Abstract
Background
Clinical and experimental data suggest an association between the presence of bacterial and/or fungal infection and the development of different types of cancer, independently of chemotherapy-induced leukopenia. This has also been postulated for the development of lung cancer, however the prevalence and the exact species of the bacteria and fungi implicated, have not yet been described.
Aim
To determine the presence of bacterial and fungal microflora in surgically extracted samples of patients with lung cancer.
Materials and methods
In this single-center prospective, observational study, tissue samples were surgically extracted from 32 consecutive patients with lung cancer, and reverse-transcription polymerase chain reaction (RT-PCR) was used to identify the presence of bacteria and fungi strains.
Results
The analysis of the electrophoresis data pointed out diversity between the samples and the strains that were identified. Mycoplasma strains were identified in all samples. Strains that appeared more often were Staphylococcus epidermidis, Streptococcus mitis and Bacillus strains, followed in descending frequency by Chlamydia, Candida, Listeria, and Haemophilus influenza. In individual patients Legionella pneumophila and Candida tropicalis were detected.
Conclusions
A diversity of pathogens could be identified in surgically extracted tissue samples of patients with lung cancer, with mycoplasma strains being present in all samples. These results point to an etiologic role for chronic infection in lung carcinogenesis. Confirmation of these observations and additional studies are needed to further characterize the etiologic role of inflammation in lung carcinogenesis.
Keywords: lung cancer, bacteria, fungi, reverse-transcription polymerase chain reaction
Introduction
Lung cancer is the most common cancer worldwide, with 1.35 million incident cases annually, and consists one of the leading causes of mortality worldwide [1]. In addition to cigarette smoking, the major lung cancer risk factor [1], recent studies underscore an etiologic role for chronic pulmonary infection in lung carcinogenesis, acting either independently or as a cofactor to tobacco smoke in increasing lung cancer risk [2-5]. Experimental and clinical data correlate cancer development with the presence of certain pathogens, independently of chemotherapy-induced leucopenia [6-8]. Indeed, mycoplasma is one of the most often observed pathogen in lung carcinomas [9], and it has been postulated that mycoplasma-infected cells have a higher ability to metastasize in vivo than non-mycoplasma-infected cells [10]. Very similarly, the bacterium Chlamydia pneumoniae, a common cause of community-acquired pneumonia, has been implicated in lung carcinogenesis [11-16]. Staphylococcus strains likewise have been observed in many cases of patients with lung cancer [6,7,17-19]. Other studies have demonstrated the presence of colonies in respiratory tract in patients with cancer [19]; Haemophilus influenza [6,7,19-21] and Candida albicans [7,20-22] have been found in patients with lower respiratory tract malignancies. Legionella pneymophila has been diagnosed in patients with cancer [23], as well as strains of Bacillus [7], Listeria [24], and Streptococcus [6,7,17,19,25].
Importantly, previous retrospective and prospective studies have relied on serologic characterization of chronic bacterial and fungal infections [14]. To the best of our knowledge, the prevalence of bacterial and/or fungal infection in surgically extracted samples of patients with lung cancer has not been previously reported. The aim of the present study, therefore, was to determine the presence of bacterial and fungal microflora in surgically removed tissue samples of patients with lung cancer, by using PCR methods and special primers.
Materials and methods
In this single-center prospective, observational study, tissue samples were surgically removed from 32 consecutive patients with lung cancer. The samples were maintained in RPMI culture medium (Sigma, R0883, Germany). The tissue was dissociated and 2 ml Trypsin - 0,25% EDTA (Invitrogen, 25200-072, California) was added in order to detach the cells. The trypsin has been inactivated by using FBS (Invitrogen, 10106-169, California) and cells were centrifuged at 1,200 rpm for 10 min. Then cells were incubated in 25 cm2 flasks (Orange Scientific, 5520200, Belgium) at 37°C, in a 5% CO2 atmosphere, until well developed. RNA was extracted using TRIZOL (Invitrogen, 15596-026, California) and was used as a template to generate cDNA using the First strand cDNA synthesis kit (Fermentas, K1612, Canada). The First strand cDNA was used as a template for the Gradient-PCR reaction, which was performed using GoTaq Flexi polymerase (Promega, M8305, USA). Primers have been designed with Gene Expression 1.1 software. The PCR conditions were set as follows: initial denaturation at 95°C for 10 min to activate the polymerase, 35 cycles of denaturation at 94°C for 45 sec, followed by annealing at 52-58°C for 45 sec and an extension step at 72°C for 60 sec. A final extension step was performed at 72°C for 10 min. The PCR products were then separated on 1.5% agarose gel (Merck, 1012360500, USA) stained with GelGreen (Gentaur, 41005, Belgium), and finally observed under UV-light. A 100-bp ladder (Promega, G2101, USA) was used as marker.
