Abstract
Of 220 Shiga toxin-producing Escherichia coli (STEC) strains collected in central France from healthy cattle, food samples, and asymptomatic children, 12 possessed the eae gene included in the locus of enterocyte effacement (LEE) pathogenicity island. Based on gene typing, we observed 7 different eae espA espB tir pathotypes among the 12 STEC strains and described the new espAβv variant. As previously observed, the O157 serogroup is associated with eaeγ, O26 is associated with eaeβ, and O103 is associated with eaeɛ. However, the unexpected eaeζ allele was detected in 5 of the 12 isolates. PCR amplification and pulsed-field gel electrophoresis using the I-CeuI endonuclease followed by Southern hybridization indicated that the LEE was inserted in the vicinity of the selC (three isolates), pheU (two isolates), or pheV (six isolates) tRNA gene. Six isolates harbored two or three of these tRNA loci altered by the insertion of integrase genes (CP4-int and/or int-phe), suggesting the insertion of additional foreign DNA fragments at these sites. In spite of great genetic diversity of LEE pathotypes and LEE insertion sites, bovine strains harbor alleles of LEE genes that are frequently found in clinical STEC strains isolated from outbreaks and sporadic cases around the world, underscoring the potential risk of the bovine strains on human health.
Enterohemorrhagic Escherichia coli (EHEC) strains are the most common cause of postdiarrheal hemolytic-uremic syndrome (HUS) in humans. The major serotype implicated is O157:H7, particularly in the United States, the United Kingdom, and Japan (13, 21, 33). However, several non-O157:H7 serotypes have emerged and been implicated both in outbreaks and in sporadic cases of infection, especially in continental Europe, but also in Australia and North and South America (7, 38, 42). Shiga toxins (Stxs) are the major virulence factors produced by EHEC strains and are responsible for the principal manifestations of HUS (25). The locus of enterocyte effacement (LEE) is a pathogenicity island (PAI) which encodes genes involved in effacement of intestinal epithelial cell microvilli and in intimate adherence between bacteria and the epithelial cell membrane (15). The LEE is present in Shiga toxin-producing E. coli (STEC) and EPEC strains, but its size varies from 35 kb in EPEC E2348/69 to 85 kb in rabbit STEC 84/110-1 (18, 37). The LEE contains five major operons: the LEE1, LEE2, and LEE3 operons contain the esc and sep genes, which encode a type III secretion system involved in extracellular secretion of effector proteins; the LEE4 operon encodes the EspA, EspB, and EspD proteins secreted by the type III secretion apparatus; and the LEE5 operon contains eae, encoding intimin, tir, encoding a translocated receptor for intimin, and cesT, encoding a molecular chaperone for Tir (5, 20, 30). Intimin binding to Tir mediates intimate attachment of bacteria to epithelial cells. The esc and sep genes are highly conserved among STEC strains, whereas the secreted proteins EspA and EspB, as well as Tir and Eae, are highly variable. Ten variants of Eae and three variants of Tir, EspA, and EspB have been described (1, 8, 24, 44). Previous results suggest that the prevalence of specific combinations of Eae, Tir, EspA, and EspB variants, called pathotypes, varies depending on the host and the pathogenicity potential of the strains (8, 10, 12). All EHEC O157:H7 clinical isolates possess the LEE, as do the vast majority of pathogenic non-O157:H7 EHEC isolates. The insertion site of the LEE in the chromosome is different depending on the clonal phylogeny of the strains. Two STEC clones have been defined by multilocus enzyme electrophoresis of 20 housekeeping proteins (41). In the EHEC1 clone, containing the O157:H7 serotype, the LEE is inserted at 82 min on the E. coli K12 chromosome, just downstream of the selC locus encoding the tRNA for selenocysteine. In the EHEC2 clone, containing the O111 and O26 serogroups, the LEE is inserted at 94 min on the K12 chromosome, at the vicinity of the pheU gene encoding the tRNA for phenylalanine (34, 43). Recently, a new PAI containing a LEE was detected in STEC strains and localized in the vicinity of the pheV tRNA gene at 67 min on the E. coli K12 chromosome (14, 37).
Healthy cattle appear to be the main reservoir of STEC strains, which are transmitted to humans through food contaminated by fecal material. A recent 1-year survey in central France showed that 70% of healthy cattle at the slaughterhouse and 10% of food samples (meat and raw-milk cheese) were positive for the stx genes (26). Two hundred ten isolates were recovered from these samples. In addition, 10 isolates were recovered during the same period and in the same area from stool samples of asymptomatic hospitalized children (26). For this study, we investigated the presence of genes belonging to the five LEE operons in the 220 isolates of this STEC collection, located the chromosomal insertion site of the LEE, and determined the eae tir espA espB pathotypes of the isolates to evaluate the potential pathogenicity of these STEC strains for humans. We propose a convenient multiplex assay to discriminate between five eae alleles and described the characterization of a new espA variant.
MATERIALS AND METHODS
Bacterial strains.
The STEC isolates used in this study are part of a collection established during a 1-year survey in central France by Pradel et al. (26). The E2348/69, RDEC-I, and EDL933 reference strains were used as positive controls for α, β, and γ variants, respectively, of intimin, Tir, EspA, and EspB. PMK5 was used as the prototype strain for intimin ɛ (24). The E2348/69 (eaeα1), EF26 (eaeα2), EDL933 (eaeγ1), ED31 (eaeγ2), RDEC-1 (eaeβ1), ICC95 (eaeβ2), and PMK5 (eaeɛ) E. coli strains were used as positive controls for PstI restriction fragment length polymorphism analysis of intimin. Shigella flexneri M90T was used as a positive control for the insertion of the CP4-int gene downstream of selC. DH5α (E. coli K12 strain) was used as a negative control.
Colony hybridization.
