Abstract
This paper describes the use of the alr gene, encoding alanine racemase, as a promoter-screening tool for the identification of conditional promoters in Lactobacillus plantarum. Random fragments of the L. plantarum WCFS1 genome were cloned upstream of the promoterless alr gene of Lactococcus lactis in a low-copy-number plasmid vector. The resulting plasmid library was introduced into an L. plantarum Δalr strain (MD007), and 40,000 clones were selected. The genome coverage of the library was estimated to be 98%, based on nucleotide insert sequence and restriction analyses of the inserts of randomly selected clones. The library was screened for clones that were capable of complementing the d-alanine auxotroph phenotype of MD007 in media containing up to 10, 100, or 300 μg of the competitive Alr inhibitor d-cycloserine per ml. Western blot analysis with polyclonal antibodies raised against lactococcal Alr revealed that the Alr production level required for growth increased in the presence of increasing concentrations of d-cycloserine, adding a quantitative factor to the primarily qualitative nature of the alr complementation screen. Screening of the alr complementation library for clones that could grow only in the presence of 0.8 M NaCl resulted in the identification of eight clones that upon Western blot analysis showed significantly higher Alr production under high-salt conditions than under low-salt conditions. These results established the effectiveness of the alanine racemase complementation screening method for the identification of promoters on their conditional or constitutive activity.
Lactic acid bacteria (LAB) are a group of gram-positive bacteria that are of major economic importance. LAB are applied extensively in both the production and preservation of a wide variety of food products. During industrial processes, LAB often face stressful conditions, such as changes in temperature, acidity, osmolarity, and oxidative conditions. To improve industrial performance or implement desirable properties in LAB, it is of importance to gain insight into the complex regulatory processes that occur in these microbes during stress. Moreover, various LAB have developed into bacterial model systems because of their easy genetic accessibility, availability of genetic tools, and their relatively small and known genomes (4, 26). Hence, insight into gene expression and its control allows for further understanding of genome-wide regulation and the development of tools for the in situ and controlled expression of desirable functions. This justifies the increasing effort that is put in the analysis of gene expression signals and the characterization of regulatory elements in LAB (11, 12, 18, 22, 24).
Promoter-probe vectors are suitable genetic tools for the identification and characterization of promoters and the effective evaluation of their activity. Promoters in LAB have been studied mainly by insertion of chromosomal DNA fragments upstream of a promoterless reporter gene, such as the antibiotic resistance marker chloramphenicol acetyltransferase (Cat). The genes cat-194 from Staphylococcus aureus (1) and cat-86 from Bacillus pumilis (37), both encoding Cat, have been exploited as promoter-probes for the identification of streptococcal and lactococcal promoters, respectively. These systems are based on the selection of promoter activities by monitoring growth in the presence of chloramphenicol. In addition, promoter activities can be quantified by measuring Cat activity in a simple assay. Unfortunately, only relatively strong promoters were identified in these screening efforts, indicating that the Cat production levels required for selection are high (1, 37).
A second group of frequently used reporter genes encode sugar hydrolases. These genes generally allow qualitative detection of the encoded enzyme activity by addition of a chromogenic substrate to plates and quantitative assessment of promoter strength by a simple assay. Examples of sugar hydrolases employed in promoter-screening procedures in LAB include the Lactococcus lactis lacG gene (35), encoding phospho-β-galactosidase, and the lacZ genes from Leuconostoc mesenteroides (24), Streptococcus thermophilus (18), and Escherichia coli (33), encoding β-galactosidases. The application possibilities of these genes in promoter screens remain limited to LAB that do not express endogenous genes encoding these (phospho)-β-galactosidases. Since many LAB are generally known to effectively ferment lactose, it is not surprising that several (phospho)-β-galactosidases have been found in these bacteria (13, 19, 29). Alternatively, the β-glucuronidase-encoding gusA genes originating from E. coli and Lactobacillus gasseri have been used to study promoter activities in LAB (30, 31). Although sugar hydrolases have been successfully applied in promoter-screening strategies in several species of LAB, the selection of promoter activities by using these systems is not based on conditional growth, resulting in laborious procedures and involving evaluation of enzyme activity levels in large numbers of colonies under various conditions (33).
A third group of reporter systems involves the emission of light. A mutant green fluorescent protein from Aequorea victoria was optimized for bacterial expression (10, 21). However, very high gfp expression levels appeared to be required for effective detection of light-emitting LAB, thereby limiting the applicability of this system in these bacteria (21). Alternative strategies with the luciferase (luxAB) genes from Vibrio harveyi and Vibrio fischeri were shown to be more sensitive (9, 16). Unfortunately, nonyl aldehyde is required as a substrate for luciferase in the light emission reaction. Therefore, enzyme activities can be visualized only by using disrupted cells. In analogy with the sugar hydrolase-based systems, screens involving light emission are not based on conditional growth, disallowing the efficient use of this group of promoter-probes for genome-wide screens for conditionally active promoters.
In previous work we established the use of alr as a food-grade selection marker (5). Here we describe the utilization of the same gene as a promoter-probe to study global gene expression and its control in L. plantarum. It is demonstrated that the allosteric Alr inhibitor d-cycloserine can be used during the screening procedures to add a quantitative factor to the primarily qualitative nature of the screen. Moreover, the experiments demonstrate that alr allows rapid and highly selective identification of conditionally active, high-salt-inducible promoters.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids that were used in this study are listed in Table 1. E. coli strains MC1061 (7) and BL21 (Novagen, Madison, Wis.) were used as cloning hosts during construction of pNZ7119 and the overproduction of alanine racemase from pNZ7122, respectively. E. coli was grown aerobically in TY medium (32). L. lactis MG1363 (20) was used as a cloning host during construction of pNZ7120, pNZ7121, and the L. plantarum promoter library. L. lactis MG1363 was grown without aeration at 30°C in M17 (Merck, Darmstadt, Germany), supplemented with 0.5% (wt/vol) glucose (GM17). L. plantarum WCFS1 (26) and its alr mutant derivative MD007 (5) were grown at 37°C in MRS (Difco, Surrey, United Kingdom) without aeration. Various concentrations of d-cycloserine (from freshly prepared, filter-sterilized stock solutions) or 200 μg of d-alanine/ml was added to the media when indicated. When appropriate, antibiotics were added to the media: for E. coli ampicillin (50 μg/ml) was added; for L. lactis and L. plantarum erythromycin (5 μg/ml) was added.
