [Google Scholar]
- 20.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effects on populations of aquatic bacteria. FEMS. Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 21.Hahn, M. W., H. Lünsdorf, Q. L. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinbokel, J. F. 1978. Studies on the functional role of tintinnids in the Southern California Bight. I. Grazing and growth rates in laboratory cultures. Mar. Biol. 47:177-189. [Google Scholar]
- 23.Jürgens, K., and H. Güde. 1994. The potential importance of grazing-resistant bacteria in planktonic systems. Mar. Ecol. Prog. Ser. 112:169-188. [Google Scholar]
- 24.Jürgens, K., H. Arndt, and K. O. Rothhaupt. 1994. Zooplankton mediated changes of bacterial community structure. Microb. Ecol. 27:27-42. [DOI] [PubMed] [Google Scholar]
- 25.Jürgens, K., and W. R. De Mott. 1995. Behavioral flexibility in prey selection by bacterivorous nanoflagellates. Limnol. Oceanogr. 40:1503-1507. [Google Scholar]
- 26.Matz, C., J. Boenigk, H. Arndt, and K. Jürgens. 2002. Role of bacterial phenotypic traits in selective feeding of the heterotrophic nanoflagellate Spumella sp. Aquat. Microb. Ecol. 27:137-148. [Google Scholar]
- 27.Monger, B. C., and M. R. Landry. 1990. Direct-interception feeding by marine zooflagellates: the importance of surface and hydrodynamic forces. Mar. Ecol. Prog. Ser. 65:123-140. [Google Scholar]
- 28.Monger, B. C., and M. R. Landry. 1991. Prey-size dependency of grazing by free-living marine flagellates. Mar. Ecol. Prog. Ser. 74:239-248. [Google Scholar]
- 29.Nisbet, B. 1987. Nutrition and feeding strategy in protozoa. Croom Helm, London, United Kingdom.
- 30.Pernthaler, J., B. Sattler, K. Šimek, A. Schwarzenbacher, and R. Psenner. 1996. Top-down effects on the size-biomass distribution of a freshwater bacterioplankton community. Aquat. Microb. Ecol. 10:255-263. [Google Scholar]
- 31.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-947. [Google Scholar]
- 32.Posch, T., K. Šimek, J. Vrba, J. Pernthaler, J. Nedoma, B. Sattler, B. Sonntag, and R. Psenner. 1999. Predator-induced changes of bacterial size-structure and productivity studied on an experimental microbial community. Aquat. Microb. Ecol. 18:235-246. [Google Scholar]
- 33.Robert, R. S., and F. J. Rohlf. 1998. The principles and practice of statistics in biological research. W. H. Freeman & Co., New York, N.Y.
- 34.Salat, J., and C. Marrase. 1994. Exponential and linear estimation of grazing on bacteria: Effects of changes in the proportion of marked cells. Mar. Ecol. Prog. Ser. 104:205-209. [Google Scholar]
- 35.Šimek, K., and T. H. Chrzanowski. 1992. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl. Environ. Microbiol. 58:3715-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Šimek, K., J. Vrba, J. Pernthaler, T. Posch, P. Hartman, J. Nedoma, and R. Psenner. 1997. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl. Environ. Microbiol. 63:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Šimek, K., P. Kojecka, J. Nedoma, P. Hartman, J. Vrba, and J. R. Dolan. 1999. Shifts in the bacterial community composition associated with different microzooplankton size fractions in a eutrophic reservoir. Limnol. Oceanogr. 44:1634-1644. [Google Scholar]
- 38.Sime-Ngando, T., G. Bouredier, C. Amblard, and B. Pinel-Alloul. 1991. Short-term variations in specific biovolumes of different bacterial forms in aquatic ecosystems. Microb. Ecol. 21:211-226. [DOI] [PubMed] [Google Scholar]
- 39.Sommaruga, R., and R. Psenner. 1995. Permanent presence of grazing resistant bacteria in a hypertrophic lake. Appl. Environ. Microbiol. 61:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spring, S., U. Jaeckel, M. Wagner, and P. Kaempfer. Ottowia thiooxidans gen. nov., sp. nov., a novel facultatively anaerobic, N2O producing bacterium isolated from activated sludge and transfer of Aquaspirillum gracile to Hylemonella gracilis gen. nov. comb. nov. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]