Bacteria were grown overnight in Luria-Bertani (LB) broth at 37°C. Five microliters of bacterial culture was spotted onto a nylon membrane (Hybond N+; Amersham Biotech) and incubated for 5 h on LB agar at 37°C. Lysis, denaturation, and neutralization were performed as described previously (29). The filter was probed at 42°C in a DIG Easy Hyb granule (Roche Diagnostics) with digoxigenin (DIG)-labeled DNA generated from the E2348/69 E. coli strain by use of the PCR DIG probe synthesis kit (Roche Diagnostics) and the primers indicated in Table 1. Hybridization was detected by chemiluminescence using CDP-Star as a substrate for alkaline phosphatase according to the manufacturer's instructions (Roche Diagnostics).
TABLE 1.
PCR primers and conditions
| Function of primer set | Primer | Sequence | Annealing temp (°C) | Amplicon size (bp) | Accession no.a | Reference |
|---|---|---|---|---|---|---|
| escR probe | escR-F | GCATTCAACAGGTACCACCAAAC | 52 | 472 | AF022236 | This study |
| escR-F | CCAGCCTCCAACAAGAATG | AF022236 | This study | |||
| escS probe | escS-F | CATAGCGGCCTCTGTTATCG | 52 | 190 | AF022236 | This study |
| escS-R | CGTTCACCTTCGGAATCAT | AF022236 | This study | |||
| escU probe | escU-F | GAAGAGGTAATGGCTGCAGTG | 52 | 606 | AF022236 | This study |
| escU-R | CAGTATCCTTGGCTTCTCG | AF022236 | This study | |||
| escJ probe | escJ-F | CGTCCTGTCCTGAGGATGAC | 52 | 460 | AF022236 | This study |
| escJ-R | GCAAGCACTGTTGCTATCCA | AF022236 | This study | |||
| sepQ probe | sepQ-F | GCCTTACCGGAAGGTGATG | 53 | 383 | AF022236 | This study |
| sepQ-R | CGGATCTGCAGCCAGAAC | AF022236 | This study | |||
| espA probe | espA-F | GTTATTTACCAAGGGATATTCCT | 57 | 576 | AF022236 | This study |
| espA-R | GATACATCAACTACAGCATCAG | AF022236 | This study | |||
| espB probe | espB-F | CTGTTTTGAGCAGCACGACTG | 57 | 531 | AF022236 | This study |
| espB-R | GCAACATCATCAGTGATACCG | AF022236 | This study | |||
| cesT probe | cesT-F | CGTTATCTGATGCCAATGACG | 53 | 242 | AF022236 | This study |
| cesT R | CCATCGACTTAACGACGACTT | AF022236 | This study | |||
| selC analysis | K260 | GAGCGAATATTCCGATATCTGGTT | 52 | 620, 720b | 18 | |
| K272 | GAACCATTAGCGTTAATCAGCAG | AF071034 | This study | |||
| pheU analysis | K913c | CATCGGCTGGCGGAAGATAT | 57 | 34 | ||
| K916 | GGACGGAGTAACAAGCCATTC | 908 | AE000486 | This study | ||
| K917 | GTGCCAGAGTCTTCTCTTTAC | 599 | AF453441 | This study | ||
| pheV analysis | pheV-F | CGATGATGGCAGACTTCAGC | 57 | 583 | AE000379 | This study |
| pheV-R | GGATGAAAAATATGCGCTGCG | AE000379 | This study | |||
| espAβv sequencing | yba-F | GACTGGCTACTGGAGAGCC | 54 | 949 | AF022236 | This study |
| yba-R | GATTTAACCAGTTGTAAATC | AF022236 | This study | |||
| eaeζ and eaeɛ detection | B73d | TACTGAGATTAAGGCTGATAA | 50 | 8 | ||
| B170 | CATGTATCGAATACATCAGC | 656 | AF116899 | This study | ||
| B171 | GCGCCATTAGCGGAGGTTCC | 341 | AJ298279 | This study | ||
| eaeζ subtyping | SK1 | CCCGAATTCGGCACAAGCATAAGC | 61 | 2401 | 31 | |
| LP6 | ACTTGACCAGTGGAATCCACCG | AF449416 | This study | |||
| espAβv detection | B167 | GATACATCAACTGCAACATC | 48 | 375 | AY223511 | This study |
| B168 | GTCTTTTACATCACTGACC | AY223511 | This study | |||
| yicK probe | yicK-F | CACCACTAAAGAACGCGTTG | 54 | 682 | AE000443 | This study |
| yicK-R | GTGAGTAGTAGCGTTTAGCG | AE000443 | This study | |||
| cadC probe | cadC-F | CCAATACCGTGCTCAACAATG | 54 | 700 | AE000486 | This study |
| cadC-R | GAACAGAAGTCTGGAATATACC | AE000486 | This study | |||
| yqgA probe | yqgA-F | CAACGCTAGTGCAGTCTTAC | 54 | 582 | AE000379 | This study |
| yqgA-R | GGTTGCCAGCAGCAATAAAC | AE000379 | This study |
Accession numbers of the DNA sequences used to design the primers.
The amplicons obtained from EDL933 and S. flexneri M90T were 620 and 720 bp, respectively.
K913 is the forward primer used with K916 or K917.
B73 is the forward primer used with B170 and B171 in multiplex PCR to amplify eaeζ and eaeɛ, respectively.
Typing of eae, tir, espA, and espB genes and eae subtyping.
Amplicons were obtained by use of AccuTaq LA DNA polymerase (Sigma) from genomic DNA purified by the DNeasy tissue kit (Qiagen). Multiplex PCR systems to discriminate the three types (α, β, and γ) of the eae, espA, espB, and tir genes were used as previously described (8). The B167 and B168 primers (Table 1) were designed to specifically detect the espAβv variant. The different eae subtypes (α1, α2, β1, β2, γ1, and γ2) were discriminated by a restriction fragment length polymorphism-PCR method that was described previously: eae genes were amplified with SK1 as the universal forward primer and reverse primers specific to eaeα, eaeβ, and eaeγ, and the amplicons were digested with the PstI restriction endonuclease (24, 31). The LP6 primer (Table 1) was designed as a reverse primer to be used in combination with SK1 to detect putative eaeζ subtypes by the same method. DNAs extracted from positive and negative controls were included in every run of PCR.