TABLE 1.
Strains, plasmids, and primers used in this study and their relevant characteristics and references
| Material | Relevant feature(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| MC1061 | Cloning host | 7 |
| BL21 | Cloning host for the utilization of T7 promoter-regulated expression | Novagen |
| L. lactis | ||
| MG1363 | Cloning host | 20 |
| L. plantarium | ||
| WCFS1 | Wild type for which the genome sequence is available | 26 |
| MD007 | L. plantarum WCFS1 Δalr, d-alanine auxotroph | 5 |
| Plasmids | ||
| pNZ7110 | Apr,b pUC18 derivative containing cre and TpepN | 5 |
| pNZ7119 | Apr, pNZ7110 derivative with cre replaced by L. lactis alr | This work |
| pIL252 | Emr | 34 |
| pNZ7120 | Emr, pIL252 derivative containing L. lactis alr from pNZ7119 | This work |
| pNZ7121 | Emr, pNZ7120 derivative containing L. plantarum PIdhL promoter upstream of alr | This work |
| pET-16B | Apr, vector for His-tagged overexpression of heterologous proteins in E. coli | Novagen |
| pNZ7122 | Apr, pET16-B derivative containing His-tagged fragment of alr | This work |
| Primers | ||
| PB1 | 5′-ACCGCTACGGATCACATC-3′ | |
| PB2 | 5′-CTCGGGGCAGTAAGACTA-3′ | |
| PB3 | 5′-GTGGTGAAGTTTTCATGG-3′ | |
| LLALR-TF-Fa | 5′-AGAGAAAGGTTTAAATCATGAAAAC-3′ | |
| LLALR-TF-Ra | 5′-ACGTTCTAGATTATTTATAGATTCTTTTGATTC-3′ | |
| P1dhL-Fa | 5′-GAAGATCTTCAATCTTCTCACCGTCTTG-3′ | |
| P1dhL-Ra | 5′-GAAGATCTTCAATAAGTCATCCTCTCGT-3′ | |
| SH4a | 5′-CCGCTCGAGGAAATTTGGCCAGCAGTGAAAGC-3′ | |
| SH5a | 5′-CGCGGATCCCTTATGGGATTTATCTTCC-3′ |
Underlined sequences indicate restriction sites subsequently used in cloning procedures.
Apr, encodes ampicillin resistance; Emr, encodes erythromycin resistance.
DNA techniques and DNA sequence analysis.
Plasmid DNA was isolated from E. coli on a small scale by using the alkaline lysis method (3, 32). Large-scale plasmid DNA isolations were performed by using Jetstar columns according to the manufacturer's instructions (Genomed GmbH, Bad Oberhausen, Germany). DNA isolation and transformation in L. lactis and L. plantarum were performed as described previously (14, 17, 25). Standard procedures were applied for DNA manipulations with E. coli (32). Restriction endonucleases, Taq and Pwo polymerase, T4 DNA ligase, and calf intestine alkaline phosphatase were used by following the recommendations of the manufacturer (Promega, Leiden, The Netherlands, and Boehringer, Mannheim, Germany). Primers were purchased from Pharmacia Biotech (Roosendaal, The Netherlands). The sequences of the inserts present in the pNZ7120 derivatives (see below) were amplified by PCR using primers PB1 and PB2 (Table 1), followed by mini-Elute PCR purification (Qiagen, Westburg, Germany). Partial insert sequences were determined by using approximately 25 ng of the PCR product and 1 pmol of fluorescently labeled primer PB3 (Table 1) in the Thermo Sequenase cycle sequence protocol provided by the manufacturer (Pharmacia Biotech). Sequence reaction products were analyzed by using an ALFred DNA sequencer (Pharmacia Biotech). The determined insert sequences were assigned to chromosomal loci by using Blast-N and the L. plantarum genome sequence (26) as the database.
Construction of promoter-probe vector pNZ7120.
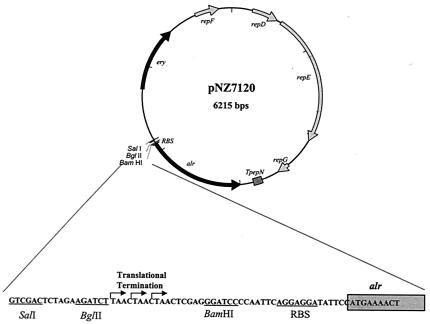
Plasmid pGIP012 (5) was used as a template to amplify the alr gene from L. lactis MG1363 (20) by using Pwo polymerase and the primers LLALR-TF-F and LLALR-TF-R (Table 1). The resulting 1.1-kb amplicon was digested with BspHI and XbaI (both restriction sites introduced with the primers) and was cloned into NcoI-NheI-digested pNZ7110 (5). The resulting plasmid was designated pNZ7119 and was checked by automatic double-strand sequence analysis by using an ALFred DNA sequencer. Sequence reactions were performed with an Autoread kit, were initiated by using Cy5-labeled universal and reverse primers, and were continued with synthetic primers in combination with Cy13-dATP, following the instructions of the manufacturer (Pharmacia Biotech). A single point mutation was found in the coding sequence of alr, which was probably introduced during the Pwo polymerase reaction. However, this mutation did not result in a change in the alanine racemase protein sequence. pNZ7119 was digested with Ecl136II and SalI, and the resulting 1.5-kb fragment, containing the alr coding sequence preceded by the pNZ7110-derived ribosome binding site (5), was ligated into SalI-SmaI-digested pIL252 (34), resulting in pNZ7120 (Fig. 1).
FIG. 1.