This study was in compliance with the Helsinki Declaration. All participants gave informed consent and the study was approved by the institutional board review.
Results
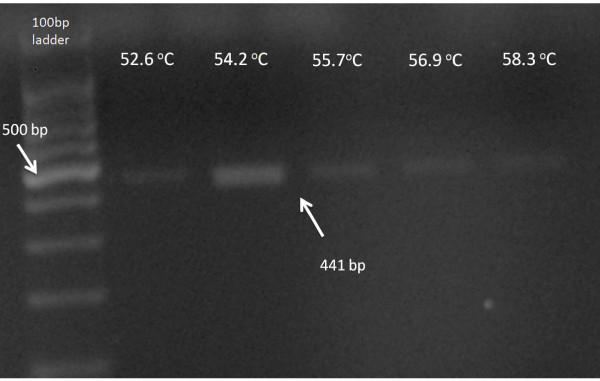
Table 1 shows the primer pairs that were used in PCR to identify the specific pathogen strains. Table 2 presents the frequency of different species and strains in the samples that were examined. The analysis of the electrophoresis data pointed out diversity between the samples and the strains that were identified in them. Mycoplasma strains were identified in all samples (Figure 1 demonstrates electrophoresis results for Mycoplasma strains). Strains that appeared more often were Staphylococcus epidermidis, Streptococcus mitis and Bacillus strains, followed in descending frequency by Chlamydia, Candida, Listeria, and Haemophilus influenza. In individual patients Legionella pneumophila and Candida tropicalis were detected.
Table 1.
Primer pairs that have been used in PCR
| Organism | Species | Forward Primer (5'-3') | Reverse Primer (5'-3') | PCR Product (bp) |
|---|---|---|---|---|
| Treponema | pallidum | AATGCGGTGGCGTAGCGATAC | TTTTGCGGTTTGCTCCACTTC | 275 |
| denticola | AGGGATATGGCAGCGTAGCAATA | CGTCCTCCCTTACGGGTTAGACT | 453 | |
| vincentii | GCGGTATGTAAGCCTGGTGTGAA | TTTGCTTTGGCACTGAAGCTCTT | 277 | |
| Neisseria | meningitidis | AAGTCGGACGGCAGCACAGA | TCAGCCGCTGATATTAGCAACAG | 421 |
| Legionella | pneumophila | AAGATTAGCCTGCGTCCGATTAG | AACCCTCCTCCCCACTGAAAGT | 232 |
| Borrelia | burgdorferi | CATGCAAGTCAAACGGGATGTA | GACCTTCTTCATTCACGCAGTG | 361 |
| americana | ||||
| valaisiana | ||||
| garinii | ||||
| recurrentis | ||||
| hispanica | ||||
| duttonii | ||||
| lusitaniae | ||||
| spielmanii | ||||
| Listeria | grayi | TCTTGACATCCTTTGACCACTCTG | TGCACCGGCAGTCACTTTAGAG | 157 |
| innocua | ||||
| monocytogenes | ||||
| welshimeri | ||||
| Helicobacter | pylori | GATTGGCTCCACTTCGCAGTA | GGCGACCTGCTGGAACATT | 560 |
| pullorum | ||||
| equorum | ||||
| canadensis | ||||
| Staphylococcus | aureus | AGGCGACTTTCTGGTCTGTAACTG | CCGAAGGGGAAGGCTCTATCT | 307 |
| Haemophilus | parasuis | CCTTGGGAAAATACTGACGCTCAT | TCCCGAAGGCACACTCTCAT | 297 |