Detection of five eae alleles by multiplex PCR.
The multiplex PCR described by China et al. to type eaeα, eaeβ, and eaeγ (8) was modified to detect five eae alleles (α, β, γ, ɛ, and ζ) in just one run of PCR. Briefly, the B170 and B171 primers (Table 1) designed to amplify eaeζ and eaeɛ, respectively, were added to the previously described primer cocktail (B73, B74, B137, and B138). PCRs were carried out in a total volume of 50 μl containing 1 μl of DNA template, 0.6 μM (each) primers, 200 μM (each) deoxynucleoside triphosphates, 5 μl of enzyme buffer, and 0.4 U of AmpliTaq DNA polymerase (Q-Biogen). PCR amplification consisted of 25 cycles of denaturation at 94°C for 2 min, annealing for 1 min at 50°C, and extension at 72°C for 1 min on a GeneAmp 2400 thermal cycler apparatus (Perkin-Elmer). The specificity of the new multiplex PCR system was demonstrated by using DNAs extracted from each of the reference E. coli strains (E2348/69 [eaeα], EDL933 [eaeγ], RDEC-I [eaeβ], PMK5 [eaeɛ], and NV92 [eaeζ]).
Analysis of the LEE insertion site by PCR amplification.
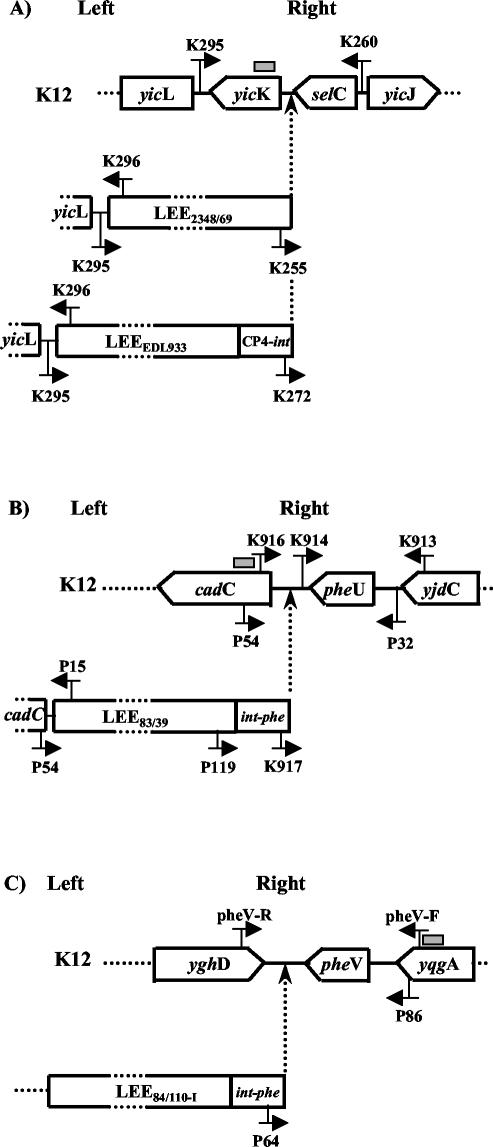
Primers K260, K255, K295, and K296 (for selC site analysis) were described by McDaniel et al. (18). Primers K913, K914 (34), P32, P15, P54, and P119 (37) were used for pheU site analysis. Primers P86 and P64 (37) were used for pheV site analysis. Additional oligonucleotide primers were designed to analyze the putative insertion site of the LEE next to the selC, pheU, or pheV tRNA gene (Table 1). The locations of these primers are shown in Fig. 1.
FIG. 1.
Schematic representation of the positions of the primers used to analyze LEE insertion. Insertion of LEE at selC (A), pheU (B), and pheV (C) tRNA sites was analyzed. LEEE2348/69 and LEEEDL933 are located at 82 min on the E. coli genome. LEE83/39 and LEE84/110-1 are located at 94 and 67 min, respectively, on the E. coli genome. The positions of the probes used for Southern hybridization (PFGE analysis) are indicated by gray boxes. Nucleotide sequences are available at the GenBank database as follows: LEEE2348/69, AF022236; LEEEDL933, AF071034; LEE83/39, AF453441; LEE84/110-1, AF453442; selC, AE000443; pheU, AE000486; pheV, AE000486.
PFGE and Southern hybridization.
Pulsed-field gel electrophoresis (PFGE) analysis was performed on a CHEF-DR III pulsed-field electrophoresis system (Bio-Rad). Bacteria were grown with vigorous shaking in LB broth to an optical density at 600 nm of 1.0. Cells were mixed with 1% (vol/vol) low-melting-temperature agarose (Gibco BRL) and allowed to solidify into disposable plug molds (Bio-Rad). After lysis overnight at 55°C in lysing solution (10 mM Tris [pH 9.0], 100 mM EDTA, 10 mM EGTA, 1% sodium dodecyl sulfate, 1 mg of proteinase K per ml), the plugs were equilibrated for 30 min with the appropriate digestion buffer and incubated overnight at 37°C with 8 U of I-CeuI restriction endonuclease (BioLabs). After digestion, the agarose plugs were equilibrated in Tris-EDTA (TE) buffer for 30 min at room temperature before PFGE. Gels containing 1% agarose were electrophoresed for 4 h (13°C; 6 V cm−1; pulse times ranging from 20 to 60 s), followed by a further 12 h under different conditions (13°C; 6 V cm−1; pulse times ranging from 60 to 100 s), and then stained with ethidium bromide. The gels were submerged in depurination solution (250 mM HCl) three times for 30 min, in denaturation solution (0.5 N NaOH, 1.5 M NaCl) three times for 30 min, and in neutralization solution (0.5 M Tris-HCl, 3 M NaCl, pH 7) twice for 15 min at room temperature. The electrophoretically separated DNA fragments were transferred overnight by capillary force onto a positively charged nylon membrane (Roche Diagnostics). The hybridization was performed at 42°C using DIG Easy Hyb granules (Roche Diagnostics). Probes were labeled with alkali-labile DIG-dUTP (PCR DIG probe synthesis kit; Roche Diagnostics) and purified by use of the Strataprep kit (Stratagene). Chemiluminescence detection with CDP-Star (NEN) was done by exposure of membranes to Hyper film ECL (Amersham Pharmacia Biotech). The eae1 probe was generated by PCR amplification as previously described (37). The primer pairs yicK-F-yicK-R, cadC-F-cadC-R, and yqgA-F-yqgA-R (Table 1; Fig. 1) were designed to amplify probes specific to the yicK, cadC, and yqgA genes, respectively. All of the probes were generated from genomic DNA purified from EDL933. To remove the probe from Southern hybridization, the membranes were incubated twice for 10 min in alkaline probe-stripping solution (0.2 N NaOH, 0.1% sodium dodecyl sulfate) at 37°C and then rinsed thoroughly in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Sequence analysis of espANV92.