Plasmid map of the promoter-probe vector pNZ7120. The vector is a derivative of the low-copy-number vector pIL252 (34) and carries an erythromycin resistance gene (ery) for selection in L. plantarum. pNZ7120 harbors a promoterless copy of the L. lactis MG1363 alr gene, which is transcribed in the orientation opposite to that of all other genes encoded on pNZ7120. The DNA sequence visualizes the multiple-cloning site and the lactococcal prtP ribosome binding site (18). Furthermore, stop codons in all three reading frames (three black arrows) upstream of alr prevent translational fusions when chromosomal fragments are cloned in the BglII or SalI site of pNZ7120. The gray box indicates the 5′ end of the alr gene.
Vector validation and genomic library construction.
The promoter region of the ldhL1 gene of L. plantarum WCFS1 was amplified by using chromosomal DNA of this strain (26) as template and the primers PldhL-F and PldhL-R (Table 1). The 0.5-kb amplicon was digested with BglII and was cloned into similarly digested pNZ7120. The resulting plasmid was designated pNZ7121 and contains the alr gene under control of the ldhL1 promoter.
To construct an L. plantarum WCFS1 library in pNZ7120, chromosomal DNA of this strain was partially digested with Sau3AI. The partial digests were size fractionated on a 1% agarose gel, and fragments ranging from 1 to 2 kb were purified by using Sephaglas Bandprep (Pharmacia Biotech). These purified fragments were cloned into BglII-digested and calf intestine alkaline phosphatase-dephosphorylated pNZ7120. Ligation mixtures were transformed to L. lactis MG1363 (20), and approximately 50,000 of the obtained colonies were collectively resuspended in GM17 medium. Plasmid DNA was isolated from these pooled cells and was introduced into L. plantarum Δalr (MD007) cells. Transformants were selected on MRS medium containing d-alanine and erythromycin to obtain all plasmid-containing cells, independent of alr expression. Approximately 40,000 colonies were collectively pooled in MRS containing 15% glycerol and were stored at −80°C in small aliquots.
Alr antibodies and Western blotting.
Plasmid pNZ7119 was used as a template to amplify a 1,002-bp fragment of alr, using SH4 and SH5 as primers (Table 1) in the PCR. The resulting amplicon was digested with XhoI and BamHI and was cloned into similarly digested pET16-B (Novagen). The resulting plasmid pNZ7122 was introduced in E. coli BL21 (Novagen), and these cells were used for isopropyl-β-d-thiogalactopyranoside-induced overproduction of a His-tagged 334-amino-acid C-terminal fragment of the lactococcal alanine racemase protein according to the induction protocol of the provider of the vector. Subsequently, cells were pelleted, stored overnight at −20°C, and resuspended in 20 ml of French press buffer (25% sucrose, 50 mM Tris-HCl, pH 8, and 1 mM phenylmethylsulfonyl fluoride). By using a French press (10,000 lb/in2, 4°C, two successive treatments) (Sim Aminco, Abcoude, The Netherlands), cell extracts were prepared and the suspension was centrifuged at 3,500 × g for 30 min at 4°C. The pellet was resuspended in 10 ml of buffer (50 mM NaPi, pH 7.6, 1 mM phenylmethylsulfonyl fluoride, 0.5% Triton X-100, 4 M urea, and 0.5% Tween 20) and was recentrifuged. The supernatant was collected and was used for the purification of the Alr protein. A Hitrap Chelating column was preloaded with nickel as described in the protocol of the manufacturer (Amersham Biosciences). During purification on the Aktaexplorer (Amersham Pharmacia Biotech, Uppsala, Sweden), buffers containing 20 mM NaPi (pH = 7.4), 0.5 M NaCl, 4 M urea, and various concentrations of imidazole were used. Five column volumes containing 10 mM imidazole were used for equilibration. Subsequently, 25 ml of cell extract containing 3.5 mg of total protein/ml was loaded on the column, followed by washing with 30 column volumes containing 10 mM immidazole and with 10 column volumes containing 59 mM imidazole. Finally, elution was performed with 18 column volumes containing 500 mM imidazole. The elution fractions were analyzed on a sodium dodecyl sulfate-15% polyacrylamide gel, and Alr purity was estimated to be >95%. The elution fractions with the highest purity and protein concentration (total yield of 3.6 mg) were pooled and were used to raise polyclonal antibodies against Alr in a rabbit (Eurogentec, Seraing, Belgium).
The selectivity and sensitivity of these antibodies were demonstrated by the specific detection of Alr in wild-type L. lactis MG1363 (20) in a Western blot experiment. Cells were cultured at 30°C, and harvested cells were disrupted by using 1 g of zirconium glass beads in a Fastprep (Qbiogene Inc., Illkirch Cedex, France) by two treatments of 40 s interspaced by 1 min of cooling of the samples on ice. Proteins in the crude cell extract were separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis. Proteins were transferred onto nitrocellulose membranes (Jepson Bolton, Watford, United Kingdom) by electroblotting (LKB 2051 Midget Multiblot), and L. lactis Alr was probed by using 1,000-fold-diluted Alr antibodies, followed by detection with secondary goat anti-rabbit antibodies conjugated to peroxidase (Pierce, Rockford, Ill.). Reactivity was visualized by using the Supersignal West Pico chemiluminescent substrate according to the manufacturer's protocol (Perbio Science, Etten Leur, The Netherlands). To determine Alr production levels in clones of interest from the alr complementation library, these clones were grown in MRS with d-alanine and erythromycin, and subsequently 10−5 dilutions were plated. Full-grown colonies were harvested in 3 ml of water, and equal amounts of cells were disrupted and further analyzed as described above.
RESULTS
Promoter-probe vector pNZ7120.