| Chlamydia | muridarum | TGTTTAGTGGCGGAAGGGTTAG | CCGTCCATTGCGAAAGATTC | 304 |
| trachomatis | ||||
| Bacillus | pumilus | TGCAAGTCGAGCGGACAGA | TCCCAGTCTTACAGGCAGGTTAC | 91 |
| aerophilus | ||||
| licheniformis | ||||
| amyloliquefaciens | ||||
| subtilis | ||||
| Bacillus II | anthracis | CGGCTTCGGCTGTCACTTATG | TCAGCACTAAAGGGCGGAAAC | 655 |
| cereus | ||||
| thuringiensis | ||||
| Mycoplasma | pneumoniae | GAGGCGAACGGGTGAGTAACA | CGCGACTGCTGGCACATAGT | 441 |
| pirum | ||||
| gallicepticum | ||||
| genitalium | ||||
| amphoriforme | ||||
| Leptospira | borgpetersenii | GGATAGCCCCGAGAGGTCATA | CCATCATCACATCGCTGCTTAT | 299 |
| Leptospira | meyeri | CGAATGTGACGGTTCCTGGTAG | TTCGCCCATTGAGCAAGATT | 210 |
| biflexa | ||||
| Staphylococcus | epidermidis | GTGAAAGACGGTTTTGCTGTCAC | CGGATAACGCTTGCCACCTAC | 359 |
| Streptococcus | mitis | GGAGCTTGCTCTTCTGGATGAG | GAGCCGTTACCCCACCAACT | 197 |
| Leptospira | interrogans | CAGCCTGCACTTGAAACTATGTG | ATAGTCCCCAGGCGGTCTACT | 266 |
| Brachyspira | hyodysenteriae | TGCCGTAGAGTGGGGGATAA | CCGCAGGCTCATCGTAAAG | 109 |
| aalborgi | ||||
| intemedia | ||||
| alvinipulli | ||||
| innocens | ||||
| suanatina | ||||
| Haemophilus | influenzae | CTTGCTTTCTTGCTGACGAGTG | TCTCAGTCCCGCACTTTCATC | 129 |
| Candida I | albicans | CCAGCCGAGCCTTTCCTTCT | TACCCCCGACCGTCCCTATT | 187 |
| parapsilosis | ||||
| dubliensis | ||||
| Candida | tropicalis | CGGTCGGGGGTATCAGTATTC | ATACTCGCTGGCTCCGTCAGT | 622 |
Table 2.
Prevalence by different species and strains
| Pathogen | Strain | Prevalence (%) |
|---|---|---|
| Legionella | pneumophila | 3.125 |
| Listeria | grayi | 9.375 |
| innocua | ||
| monocytogenes | ||
| welshimeri | ||
| Chlamydia | muridarum | 12.5 |
| trachomatis | ||
| Bacillus | pumilus | 28.125 |
| aerophilus | ||
| licheniformis | ||
| amyloliquefaciens | ||
| subtilis | ||
| Staphylococcus | epidermidis | 25 |
| Streptococcus | mitis | 21.875 |
| Haemophilus | influenzae | 6.25 |
| Mycoplasma | pneumonia | 100 |
| pirum | ||
| gallicepticum | ||
| genitalium | ||
| amphoriforme | ||
| Candida | albicans | 12.5 |
| parapsilosis | ||
| dubliensis | ||
| Candida | tropicalis | 3.125 |
Figure 1.
Electrophoresis results for Mycoplasma strains.