PCR amplification was performed from genomic DNA purified by use of the DNeasy tissue kit (Qiagen). The espANV92 gene was amplified with primers yba-F and yba-R (Table 1), located upstream and downstream, respectively, of the espA gene. The primers were designed according to the LEEE2348/69 and LEEEDL933 nucleotide sequences (accession no. AF022236 and AF071034, respectively). PCR amplifications were performed with AccuTaq LA DNA polymerase (Sigma) as recommended by the manufacturer. Amplification products were purified by use of the Strataprep kit (Stratagene). Sequencing of espANV92 was initiated with primer yba-F or yba-R and the PCR product as template. Additional internal primers were designed to sequence the whole gene. The nucleotide sequence was determined for both strands by the dideoxynucleotide triphosphate chain termination method of Sanger, with the Dye Deoxy Terminator cycle sequence kit and an ABI 373A DNA automatic sequencer (Applied Biosystems). Sequence comparisons were performed by using the database at the National Center for Biotechnology Information (National Institutes of Health, Bethesda, Md.), using the FASTA and BLAST search algorithms and CLUSTAL W multiple sequence alignment software. In silico E. coli K12 sequence analyses were performed with Colibri software (http://genolist.pasteur.fr).
Nucleotide sequence accession number.
The nucleotide sequence of espANV92 has been submitted to the GenBank database under accession no. AY223511.
RESULTS
Detection of LEE genes among the STEC library.
Of the 220 isolates collected during a 1-year survey in central France from healthy cattle, food samples, and asymptomatic children, only 12 (3 of 10 isolates from children and 9 of 196 isolates from cattle) possessed the eae gene (26). To investigate whether these isolates harbored an entire LEE and whether the eae-negative strains harbored an incomplete LEE, we tested the capacity of their genomic DNAs to hybridize with LEE probes representative of each of the five LEE operons in a colony blot hybridization assay. The escR, escS, and escU genes were chosen for the LEE1 operon, escJ was used for the LEE2 operon, sepQ was used for the LEE3 operon, espA and espB were used for the LEE4 operon, and cesT was used for the LEE5 operon. All of the eae-positive isolates (n = 12) hybridized with all of the probes tested, whereas none of the eae-negative isolates (n = 208) hybridized with any of the probes. These results indicate that the LEE was either entirely present or entirely absent.
Typing of the eae, tir, espA, and espB genes.
We observed a high level of heterogeneity for the pathotypes, since at least seven different pathotypes were represented by the 12 strains (Table 2). Remarkably, four of the five isolates containing the eaeζ gene belonged to the pathotype tirα espAα espBα. The O157:H7 bovine isolate (NV95) was of pathotype eaeγ1 tirγ espAγ espBγ, as did the reference O157:H7 EHEC strain EDL933. One isolate from a child and one isolate from cattle were of type eaeβ1 tirβ espAβ espBβ, as was initially described for the RDEC-I reference strain (8, 24).
TABLE 2.
Typing of LEE genes
| Strain | Origina | Serotype | Subtype of gene
|
|||
|---|---|---|---|---|---|---|
| eae | tir | espA | espB | |||
| NV100 | B | OR:H− | ζ | α | α | α |
| NV163 | B | O84:H− | ζ | α | α | α |
| NV169 | B | O150:H− | ζ | α | α | α |
| NV220 | B | O103:H2 | ɛ | β | βv | β |
| NV258 | B | OR:H+ | ζ | α | α | α |
| NV270 | B | O98:H− | γ2 | α | α | α |
| NV2 | C | O49:H− | β2 | α | α | α |
| NV10 | C | O26:H+ | β1 | β | β | β |
| NV19 | C | O127:H+ | γ2 | α | α | α |
| NV92 | B | O49:H+ | ζ | α | βv | α |
| NV95 | B | O157:H7 | γ1 | γ | γ | γ |
| NV281 | B | OX177:H− | β1 | β | β | β |
B, STEC isolated from healthy cattle; C, STEC isolated from asymptomatic children.
Genetic characterization of a new espA variant.
The NV92 (O49:H+) and NV220 (O103:H2) espA genes were found to be untypeable. Therefore, the nucleotide sequence of the whole espA gene from NV92 was determined. The predicted protein showed 92% identity with EspAβ of RDEC-1, 82% identity with EspAα of E2348/69, and 80% identity with EspAγ of EDL933. In view of these identity percentages, we propose the name EspAβv for this variant. EspAβv also appeared to be closely related to the EspA protein encoded by the LEE of two human O111:H2 and O128:H2 EPEC strains isolated, respectively, in Chile and Brazil, and to that of the 4221 EPEC strain isolated from a dog (accession no. AJ225018, AJ225021, and U65681) (3, 4, 23). In addition to the one from NV92, a specific espAβv DNA product was PCR amplified from NV220 (O103:H2) and from the O103:H2 PMK5 reference strain.
PCR analysis of the LEE insertion at selC.