The main goal of the work described here was to identify and analyze conditionally active promoters in L. plantarum by using a promoter-probe. The gusA gene, encoding β-glucuronidase, has been used to study promoters originating from the genome of L. lactis (30). Since no genes encoding β-glucuronidase were found in the L. plantarum WCFS1 genome (26), the potential of the E. coli gusA gene as a promoter-probe in L. plantarum was initially evaluated. However, L. plantarum harboring a plasmid carrying gusA under control of a constitutive promoter showed severe fluctuation of blue coloring on 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid-containing plates, preventing the effective application of this gene as a promoter-probe vector in L. plantarum (data not shown). Since we already exploited the alr gene as a food-grade selection marker (5), we investigated the applicability of this gene as an alternative method for the selection of conditionally active promoters. Hence, based on the low-copy-number plasmid pIL252 (34), a vector was constructed carrying a promoterless copy of the alr gene of L. lactis MG1363 (Fig. 1). Both the empty vector (pNZ7120) and a derivative containing the alr gene under control of the ldhL1 promoter from L. plantarum WCFS1 (pNZ7121) were introduced in L. plantarum Δalr (MD007) cells and were plated on MRS medium with or without d-alanine. On MRS plates with d-alanine, similar numbers of colonies were obtained for MD007 harboring either pNZ7120 (no promoter) or pNZ7121 (ldhL1 promoter), while on plates lacking d-alanine colonies were obtained only with cells harboring pNZ7121. These results indicate that pNZ7120 is a suitable promoter-probe vector for the detection of promoter activities in L. plantarum.
Construction and functionality of an alr complementation library in pNZ7120.
To use pNZ7120 as a promoter-probe for the detection of promoters, a library of partially digested Sau3AI fragments derived from L. plantarum WCFS1 genomic DNA was constructed via the intermediate host L. lactis MG1363. Approximately 40,000 colonies were obtained in L. plantarum MD007, and the library was analyzed for completeness in several ways. First, 100 colonies were randomly picked from MRS plates and their plasmids were used as a template for PCR analysis. This demonstrated that all investigated clones contain an insert, of which the average size was estimated to be 1.3 kb (data not shown). To assess insert redundancy, restriction patterns of all amplicons obtained with Sau3AI or HhaI were compared. This indicated that redundancy in the library was below 10%. Moreover, 30 of these amplicons were used for partial sequence analysis, which revealed them to be randomly derived from different chromosomal regions of the L. plantarum genome. These results support the randomness of the library, and genome coverage was estimated to be approximately 98% (data not shown).
To verify the functionality of the alr-based selection system, the library was plated with and without d-alanine, revealing that approximately 25% of the total library was able to grow on MRS plates lacking d-alanine (Table 2). These clones likely harbored a plasmid that contains a fragment of L. plantarum chromosomal DNA with a promoter element properly oriented for the expression of alr to a level sufficient for complementation of the d-alanine auxotroph phenotype of MD007. This expectation was investigated by insert sequence analysis of 16 randomly picked clones. All these inserts contained a 3′-truncated open reading frame (ORF) according to the annotation of the L. plantarum genome sequence (26), and their preceding upstream regions potentially carried a properly oriented promoter element responsible for alr expression (data not shown). In contrast, 16 randomly picked clones from a plate containing d-alanine did not display any preference of insert orientation relative to alr (data not shown). These experiments indicate that alr can be used as a promoter-probe for the selection of active promoters from the alr complementation library.
TABLE 2.
Decreasing numbers of CFU when clones are selected from the L. plantarum alr complementation library that are capable of growth on media containing increasing concentrations of d-cycloserine, without the addition of d-alaninea
| Concn (μg/ml) of:
|
% CFU | |
|---|---|---|
| d-Cycloserine | d-Alanine | |
| 0 | 200 | 100 |
| 0 | 0 | 24.6 |
| 10 | 0 | 14.6 |
| 20 | 0 | 13.0 |
| 50 | 0 | 10.9 |
| 100 | 0 | 9.56 |
| 200 | 0 | 6.97 |
| 400 | 0 | 3.96 |
| 600 | 0 | 2.46 |
| 900 | 0 | <0.01 |
Percentages are relative to the number of colonies on a control plate containing d-alanine but lacking d-cycloserine.
Quantitative selection of promoters with d-cycloserine.
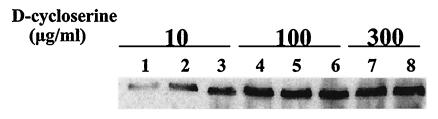
d-Cycloserine has been described as an allosteric Alr inhibitor (5, 6, 28). Previous experiments have demonstrated that higher levels of Alr result in higher levels of d-cycloserine resistance (5). This observation suggests that d-cycloserine can be used for the selection of clones from the alr complementation library that produce Alr at a relatively high level. To evaluate this possibility, the library was plated on MRS containing a range of d-cycloserine concentrations. After 3 days of growth, plates with higher concentrations of d-cycloserine contained fewer colonies (Table 2), suggesting that these colonies represent clones harboring pNZ7120 derivatives that produced Alr at a level sufficient to overcome the inhibitory effect of this compound. To further investigate this observation, dilutions of the alr complementation library were plated and grown on MRS lacking d-cycloserine. Subsequently, 384 colonies were randomly selected and were cultured overnight in a microtiter plate containing MRS lacking d-cycloserine. These cultures were used for replica plating to MRS plates containing 0, 10, 100, or 300 μg of d-cycloserine/ml. Growth on the plates containing d-cycloserine was compared to that on the control plate lacking d-cycloserine, leading to the identification of clones capable of growth on plates containing up to 10, 100, or 300 μg of d-cycloserine/ml. From these three groups, 28 clones were randomly selected, and insert sequence analysis revealed the corresponding chromosomal loci, the corresponding L. plantarum genes, and their potential promoters (Table 3). The Alr production levels in individual clones were investigated in eight randomly picked clones from the three groups. These clones were plated on MRS lacking d-cycloserine, and after 48 h full-grown colonies were harvested and equal numbers of cells were used to prepare crude cell extracts. To investigate the Alr production in each d-cycloserine resistance group, the cell extracts were analyzed by Western blotting with Alr-specific antibodies (Fig. 2). All clones that displayed MD007 complementation at 100 and 300 μg of d-cycloserine/ml showed a clearly higher Alr production level than did the clones that manifested growth only at 10 μg of d-cycloserine/ml. However, when this approach was used, no clear differences in Alr production levels could be visualized in comparing the clones that displayed MD007 complementation at 100 and 300 μg of d-cycloserine/ml. Nevertheless, these experiments demonstrate that L. plantarum genomic fragments displaying high promoter activity can be selected from the alr complementation library by using high concentrations of d-cycloserine, adding a quantitative factor to the primary qualitative nature of the alr complementation screen.