Discussion
Lung cancer is the most common cancer worldwide and is a leading causes of mortality worldwide [1]. Many recent studies have underscored the etiologic role of chronic pulmonary infection in lung carcinogenesis, concluding that inflammation increases the risk for incident lung cancer [2-5]. Numerous studies on lung cancer have pointed out the appearance of Mycoplasma strains in patients and suggest association of infection with tumorigenesis; it has been postulated that mycoplasma-infected cells have a higher ability to metastasize in vivo than non-mycoplasma-infected cells [10]. Candida species have been isolated from patients with lower respiratory tract infection [7,20-22]. Haemophilus influenza [6,7,19-21], Staphylococcus epidermidis [6,7,17-19], Streptococcus species [6,7,17,19,25], Legionella pneymophila [23], as well as strains of Bacillus [7], Listeria [24] and Streptococcus [6,7,17,19,25] have been also identified in patients with different pulmonary diseases. Very similarly, the bacterium Chlamydia pneumoniae, a common cause of community-acquired pneumonia, has been implicated in lung carcinogenesis [11-16]. A recent meta-analysis by Zhan et al. [16] of 12 studies involving 2595 lung cancer cases and 2585 controls from four prospective studies and eight retrospective studies, was conducted to analyze the association between C. pneumoniae infection and risk of lung cancer. Overall, people exposed to C. pneumoniae infection had an odds ratio (OR) of 1.48 (95% confidence interval (CI), 1.32-1.67) for lung cancer risk, relative to those not exposed. Of interest, a higher titre was an even better risk prognosticator (OR for IgA ≥64 cutoff group, 2.35; 95% CI, 1.88-2.93; OR for IgA ≥16 cutoff group, 1.22; 95% CI, 1.06-1.41).
These data strongly support the idea that lung cancer is a biofilm associated chronic infection. Biofilms are microorganism populations organized in a form of colonies using self-produced extracellular matrix that works as infrastructure material. The vast majority of the micro-"colonists" establish biofilms on any inert or diseased biological surface. They adhere to each other, divide, cooperate, and, progressively, their bio-mass grows, matures and finally disperses. It resembles malignant behavior (tumors composed by cancer cells and by stroma cells-monocytes, lymphocytes, microvessels, can metastasize). Therefore, many researchers imply that lung malignancies are communities of diverse pathogens resistant to antibiotics.
One of the major limitations in most of the previous studies was the use of serologic characterization to identify chronic bacterial or fungal infections [14]. This has resulted in conflicting results and great variability in relative risk estimations among seropositive individuals [14,15,26-29]. This wide variability could also reflect the retrospective nature of most of the studies, the small sample sizes, or inadequate adjustment for confounding factors [14]. New techniques, such as PCR-RFLPs, Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) and microcolony methods allow examination and analysis of microbial communities [30,31]. Analyzing the constituents of microbial biofilms responsible for lung disease may help us discover novel strategies to control malignancies.
To the best of our knowledge, the prevalence of bacterial and/or fungal infection in surgically extracted samples of patients with lung cancer has not been previously reported. Therefore, the main purpose of the present study was to determine the presence of bacterial and fungal microflora in surgically removed tissue samples of patients with lung cancer, by using PCR methods and special primers. In this study, specific primers were designed in order to amplify as many different strains of microorganisms. Pairs of primers that were designed were capable of amplifying Treponema, Neisseria, Legionella, Borrelia, Listeria, Helicobacter, Staphylococcys, Haemophilus, Bacillus, Leptospira, Streptococcus, Mycoplasma, Candida and Brachyspira species. It is worth noting that Mycoplasma species were observed in all samples. Staphylococcus epidermidis and Streptococcus mitis were almost seen in one quarter of patients. Neither Treponema strains nor Leptospira, Helicobacter, and Staphylococcus aureus strains were observed in this study.
Conclusion
A diversity of pathogens could be identified in surgically extracted tissue samples of patients with lung cancer, with mycoplasma strains being present in all samples. These results point to an etiologic role for chronic infection in lung carcinogenesis. Confirmation of these observations and additional studies are needed to further characterize the etiologic role of inflammation in lung carcinogenesis, thus making it possible to apply new therapeutic modalities.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PA carried out the molecular studies and drafted the manuscript. AT participated in the design of the study and collected all tissue samples. IP participated in the design of the study and coordination. MT carried out the molecular studies and drafted the manuscript. MC carried out the molecular studies and drafted the manuscript. GG performed the statistical analysis and drafted the manuscript.