An absence of amplification was observed for six strains with primers K260 and K295, suggesting a modified selC region (Fig. 1; Table 3). However, only NV2 (O49:H−), isolated from a child, possessed the right and left junctions of LEEE2348/69. NV92 (O49:H+) and NV95 (O157:H7), isolated from cattle, possessed the left side but not the right border of LEEE2348/69, suggesting an altered or truncated right junction. Thus, we designed the primer K272, which is located at the 5′end of the CP4-like integrase gene (CP4-int) present at the right end of LEEEDL933 but not of LEEE2348/69 (Table 1; Fig. 1). PCR products obtained by use of primers K260 and K272 were obtained only from NV92, NV10, and NV281, but the size of the amplicon was 100 bp longer than expected (720 instead of 620 bp in EDL933) (Table 3). Sequence analysis of the amplicon obtained from NV92 showed that the DNA region located between selC and CP4-int (301 bp) was closely related to the corresponding region of two other selC-associated PAIs: the locus of proteolysis activity present on the 4797/97 STEC strain and SHI-2 present on the M90T S. flexneri strain (accession no. AJ278144 and AF141323, respectively) (32). Accordingly, a 720-bp amplicon was also obtained from the S. flexneri strain M90T (data not shown). Comparison with the corresponding region of LEEEDL933 showed 82% identity over the first 124 bp and a 106-bp deletion in LEEEDL933. This deletion clearly explained the difference in size of the amplicon obtained by PCR with primers K260 and K272. In summary, the selC locus was altered in 6 of 12 strains. However, only two strains harbored a LEE with right and left sides similar to those of LEEE2348/69 (NV2 [O49:H−]) or LEEEDL933 (NV92 [O49:H+]). The LEE of NV95 (O157:H7) seemed to be inserted at selC but had an altered right side. In NV10 (O26:H+) and NV281 (OX177:H−), a CP4-int gene was inserted at selC but there was no evidence that the LEE was inserted at that location.
TABLE 3.
Analysis of LEE insertion site
| Strain | PCR analysis of the selC sitea
|
PCR analysis of the pheU site
|
PCR analysis of the pheV site
|
Gene present by PFGE analysisd | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| selC site | LEE right side | LEE left side | CP4-int | pheU site | LEE right side | LEE left sideb | int-phe | pheV site | int-phec | ||
| NV100 | I | NR | NR | NR | D | − | − | + | D | + | pheV |
| NV163 | I | NR | NR | NR | D | − | − | + | D | + | pheV or pheU |
| NV169 | I | NR | NR | NR | I | NR | NR | − | D | + (2.4 kb) | pheV |
| NV220 | I | NR | NR | NR | D | − | − | − | D | + (2.4 kb) | pheV |
| NV258 | I | NR | NR | NR | D | − | − | + | D | + | pheV |
| NV270 | I | NR | NR | NR | I | NR | NR | − | D | + | pheV |
| NV2 | D | + | + | − | I | NR | NR | − | I | NR | selC |
| NV10 | D | − | − | + | D | + | + (2.3 kb) | + | D | + | pheU |
| NV19 | D | − | − | − | I | NR | NR | − | D | + | pheV |
| NV92 | D | − | + | + | D | − | − | + | D | + | selC |
| NV95 | D | − | + | − | I | NR | NR | − | D | + | selC |
| NV281 | D | − | − | + | D | + | + (6.0 kb) | + | D | + | ND |
PCRs were performed with the primers shown in Fig. 1. I, intact site; D, disrupted site; NR, not relevant due to an intact selC, pheU, or pheV site; ND, not determined; +, amplification; −, no amplification.
Amplicon sizes distinct from that obtained with the RDEC-I reference strain are indicated in parentheses.
Amplicon sizes distinct from that obtained with the 84110-1 reference strain are indicated in parentheses.
tRNA locus present in the DNA fragment containing the LEE.
PCR analysis of the LEE insertion at pheU.
The pheU region was found to be intact in the 12 strains by use of primers K913 and K914. A putative insertion of the LEE between cadC and pheU, as recently described for the LEEs of REPEC 83-39 (LEE83-39) and RDEC-I (LEERDEC-I) (37), was then investigated by use of primers K916 and K913 (Fig. 1). The absence of amplification for 7 of the 12 isolates (NV100, NV163, NV220, NV258, NV10, NV92, and NV281) suggested that the pheU region was modified in these strains (Table 3). By use of primers K917 and K913 (Fig. 1), all of the STEC isolates with a disrupted pheU region, except NV220, were shown to harbor an int-phe gene immediately downstream of pheU. However, only two of them (NV10 and NV281) were positive for the LEE right border amplified with primers P119 and P32. With primers P54 and P15, the size of the LEE left side amplicon (2.3 kb for NV10 and 6.0 kb for NV281) appeared different from that of the RDEC-I reference strain (3.0 kb) (Table 3). Partial sequencing of the 3′ and 5′ ends (up to 750 bp) of the NV10 and NV281 amplicons showed 95 to 99% identity with the corresponding DNA sequences of the LEE83/39 left junction, demonstrating the P54-P15 amplification specificity. In summary, seven strains presented an altered pheU region. The LEE was probably inserted at this site in two of them (NV10 [O26:H+] and NV281 [OX177:H−]). The int-phe gene was inserted downstream of pheU in four of the strains (NV100 [OR:H−], NV163 [O84:H−], NV258 [OR:H+], and NV92 [O49:H+]), but there was no evidence that the LEE was integrated at this location.
PCR analysis of the LEE insertion at pheV.
The pheV region appeared to be altered in 11 of the 12 isolates (Table 3). With primers P64 and P86 (Fig. 1), an amplicon of the expected size (1.1 kb) was obtained from nine of them, demonstrating the presence of int-phe near pheV. In addition, an amplicon of approximately 2.4 kb was obtained from both NV169 and NV220. Partial DNA sequencing of the amplicons (up to 710 bp) showed that their 3′ and 5′ ends were closely related to the LEE84/110-I right border (97 to 99% identity). Furthermore, an IS629-related insertion sequence was inserted within the int-phe gene of these isolates, indicating the presence of additional sequences at the LEE right border. In summary, all of the strains except NV2 had an int-phe gene inserted downstream of pheV, but the presence of the LEE at this site was not demonstrated.