TABLE 3.
Clones in the alr complementation library that are capable of growth in the presence of a maximum concentration of 10, 100, or 300 μg of d-cyloserine/ml, without the addition of d-alanine
| [d-Cycloserine] (μg/ml) | Clone name | Insert start coordinateb | Estimated insert size (kb) | ORF in insertc | Function of homologue(s) |
|---|---|---|---|---|---|
| 10 | pNZ7120-csl1 | 1037318 | 1.7 | lp_1141 (purK2) | Phosphoribosylaminoimidazole carboxylase, ATPase subunit |
| 10 | pNZ7120-csl2 | 1216019 | 2.0 | lp_1320 | Conserved hypothetical protein |
| 10 | pNZ7120-csl3 | 1000151 | 2.5 | lp_1101 (ldhl2) | Lactate dehydrogenase |
| 10 | pNZ7120-csl4 | 2617565 | 2.5 | lp_2937 | Transcription regulator |
| 10 | pNZ7120-csl5a | 1309715 | 3.0 | lp_1430 | Glycosyltransferase |
| 10 | pNZ7120-csl6a | 1904621 | 2.6 | lp_2109 (urvC) | Excinuclease ABC, subunit C |
| 10 | pNZ7120-csl7 | 416640 | 2.5 | lp_0464 | Zinc/iron ABC transporter |
| 10 | pNZ7120-csl8a | 3008044 | 3.0 | lp_3392 | Transport protein |
| 100 | pNZ7120-csm1 | 470883 | 1.7 | lp_0525 (kupI) | Potassium uptake protein |
| 100 | pNZ7120-csm2 | 3035271 | 1.7 | lp_3418 (pck) | Phosphoenolpyruvate carboxykinase |
| 100 | pNZ7120-csm3 | 1038952 | 1.2 | lp_1144 (pcrA) | ATP-dependent DNA helicase |
| 100 | pNZ7120-csm4 | 3308262 | 1.6 | lp_3688a (rpmH) | Ribosomal protein L34 |
| 100 | pNZ7120-csm5 | 2948260 | 1.6 | lp_3312 | Conserved hypothetical protein |
| 100 | pNZ7120-csm6 | 2373288 | 1.8 | lp_2671 | Integral membrane protein |
| 100 | pNZ7120-csm7a | 2019577 | 2 | lp_2231a | Hypothetical protein |
| 100 | pNZ7120-csm8a | 2855464 | 1.5 | lp_3204 (nupC) | Nucleoside transport protein |
| 100 | pNZ7120-csm9a | 1454904 | 3 | lp_1599 (folD) | Methylenetetrahydrofolate dehydrogenase and cyclohydrolase |
| 100 | pNZ7120-csm10 | 2039898 | 2.5 | lp_2256 (ccpA) | Catabolite control protein A |
| 100 | pNZ7120-csm11 | 90985 | 2.8 | lp_0101 | Cobalt ABC transporter component |
| 300 | pNZ7120-csh1 | 2668545 | 1.7 | lp_3000 | ABC transporter, ATP-binding and permease protein |
| 300 | pNZ7120-csh2a | 3015121 | 2.5 | lp_3399 (cpd) | 2′,3′-Cyclic-nucleotide 2′-phosphodiesterase precursor |
| 300 | pNZ7120-csh3a | 2483803 | 2.5 | lp_2793 | Hypothetical protein |
| 300 | pNZ7120-csh4 | 1383808 | 2 | lp_1512 (dnaB) | Replication initiation and membrane attachment protein DnaB |
| 300 | pNZ7120-csh5 | 676236 | 2.2 | lp_0739 (secA) | Preprotein translocase, SecA subunit |
| 300 | pNZ7120-csh6 | 2990240 | 0.5 | lp_3363 (copB) | Copper-transporting ATPase |
| 300 | pNZ7120-csh7 | 493323 | 1.8 | lp_0542 | Conserved hypothetical protein |
| 300 | pNZ7120-csh8 | 2326766 | 2.4 | lp_2608 | Na+/H+ antiporter |
Clones used for Western blot analysis (Fig. 2).
Insert start coordinate indicates the first base pair of the L. plantarum genome sequence in the insert of the pNZ7120 derivatives directly upstream of alr.
Gene names are indicated between brackets if present in the present annotation of the L. plantarum WCFS1 genome (26).
FIG. 2.
Western blot analysis of Alr expression levels in colonies sensitive to different concentrations of d-cycloserine. Randomly selected individual clones that originally were selected on MRS plates containing 10 (lanes 1 through 3), 100 (lanes 4 through 6), or 300 (lanes 7 and 8) μg of d-cycloserine/ml.
Conditional promoter screen for 0.8 M NaCl-activated promoters.