All authors read and approved the final manuscript.
Contributor Information
Panagiotis Apostolou, Email: apostolou.panagiotis@rgcc-genlab.com.
Aggeliki Tsantsaridou, Email: ats61ath@hotmail.com.
Ioannis Papasotiriou, Email: papasotiriou.ioannis@rgcc-genlab.com.
Maria Toloudi, Email: jpapasot@otenet.gr.
Marina Chatziioannou, Email: ggiamouzis@yahoo.com.
Gregory Giamouzis, Email: ggiamou@emory.edu.
References
- Alberg AJ, Brock MV, Samet JM. Epidemiology of lung cancer: looking to the future. J Clin Oncol. 2005;23(14):3175–85. doi: 10.1200/JCO.2005.10.462. [DOI] [PubMed] [Google Scholar]
- Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7(10):778–90. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- Govindan R. Lung cancer in never smokers: a new hot area of research. Lancet Oncol. pp. 304–5. [DOI] [PubMed]
- Engels EA. Inflammation in the development of lung cancer: epidemiological evidence. Expert Rev Anticancer Ther. 2008;8(4):605–15. doi: 10.1586/14737140.8.4.605. [DOI] [PubMed] [Google Scholar]
- Ballaz S, Mulshine JL. The potential contributions of chronic inflammation to lung carcinogenesis. Clin Lung Cancer. 2003;5(1):46–62. doi: 10.3816/CLC.2003.n.021. [DOI] [PubMed] [Google Scholar]
- Dancewicz M. et al. [Bronchial bacterial colonization in patients with lung cancer] Pneumonol Alergol Pol. 2009;77(3):242–7. [PubMed] [Google Scholar]
- Szymankiewicz M, Kowalewski J, Dancewicz M. [Bacteriological and mycological analysis of material taken from lower respiratory tract in patients with malignancy] Pol Merkur Lekarski. 2006;21(123):218–22. [PubMed] [Google Scholar]
- Pieper R, Book K, Nord CE. Microbial flora associated with pulmonary neoplasms. Scand J Thorac Cardiovasc Surg. 1984;18(3):259–61. doi: 10.3109/14017438409109904. [DOI] [PubMed] [Google Scholar]
- Huang S. et al. Mycoplasma infections and different human carcinomas. World J Gastroenterol. 2001;7(2):266–9. doi: 10.3748/wjg.v7.i2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushio S. et al. Metastasis-promoting activity of a novel molecule, Ag 243-5, derived from mycoplasma, and the complete nucleotide sequence. Microbiol Immunol. 1995;39(6):393–400. doi: 10.1111/j.1348-0421.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- Chaturvedi AK. et al. Chlamydia pneumoniae infection and risk for lung cancer. Cancer Epidemiol Biomarkers Prev. 2010;19(6):1498–505. doi: 10.1158/1055-9965.EPI-09-1261. [DOI] [PubMed] [Google Scholar]
- Anttila T. et al. Chlamydia pneumoniae infection and the risk of female early-onset lung cancer. Int J Cancer. 2003;107(4):681–2. doi: 10.1002/ijc.11353. [DOI] [PubMed] [Google Scholar]
- Koyi H. et al. Chlamydia pneumoniae may be associated with lung cancer. Preliminary report on a seroepidemiological study. APMIS. 1999;107(9):828–32. [PubMed] [Google Scholar]
- Littman AJ, Jackson LA, Vaughan TL. Chlamydia pneumoniae and lung cancer: epidemiologic evidence. Cancer Epidemiol Biomarkers Prev. 2005;14(4):773–8. doi: 10.1158/1055-9965.EPI-04-0599. [DOI] [PubMed] [Google Scholar]
- Littman AJ. et al. Chlamydia pneumoniae infection and risk of lung cancer. Cancer Epidemiol Biomarkers Prev. 2004;13(10):1624–30. [PubMed] [Google Scholar]