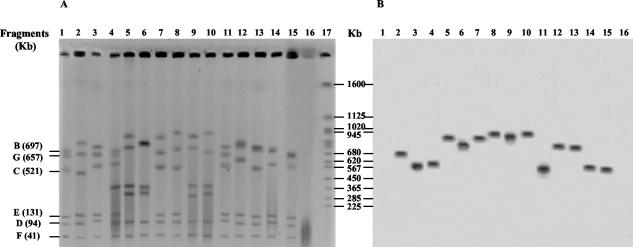
Analysis of the LEE integration site by PFGE and Southern hybridization.
Since PCR analysis did not allow determination with absolute certainty of the chromosomal location of the LEE for most of the strains, PFGE using I-CeuI and Southern hybridization analyses were undertaken. The I-CeuI restriction endonuclease cuts E. coli K12 DNA at each of the seven 23S rRNA loci (17, 37). In silico, the I-CeuI restriction pattern of the K12 genome revealed the presence of the following seven fragments: A (2,500 kb), B (697 kb), C (521 kb), D (94 kb), E (131 kb), F (41 kb), and G (657 kb). BLASTn analysis demonstrated that the pheV, selC, and pheU tRNA genes are located within fragments B, C, and G, respectively. Genomic DNAs of the 12 STEC strains and the 4 reference strains (DH5α, E2348/69, EDL933, and RDEC-I) were digested with I-CeuI and separated by PFGE. Unfortunately, despite several assays, intact DNA from NV281 was never obtained, preventing pattern analysis of this strain. Fragment A (2,500 kb) did not enter the gel under our experimental conditions. Comparison of PFGE patterns revealed a high level of heterogeneity (Fig. 2). However, NV169 and NV220, on the one hand, and NV2 and the EPEC E2348/69 strain, on the other hand, showed similar restriction patterns. To identify the DNA fragment containing the LEE, Southern hybridization was performed with the eae1 probe specific to the sequence encoding the conserved N-terminal region of intimin (Fig. 2). Because of the high level of heterogeneity observed for the PFGE patterns (Fig. 2), determination of the DNA fragments harboring pheV (fragment B), selC (fragment C), or pheU (fragment G) was uncertain. Thus, the membranes were rehybridized successively with probes generated from the yicK, cadC, and yqgA genes, adjacent to the selC, pheU, and pheV tRNA genes, respectively (data not shown). The LEE of NV2, NV92, and NV95 was carried by the DNA fragment containing yicK and selC. The LEE of NV100, NV169, NV220, NV258, NV270, and NV19 was carried by the DNA fragment containing yqgA and pheV, and the LEE of NV10 was carried by the DNA fragment containing cadC and pheU. The cadC probe did not recognize NV100 and NV258 DNAs, suggesting that a deletion in the chromosomal region containing pheU had occurred in these isolates. The LEE insertion site of NV163 could not be identified. Indeed, the eae1 probe hybridized with a fragment of approximately 750 kb harboring both cadC and yqgA, thus corresponding to the superimposition of fragments G (containing pheU) and B (containing pheV).
FIG. 2.
Determination of the integration site of the LEE by PFGE and Southern hybridization. (A) PFGE patterns of chromosomal DNA digested with I-CeuI. (B) Southern hybridization with the eae1 probe specific to the conserved 5′ end of eae. Lanes: 1, E. coli DH5α; 2, RDEC-I; 3, E2348/69; 4, EDL933; 5, NV100; 6, NV163; 7, NV169; 8, NV220; 9, NV258; 10, NV270; 11, NV2; 12, NV10; 13, NV19; 14, NV92, 15, NV95; 16, NV281, 17, Saccharomyces cerevisiae DNA size standard (Bio-Rad). The fragment sizes (in kilobases) of the E. coli K12 DNA digested with I-CeuI are shown at the left.
DISCUSSION
In a previous study, members of our laboratory screened a collection of 220 STEC strains isolated from healthy cattle, food products, and asymptomatic children for the presence of the intimin-encoding gene and found that only 12 strains were eae positive (26). For this report, we characterized the LEEs harbored by these isolates in order to evaluate their potential risk for human health.
An absolute correlation was found between the presence of eae and the presence of the other genes of the LEE. None of the LEE genes was ever detected in eae-negative strains. These data confirmed the usefulness of eae in screening for LEE-positive STEC strains. The proteins encoded by different LEEs (EspA, EspB, Tir, or intimin) show a high degree of sequence polymorphism. To date, 10 distinct variants of eae (α, β, ɛ, γ, θ, ζ, ι, κ, λ, and η) and 3 variants of espA, espB, and tir (α, β, and γ) have been described (24, 36, 44). Previous studies indicated that some serotypes are highly associated with a particular intimin variant (8, 9, 24, 28). In agreement with this observation, we found the O157, O26, and O103 strains to be associated with eaeγ, eaeβ, and eaeɛ, respectively. As expected, the variant eaeα was not found. Until now, this variant was found only in EPEC strains, never in STEC strains. Intimin β appears to be the most ubiquitous type among pathogenic eae-positive E. coli isolates of human and animal origins (10, 19, 24). However, recent studies demonstrate that intimin γ is the most frequent subtype among EPEC and STEC strains isolated from healthy calves (8, 9). Our results did not support this finding, since only two bovine isolates were found with an eaeγ allele. Interestingly, the eaeζ variant was present in five bovine strains. The eaeζ allele was recently described for O111:H9 and O111:NM EHEC strains isolated from an outbreak of diarrhea in Finland that affected >700 persons (36) and for an O84 STEC strain isolated from calves in Germany. Here we validate a convenient multiplex PCR system for identification of the α, β, γ, ɛ, and ζ eae alleles in only one run of PCR. This new genetic tool could be easily used in future studies to investigate the intimin ζ frequency among a large number of EPEC and STEC strains.