The possibility to select conditionally activated clones from the alr complementation library was investigated by studying the salt response. For this purpose the library was plated on MRS containing 0.8 M NaCl. Subsequently, 768 colonies were randomly picked, organized in microtiter plates, and grown overnight in MRS containing d-alanine. All clones were replicated on MRS plates with or without 0.8 M NaCl and containing no or up to 200 μg of d-cycloserine/ml. Growth was periodically compared between the different plates over 3 days, and 46 clones were identified that showed conditional growth only in the presence of 0.8 M NaCl. The corresponding chromosomal inserts present in the pNZ7120 derivatives were partially sequenced, revealing 23 unique loci. Subsequently, 20 of the 46 clones were found to grow only on plates containing 0.8 M NaCl over a range of different d-cycloserine concentrations that varied at least by eightfold (Table 4), indicating that the expression in the presence of NaCl is relatively high compared to expression in the absence of NaCl. Sequence comparison of their inserts showed the presence of eight different chromosomal loci (Table 4). Strikingly, an insert in pNZ7120-con1 harboring a 3′-truncated fragment of Lp_1459, encoding a gene with no homologues in the database, and its upstream sequence was found in 10 of the 20 identified clones. Similarly, a 3′-truncated fragment of Lp_1435, encoding an integral membrane protein and its upstream sequence, was found in two of the identified inserts. The latter two inserts differed in size (approximately 900 and 2,200 bp), indicating that this promoter element was independently selected twice from the library. In pNZ7120-con4 and -con6 no ORF and consequently no putative promoter region could be identified in the present genome annotation database of L. plantarum WCFS1 (26). However, manual inspection of these loci revealed the presence of a short ORF (162 and 165 bp, respectively) lacking any database homologue that could be preceded by a high-osmolarity-induced promoter (Table 4).
TABLE 4.
Clones capable of complementing the d-alanine auxotrophy of L. plantarum Δalr MD007 only in the presence of 0.8 M NaCl
| Clone name | Insert start coordinate | Estimated insert size (kb) | Redun- dancy | Gene in fragment | Putative function | Target concn of d-cycloserine (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2.5 | 5 | 10 | 25 | 50 | 100 | 200 | ||||||
| pNZ7120-con1 | 1335151 | 0.9 | 10 | lp_1459 | Unknown | C | C | C | C | C | C | − | − |
| pNZ7120-con2 | 1315041/1314390 | 0.9/2.2 | 2 | lp_1435 | Integral membrane protein | + | C | C | C | C | − | − | − |
| pNZ7120-con3 | 2905725 | 1.2 | 1 | lp_3266 (glyK) | Glycerate kinase | + | + | C | C | C | C | − | − |
| pNZ7120-con4 | 2901280 | 2.0 | 2 | ORF162b | Unknown | + | + | C | C | C | C | C | C |
| pNZ7120-con5 | 2242522 | 1.1 | 1 | lp_2509 | Permease, DMT superfamily | + | + | + | C | C | C | C | C |
| pNZ7120-con6 | 427258 | 0.7 | 1 | ORF165b | Unknown | + | + | + | + | C | C | C | C |
| pNZ7120-con7 | 2707493 | 3.0 | 1 | lp_3045 | Short-chain dehydrogenase | + | + | + | + | C | C | C | C |
| pNZ7120-con8 | 2962996 | 3.5 | 2 | lp_3330 | Unknown | + | + | + | + | C | C | C | C |
C indicates at which d-cycloserine concentrations growth was observed only in the presence of 0.8 M NaCl. + indicates at which d-cycloserine concentrations growth was detected independent of the presence of NaCl. − indicates that no growth was observed under either set of conditions.
Although, in this pNZ7120 derivative, no ORF is present on these chromosomal fragments in the present annotation database of L. plantarum WCFS1, manual inspection revealed the small ORFs indicated in the table.
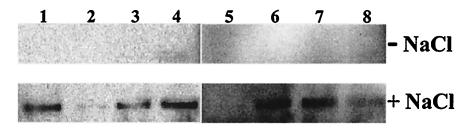
Conditional, 0.8 M NaCl-induced alr expression was further investigated in the eight clones selected for conditional growth over a range of d-cycloserine concentrations differing by at least eightfold. Overnight cultures in MRS containing d-alanine were diluted and plated on MRS lacking d-alanine and d-cycloserine, with or without 0.8 M NaCl. After 3 days colonies were harvested and cell extracts were analyzed by Western blotting with the Alr antibodies (Fig. 3). It could clearly be established that Alr expression driven by six of these eight chromosomal fragments is higher under high-salt conditions than under standard conditions. In this experiment, no Alr production could be detected under either set of conditions for the clones pNZ7120-con2 and -con5. When increased amounts of cell extract originating from these two clones were analyzed by Western blotting, 0.8 M NaCl-dependent Alr production could be visualized (data not shown). However, the observation that pNZ7120-con5 is capable of growth on plates containing 200 μg of d-cycloserine/ml (Table 4) appears to disagree with these relatively low production levels of Alr. A possible explanation could be that the promoter element harbored by pNZ7120-con5 is induced by the combination of d-cycloserine and NaCl, but not by NaCl or by d-cycloserine alone. Overall, these experiments indicate that the alr complementation library can be used for the effective selection of L. plantarum chromosomal fragments displaying conditional promoter activity.
FIG. 3.
Western blot analysis of 0.8 M NaCl-induced Alr expression in 8 clones. Alr production levels were clearly higher in all eight clones (lanes 1 to 8) when cells were grown in media to which 0.8 M NaCl was added (lower panel) than when cells were grown in standard MRS media (upper panel). It should be noted that, when larger amounts of cell extract were used, a more obvious signal for clones pNZ7120-con2 and -con5 was detected (data not shown).
DISCUSSION
Previous studies suggested that the alanine racemase-encoding gene could be used as a complementation-based promoter-probe in an alr mutant of L. plantarum (5). This application was investigated by using the lactococcal alr gene, and analysis of the 3.3-Mbp genome of L. plantarum with an alr complementation library resulted in the identification of eight chromosomal loci that show conditional promoter activity only under high-salt conditions. Intrinsic to complementation-based screening procedures, growth was used as the primary selection criterion. Therefore, the strategy described here has the same primary characteristics as the cat screening system, which is also based on growth in the presence of different amounts of chloramphenicol (1, 37). These types of growth-based screens are easy to perform and hence have advantages over screens based on sugar hydrolases (18, 24, 30, 31, 33, 35) or light-emitting systems (9, 10, 16, 21), which are laborious when applied in genome-wide promoter screens. However, only promoters with a relatively high expression level have been detected in the cat system, because selection at low chloramphenicol concentrations is difficult to distinguish from endogenous resistance. In contrast, alanine racemase is required only at low levels to achieve complementation and can therefore be used for the identification of promoters with low activities. No revertants of the d-alanine auxotroph phenotype have been encountered in L. plantarum (data not shown). Moreover, by application of the allosteric inhibitor d-cycloserine in the screening procedure, the alr system can be used to select promoters with higher activity levels. The addition of increasing concentrations of d-cycloserine allowed the direct selection of L. plantarum chromosomal fragments that drove alr expression to relatively high levels. Thereby, d-cycloserine adds a quantitative factor to this screening system. These features make the alr-based screening system suitable for identification of promoters in a relatively long dynamic range and convenient for sensitive, genome-wide promoter-screening procedures.