- Zhan P. et al. Chlamydia pneumoniae infection and lung cancer risk: a meta-analysis. Eur J Cancer. 2011;47(5):742–7. doi: 10.1016/j.ejca.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Balasaniants GS, Torkatiuk EA. [Microbial spectrum of respiratory tract in patients with different pulmonary diseases] Zh Mikrobiol Epidemiol Immunobiol. 2010. pp. 7–11. [PubMed]
- Korona-Glowniak I. et al. [Phenotypic characteristics of coagulase-negative Staphylococci colonizing pleural drains in patients with lung cancer after thoracic surgery] Med Dosw Mikrobiol. 2003;55(2):109–15. [PubMed] [Google Scholar]
- Rancic M, Ristic L, Stankovic I. [Infective complications in patients with lung cancer] Med Pregl. 2011;63(9-10):643–7. doi: 10.2298/mpns1010643r. [DOI] [PubMed] [Google Scholar]
- Jha BJ. et al. Characterization of Candida species isolated from cases of lower respiratory tract infection. Kathmandu Univ Med J (KUMJ) 2006;4(3):290–4. [PubMed] [Google Scholar]
- Laroumagne S. et al. [Incidence and characteristics of bronchial colonisation in patient with lung cancer: a retrospective study of 388 cases] Rev Mal Respir. 2011;28(3):328–35. doi: 10.1016/j.rmr.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Dendis M. et al. PCR-RFLP detection and species identification of fungal pathogens in patients with febrile neutropenia. Clin Microbiol Infect. 2003;9(12):1191–202. doi: 10.1111/j.1469-0691.2003.00719.x. [DOI] [PubMed] [Google Scholar]
- Nunnink JC, Gallagher JG, Yates JW. Legionnaires' disease in patients with cancer. Med Pediatr Oncol. 1986;14(2):81–5. doi: 10.1002/mpo.2950140205. [DOI] [PubMed] [Google Scholar]
- Khardori N. et al. Spectrum and outcome of microbiologically documented listeria monocytogenes infections in cancer patients. Cancer. 1989;64(9):1968–70. doi: 10.1002/1097-0142(19891101)64:9<1968::AID-CNCR2820640934>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Jassem E. et al. [Non-sporeforming anaerobic bacteria in bronchoalveolar lavage fluid of patients with pneumonia during the course of lung cancer] Pneumonol Alergol Pol. 1997;65(9-10):643–8. [PubMed] [Google Scholar]
- Laurila AL. et al. Serological evidence of an association between Chlamydia pneumoniae infection and lung cancer. Int J Cancer. 1997;74(1):31–4. doi: 10.1002/(SICI)1097-0215(19970220)74:1<31::AID-IJC6>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Jackson LA. et al. Association of Chlamydia pneumoniae immunoglobulin A seropositivity and risk of lung cancer. Cancer Epidemiol Biomarkers Prev. 2000;9(11):1263–6. [PubMed] [Google Scholar]
- Smith JS. et al. Lack of association between serum antibodies of Chlamydia pneumoniae infection and the risk of lung cancer. Int J Cancer. 2008;123(10):2469–71. doi: 10.1002/ijc.23814. [DOI] [PubMed] [Google Scholar]
- Koh WP. et al. Lack of association between chronic Chlamydophila pneumoniae infection and lung cancer among nonsmoking Chinese women in Singapore. Int J Cancer. 2005;114(3):502–4. doi: 10.1002/ijc.20745. [DOI] [PubMed] [Google Scholar]
- Girjes AA, Carrick FN, Lavin MF. Single DNA sequence common to all chlamydial species employed for PCR detection of these organisms. Res Microbiol. 1999;150(7):483–9. doi: 10.1016/S0923-2508(99)00108-4. [DOI] [PubMed] [Google Scholar]
- Hsieh SY. et al. Highly efficient classification and identification of human pathogenic bacteria by MALDI-TOF MS. Mol Cell Proteomics. 2008;7(2):448–56. doi: 10.1074/mcp.M700339-MCP200. [DOI] [PubMed] [Google Scholar]