A high level of heterogeneity was found for the gene subtypes of LEEs present in the 12 STEC strains, since seven different pathotypes were characterized (of them, two pathotypes were shared by isolates from children and cattle). These results differ from those of China et al., who detected only two distinct pathotypes among 71 bovine EPEC and STEC strains (8). Bovine strains harbor pathotypes (eaeβ tirβ espAβ espBβ, eaeγ tirα espAα espBα, and eaeγ tirγ espAγ espBγ) and eae alleles (γ, ɛ, and ζ) that are frequently found in clinical STEC strains isolated from outbreaks and sporadic cases around the world (8, 24, 36, 44), underscoring the potential risk of the bovine strains for human health.
We also observed a high level of heterogeneity in the LEE integration site as well as the sequences of the right and left ends of the LEE. The LEE is inserted at selC in three isolates (NV92, NV2, and NV95). The bovine O49:H+ strain NV92 possesses the CP4 integrase gene at the right end of the LEE included in the putative prophage segment that differentiates LEEEDL933 from LEEE2348/69, whereas the human O49:H− strain NV2 does not. As for all other O157:H7 isolates described so far, the LEE of the bovine O157:H7 isolate (NV95) was inserted at the selC locus. As previously reported for the REPEC 83-39 and RDEC-1 strains (37), isolates with a LEE inserted between cadC and pheU (NV10 [O26:H+] and NV281 [OX177:H−]) shared the pathotype eaeβ1 tirβ espAβ espβ and the int-phe gene at the right junction. The LEE was probably inserted at pheV in six isolates. In most cases, the right or left ends of LEEs inserted at pheV, pheU, or selC appeared different from those of the reference strains. This observation suggests that the LEE integration site could remain undetermined by the PCR amplification procedures that are commonly used. For most of our isolates, two or three tRNA loci are simultaneously occupied by CP4-int (at selC) and int-phe (at pheU and/or pheV) or by an unknown sequence. PCR amplifications using specific CP4-int or int-phe primers do not definitively demonstrate integration of the LEE at the vicinity of these tRNA loci. PFGE followed by Southern hybridization appeared to be an effective complementary method for analyzing the LEE integration site.
The selC tRNA locus is frequently used as an integration site for PAIs that possess CP4 integrase genes at their ends. In addition to the LEEEDL933, SHI-2 in S. flexneri (22, 39), the locus of proteolysis activity in STEC strain 4797/97 (32), PAI I536 in the uropathogenic E. coli (UPEC) strain 536 (11), and the selC island in the UPEC strain CFT073 (40) are inserted next to the selC tRNA gene. The selC site could also be occupied by PAIs that do not possess the CP4-int gene at their ends, such as LEEE2348/69 and SPI-3 of Salmonella enterica serovar Typhimurium (6). Similarly, pheU and pheV tRNA loci are often used as insertion sites for PAIs flanked by an int-phe integrase gene: PAI IIJ96 of UPEC strain J96 (35, 37), SHI-3 of Shigella boydii (27), and PAI IAL862 of the bovine extraintestinal pathogenic E. coli strain AL862 (16) are inserted in the vicinity of pheU, whereas the PAI of S. flexneri 2a (2), PAI IIAL862 of ExPEC strain AL862 (16), and the pheV island of UPEC strain CFT073 (40) are inserted next to pheV. In a recent study, Tauschek et al. suggested that it is the type of integrase and not the virulence factors encoded by the PAI that determines the site of integration (37). It is well documented that some pathogenic E. coli strains carry more than one PAI inserted next to different tRNA loci in the bacterial chromosome (11, 16, 40). Determination of other putative PAIs inserted in the vicinity of disrupted tRNA loci will be an interesting field for future research.
Our study reveals a high level of heterogeneity of the LEEs in (i) the chromosomal insertion site and the pathotype, (ii) the presence or absence of CP4-int and/or int-phe integrase genes, and (iii) the sequence divergence of the LEE ends. This is in agreement with phylogenetic analysis of EPEC and STEC strains suggesting that gain of the LEE occurred several times and in parallel in separate lineages (28).
Acknowledgments
We are grateful to O. Crouzet and A. Garrivier for excellent technical assistance, S. Leroy-Setrin for technical advice for PFGE experiments, S. Dutilloy for secretarial assistance, and J. P. Girardeau for critical reading of the manuscript.
REFERENCES
- 1. Adu-Bobie, J., G. Frankel, C. Bain, A. G. Goncalves, L. R. Trabulsi, G. Douce, S. Knutton, and G. Dougan. 1998. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J. Clin. Microbiol. 36:662-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Hasani, K., K. Rajakumar, D. Bulach, R. Robins-Browne, B. Adler, and H. Sakellaris. 2001. Genetic organization of the she pathogenicity island in Shigella flexneri 2a. Microb. Pathog. 30:1-8. [DOI] [PubMed] [Google Scholar]
- 3.An, H., J. M. Fairbrother, J. D. Dubreuil, and J. Harel. 1997. Cloning and characterization of the eae gene from a dog attaching and effacing Escherichia coli strain 4221. FEMS Microbiol. Lett. 148:239-245. [DOI] [PubMed] [Google Scholar]
- 4.An, H., J. M. Fairbrother, J. D. Dubreuil, and J. Harel. 1999. Cloning and characterization of the esp region from a dog attaching and effacing Escherichia coli strain 4221 and detection of EspB protein-binding to HEp-2 cells. FEMS Microbiol. Lett. 174:215-223. [DOI] [PubMed] [Google Scholar]
- 5.Beltrametti, F., A. U. Kresse, and C. A. Guzman. 1999. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J. Bacteriol. 181:3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc-Potard, A. B., and E. A. Groisman. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16:5376-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caprioli, A., and A. E. Tozzi. 1998. Epidemiology of Shiga toxin-producing Escherichia coli infections in continental Europe, p. 38-48. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 8.China, B., F. Goffaux, V. Pirson, and J. Mainil. 1999. Comparison of eae, tir, espA and espB genes of bovine and human attaching and effacing Escherichia coli by multiplex polymerase chain reaction. FEMS Microbiol. Lett. 178:177-182. [DOI] [PubMed] [Google Scholar]