The screen for high-salt-inducible promoter elements presented here demonstrates that the alr complementation library can be used for the identification of conditionally active promoters by selection of clones that grow independent of d-alanine only under a chosen set of conditions. Replication of such clones to the control set of conditions (normal laboratory conditions) allows direct selection of conditionally growing clones, indicating that alr expression is activated by the condition applied. Moreover, the clones identified in such a conditional screen can be analyzed in more detail by monitoring conditional capability of growth on plates with various concentrations of d-cycloserine, without the addition of d-alanine. The conditional screen performed here resulted in the identification of eight chromosomal L. plantarum loci that clearly possess high-salt-inducible promoter activity. These promoters are of industrial relevance, since L. plantarum is frequently encountered in artisanal and industrial fermentations of vegetables, including onions, cucumbers, olives and cabbage (8, 15), which in many cases involve high-salt conditions. Therefore, the promoter fragments identified here can be employed to construct in situ expression systems for desirable proteins, including proteolytic and flavor-forming enzymes, by simply exchanging alr for the genes encoding such functions. Moreover, screens can be designed to identify conditional promoter elements for other niches in which L. plantarum is encountered. L. plantarum WCFS1 is known to actively pass the stomach (38), indicating that screens can be designed to construct L. plantarum strains for the delivery of desirable and potentially health-promoting proteins to the gastrointestinal tract.
Of the eight most prominent salt-induced chromosomal loci (Table 4), two did not appear to contain an ORF and thus no putative promoter region in the present genome annotation database of L. plantarum WCFS1 (26). Nevertheless, manual inspection revealed small ORFs lacking any database homologue in both cases, possibly explaining the observed conditional promoter activity. Moreover, sequence comparison of all upstream regions of the 23 ORFs initially identified in the conditional screen with the Web-based sequence alignment module MEME (2) revealed four loci that contain the same conserved motif (Table 5). A subsequent analysis with this 28-bp motif and the search program MAST (2) revealed that these are the only four loci in the L. plantarum genome where this motif is present with this high level of significance, and in an identical search where MAST and the genomes of the related species L. lactis and B. subtilis (4, 27) were used, the same motif could not be found. These findings suggest that the alr system can be helpful in the identification of promoter activities that were not predicted in the primary annotation of a genome sequence, thereby allowing improvement of the annotation of such genomes with experimental data. Moreover, potential regulatory DNA sequences can be identified by in silico analysis of the identified conditional chromosomal loci.
TABLE 5.
The motif that was found in the promoter regions of four ORFs that were harbored by the pNZ7120 derivatives originating from clones that display 0.8 M NaCl-dependent growth
| Gene | Putative function | P | Motif sequence upstream of genea | Distance to ATG (bp) |
|---|---|---|---|---|
| lp_0319 | Transcription regulator | 1.3 e-12 | ATTAG G T G C G T T G T G G A C G G A C T T T T C G T G G A G TGCCG | 205 |
| lp_1435 | Integral membrane protein | 1.5 e-13 | CCCGC G T G A G C G G T G G A C C C A G T G C A A G T C T G G GACCG | 262 |
| lp_2509 | Permease, DMT superfamily | 1.3 e-12 | CAAAG G T G C G T C T G G G A A C C T A T G T T T A T G A G G AATCT | 48 |
| lp_3363 (copB) | Copper-transporting ATPase | 6.0 e-16 | CGATT G T G T G C C G G G G A C G C G G T G T T A A T G T G G TTATA | 314 |
| G T G C G C C G G G G A C C C A G T G T T A A T G T G G | ||||
| AT G T TA G G G AT C A C GC A A | ||||
| TTT CTG |
Last three rows represent the multilevel consensus sequence.
Overall, this paper describes a promoter-probe vector that allows for convenient genome-wide analysis of L. plantarum promoter elements in a long dynamic range of promoter activity levels. Moreover, the alr system was used to identify high-salt-activated promoter elements in L. plantarum. d-Alanine auxotroph mutants have been described for other bacteria (23, 36), allowing simple implementation of the alr promoter-screening system in these microbes.
Acknowledgments
We are grateful to Douwe Molenaar for bioinformatics analysis and Jolanda Lambert, Armand Hermans, and Saskia van Mil for technical assistance. We thank Masja Nierop Groot and Maria Marco for critically reading the manuscript and Pascal Hols for fruitful discussions.
Part of this work was supported by the EU project LABDEL (EU-QLRT-2000-00340).
REFERENCES
- 1.Achen, M. G., B. E. Davidson, and A. J. Hillier. 1986. Construction of plasmid vectors for the detection of streptococcal promoters. Gene 45:45-49. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, T. L., M. E. Baker, and C. P. Elkan. 1997. An artificial intelligence approach to motif discovery in protein sequences: application to steriod dehydrogenases. J. Steroid Biochem. Mol. Biol. 62:29-44. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL-1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bron, P. A., M. G. Benchimol, J. Lambert, E. Palumbo, M. Deghorain, J. Delcour, W. M. De Vos, M. Kleerebezem, and P. Hols. 2002. Use of the alr gene as a food-grade selection marker in lactic acid bacteria. Appl. Environ. Microbiol. 68:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caceres, N. E., N. B. Harris, J. F. Wellehan, Z. Feng, V. Kapur, and R. G. Barletta. 1997. Overexpression of the d-alanine racemase gene confers resistance to d-cycloserine in Mycobacterium smegmatis. J. Bacteriol. 179:5046-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 8.Cocconcelli, P. S., E. Triban, M. Basso, and V. Bottazzi. 1991. Use of DNA probes in the study of silage colonization by Lactobacillus and Pediococcus strains. J. Appl. Bacteriol. 71:296-301. [DOI] [PubMed] [Google Scholar]
- 9.Corthier, G., C. Delorme, S. D. Ehrlich, and P. Renault. 1998. Use of luciferase genes as biosensors to study bacterial physiology in the digestive tract. Appl. Environ. Microbiol. 64:2721-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315-319. [DOI] [PubMed] [Google Scholar]
- 11.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 12.Derzelle, S., B. Hallet, K. P. Francis, T. Ferain, J. Delcour, and P. Hols. 2000. Changes in cspL, cspP, and cspC mRNA abundance as a function of cold shock and growth phase in Lactobacillus plantarum. J. Bacteriol. 182:5105-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vos, W. M., I. Boerrigter, R. J. van Rooyen, B. Reiche, and W. Hengstenberg. 1990. Characterization of the lactose-specific enzymes of the phosphotransferase system in Lactococcus lactis. J. Biol. Chem. 265:22554-22560. [PubMed] [Google Scholar]
- 14.de Vos, W. M., P. Vos, H. de Haard, and I. Boerrigter. 1989. Cloning and expression of the Lactococcus lactis ssp. cremoris SK11 gene encoding an extracellular serine protease. Gene 85:169-176. [DOI] [PubMed] [Google Scholar]
- 15.Duran Quintana, M. C., P. Garcia Garcia, and A. Garrido Fernandez. 1999. Establishment of conditions for green table olive fermentation at low temperature. Int. J. Food Microbiol. 51:133-143. [DOI] [PubMed] [Google Scholar]
- 16.Eaton, T. J., C. A. Shearman, and M. J. Gasson. 1993. The use of bacterial luciferase genes as reporter genes in Lactococcus: regulation of the Lactococcus lactis subsp. lactis lactose genes. J. Gen. Microbiol. 139:1495-1501. [DOI] [PubMed] [Google Scholar]
- 17.Ferain, T., D. Garmyn, N. Bernard, P. Hols, and J. Delcour. 1994. Lactobacillus plantarum ldhL gene: overexpression and deletion. J. Bacteriol. 176:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez, M., A. Margolles, J. E. Suarez, and B. Mayo. 1999. Duplication of the beta-galactosidase gene in some Lactobacillus plantarum strains. Int. J. Food Microbiol. 48:113-123. [DOI] [PubMed] [Google Scholar]
- 20.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geoffroy, M. C., C. Guyard, B. Quatannens, S. Pavan, M. Lange, and A. Mercenier. 2000. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl. Environ. Microbiol. 66:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giard, J. C., N. Verneuil, Y. Auffray, and A. Hartke. 2002. Characterization of genes homologous to the general stress-inducible gene gls24 in Enterococcus faecalis and Lactococcus lactis. FEMS Microbiol. Lett. 206:235-239. [DOI] [PubMed] [Google Scholar]
- 23.Hols, P., M. Kleerebezem, A. N. Schanck, T. Ferain, J. Hugenholtz, J. Delcour, and W. M. de Vos. 1999. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 17:588-592. [DOI] [PubMed] [Google Scholar]
- 24.Israelsen, H., S. M. Madsen, E. Johansen, A. Vrang, and E. B. Hansen. 1995. Environmentally regulated promoters in Lactococci. Dev. Biol. Stand. 85:443-448. [PubMed] [Google Scholar]
- 25.Josson, K., T. Scheirlinck, F. Michiels, C. Platteeuw, P. Stanssens, H. Joos, P. Dhaese, M. Zabeau, and J. Mahillon. 1989. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid 21:9-20. [DOI] [PubMed] [Google Scholar]
- 26.Kleerebezem, M., J. Boekhorst, R. Van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. De Vries, B. Ursing, W. M. De Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 28.Lambert, M. P., and F. C. Neuhaus. 1972. Mechanism of d-cycloserine action: alanine racemase from Escherichia coli W. J. Bacteriol. 110:978-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, L. J., J. B. Hansen, E. K. Jagusztyn-Krynicka, and B. M. Chassy. 1982. Cloning and expression of the β-d-phosphogalactoside galactohydrolase gene of Lactobacillus casei in Escherichia coli K-12. J. Bacteriol. 152:1138-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russell, W. M., and T. R. Klaenhammer. 2001. Identification and cloning of gusA, encoding a new β-glucuronidase from Lactobacillus gasseri ADH. Appl. Environ. Microbiol. 67:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sanders, J. W., G. Venema, J. Kok, and K. Leenhouts. 1998. Identification of a sodium chloride-regulated promoter in Lactococcus lactis by single-copy chromosomal fusion with a reporter gene. Mol. Gen. Genet. 257:681-685. [DOI] [PubMed] [Google Scholar]
- 34.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 35.Simons, G., H. Buys, R. Roger, E. Koenhen, and W. M. De Vos. 1990. Construction of a promoter-probe vector for lactic acid bacteria using the lacG gene of Lactococcus lactis. J. Ind. Microbiol. 31:31-39. [Google Scholar]
- 36.Tauch, A., S. Gotker, A. Puhler, J. Kalinowski, and G. Thierbach. 2002. The alanine racemase gene alr is an alternative to antibiotic resistance genes in cloning systems for industrial Corynebacterium glutamicum strains. J. Biotechnol. 99:79-91. [DOI] [PubMed] [Google Scholar]
- 37.van der Vossen, J. M., J. Kok, and G. Venema. 1985. Construction of cloning, promoter-screening, and terminator-screening shuttle vectors for Bacillus subtilis and Streptococcus lactis. Appl. Environ. Microbiol. 50:540-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vesa, T., P. Pochart, and P. Marteau. 2000. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 14:823-828. [DOI] [PubMed] [Google Scholar]