- 9.China, B., E. Jacquemin, A. C. Devrin, V. Pirson, and J. Mainil. 1999. Heterogeneity of the eae genes in attaching/effacing Escherichia coli from cattle: comparison with human strains. Res. Microbiol. 150:323-332. [DOI] [PubMed] [Google Scholar]
- 10.Cid, D., J. A. Ruiz-Santa-Quiteria, I. Marin, R. Sanz, J. A. Orden, R. Amils, and R. de la Fuente. 2001. Association between intimin (eae) and EspB gene subtypes in attaching and effacing Escherichia coli strains isolated from diarrhoeic lambs and goat kids. Microbiology 147:2341-2353. [DOI] [PubMed] [Google Scholar]
- 11.Dobrindt, U., G. Blum-Oehler, G. Nagy, G. Schneider, A. Johann, G. Gottschalk, and J. Hacker. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70:6365-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goffaux, F., B. China, L. Janssen, and J. Mainil. 2000. Genotypic characterization of enteropathogenic Escherichia coli (EPEC) isolated in Belgium from dogs and cats. Res. Microbiol. 151:865-871. [DOI] [PubMed] [Google Scholar]
- 13.Griffin, P. M. 1998. Epidemiology of Shiga toxin-producing Escherichia coli infections in humans in the United States, p. 15-22. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 14.Jores, J., L. Rumer, S. Kiessling, J. B. Kaper, and L. H. Wieler. 2001. A novel locus of enterocyte effacement (LEE) pathogenicity island inserted at pheV in bovine Shiga toxin-producing Escherichia coli strain O103:H2. FEMS Microbiol. Lett. 204:75-79. [DOI] [PubMed] [Google Scholar]
- 15.Kaper, J. B., S. Elliott, V. Sperandino, N. T. Perna, G. F. Mayhew, and F. R. Blattner. 1998. Attaching-and-effacing intestinal histopathology and the locus of enterocyte effacement, p. 163-182. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington D.C.
- 16.Lalioui, L., and C. Le Bouguenec. 2001. afa-8 gene cluster is carried by a pathogenicity island inserted into tRNA(Phe) of human and bovine pathogenic Escherichia coli isolates. Infect. Immun. 69:937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 20.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 21.Michino, H., K. Araki, S. Minami, T. Nakayama, Y. Ejima, K. Hiroe, H. Tanaka, N. Fujita, S. Usami, M. Yonekawa, K. Sadamoto, S. Takaya, and N. Sakai. 1998. Recent outbreaks of infections caused by Escherichia coli O157:H7 in Japan, p. 73-81. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 22.Moss, J. E., T. J. Cardozo, A. Zychlinsky, and E. A. Groisman. 1999. The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol. Microbiol. 33:74-83. [DOI] [PubMed] [Google Scholar]
- 23.Neves, B. C., S. Knutton, L. R. Trabulsi, V. Sperandio, J. B. Kaper, G. Dougan, and G. Frankel. 1998. Molecular and ultrastructural characterisation of EspA from different enteropathogenic Escherichia coli serotypes. FEMS Microbiol. Lett. 169:73-80. [DOI] [PubMed] [Google Scholar]
- 24.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradel, N., V. Livrelli, C. De Champs, J. B. Palcoux, A. Reynaud, F. Scheutz, J. Sirot, B. Joly, and C. Forestier. 2000. Prevalence and characterization of Shiga toxin-producing Escherichia coli isolated from cattle, food, and children during a one-year prospective study in France. J. Clin. Microbiol. 38:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purdy, G. E., and S. M. Payne. 2001. The SHI-3 iron transport island of Shigella boydii 0-1392 carries the genes for aerobactin synthesis and transport. J. Bacteriol. 183:4176-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sanchez-SanMartin, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt, H., H. Russmann, and H. Karch. 1993. Virulence determinants in nontoxinogenic Escherichia coli O157 strains that cause infantile diarrhea. Infect. Immun. 61:4894-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt, H., W. L. Zhang, U. Hemmrich, S. Jelacic, W. Brunder, P. I. Tarr, U. Dobrindt, J. Hacker, and H. Karch. 2001. Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69:6863-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, H. R., B. Rowe, G. K. Adak, and W. J. Reilly. 1998. Shiga toxin (verotoxin)-producing Escherichia coli in the United Kingdom, p. 49-58. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 34.Sperandio, V., J. B. Kaper, M. R. Bortolini, B. C. Neves, R. Keller, and L. R. Trabulsi. 1998. Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol. Lett. 164:133-139. [DOI] [PubMed] [Google Scholar]
- 35.Swenson, D. L., N. O. Bukanov, D. E. Berg, and R. A. Welch. 1996. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect. Immun. 64:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarr, C. L., and T. S. Whittam. 2002. Molecular evolution of the intimin gene in O111 clones of pathogenic Escherichia coli. J. Bacteriol. 184:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tauschek, M., R. A. Strugnell, and R. M. Robins-Browne. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44:1533-1550. [DOI] [PubMed] [Google Scholar]
- 38.Verweyen, H. M., H. Karch, F. Allerberger, and L. B. Zimmerhackl. 1999. Enterohemorrhagic Escherichia coli (EHEC) in pediatric hemolytic-uremic syndrome: a prospective study in Germany and Austria. Infection 27:341-347. [DOI] [PubMed] [Google Scholar]
- 39.Vokes, S. A., S. A. Reeves, A. G. Torres, and S. M. Payne. 1999. The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol. Microbiol. 33:63-73. [DOI] [PubMed] [Google Scholar]
- 40.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.WHO. 1998. Zoonotic non-O157 Shiga toxin-producing Escherichia coli (STEC). Report of a WHO scientific working group meeting. World Health Organization, Geneva, Switzerland.
- 43.Wieler, L. H., T. K. McDaniel, T. S. Whittam, and J. B. Kaper. 1997. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of the strains. FEMS Microbiol. Lett. 156:49-53. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, W. L., B. Kohler, E. Oswald, L. Beutin, H. Karch, S. Morabito, A. Caprioli, S. Suerbaum, and H. Schmidt. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]