Abstract
Denaturing gradient gel electrophoresis (DGGE) of amplified fragments of genes coding for 16S rRNA was used to study the development of bacterial communities during decomposition of crop residues in agricultural soils. Ten strains were tested, and eight of these strains produced a single band. Furthermore, a mixture of strains yielded distinguishable bands. Thus, DGGE DNA band patterns were used to estimate bacterial diversity. A field experiment performed with litter in nylon bags was used to evaluate the bacterial diversity during the decomposition of readily degradable rye and more refractory wheat material in comparable luvisols and cambisols in northern, central, and southern Germany. The amount of bacterial DNA in the fresh litter was small. The DNA content increased rapidly after the litter was added to the soil, particularly in the rapidly decomposing rye material. Concurrently, diversity indices, such as the Shannon-Weaver index, evenness, and equitability, which were calculated from the number and relative abundance (intensity) of the bacterial DNA bands amplified from genes coding for 16S rRNA, increased during the course of decomposition. This general trend was not significant for evenness and equitability at any time. The indices were higher for the more degradation-resistant wheat straw than for the more easily decomposed rye grass. Thus, the DNA band patterns indicated that there was increasing bacterial diversity as decomposition proceeded and substrate quality decreased. The bacterial diversity differed for the sites in northern, central, and southern Germany, where the same litter material was buried in the soil. This shows that in addition to litter type climate, vegetation, and indigenous microbes in the surrounding soil affected the development of the bacterial communities in the litter.
Plant residues are a crucial source of nutrients in both natural and agricultural ecosystems, where synchronous plant growth and residue decomposition are essential for soil fertility. Fresh plant material (e.g., litter) represents a readily available substrate for both soil fauna and soil microorganisms. The main mineralization activity is performed by soil microbial communities, and the specific quality of the organic residues controls the decomposition rate and the related release of nutrients (21).
Litter quality generally decreases during the course of decomposition due to the loss of readily available C and the accumulation of refractory compounds (13). Simultaneously, the soil microbial biomass decreases, and the C use efficiency increases (6). The change in the quality of the organic matter induces a succession of microbial communities, which has been studied by using cultivation techniques in litter bag studies (7, 27) and in vertical soil horizons (34) in forest ecosystems. Based on their functions and ecological strategies, different dominant genera and species of microorganisms are present in biotopes (34). During the decomposition process, the r strategists dominate during the early stages and are replaced later by k strategists due to growth-limiting substrate concentrations (27, 29). Theoretically, diversity should increase during succession (1). The combination of adequate biotic diversity and heterogeneity is considered to be necessary for long-term ecological functioning and resilience of ecosystems (3). Higher biodiversity within a community is thought to reduce the spatial and temporal variations in functional activities of the communities, to mitigate the risk of loss of functions after extreme environmental conditions, and thus to preserve the average rates of related ecological processes (9).
It has been reported that only a small fraction of microorganisms in nature are cultivatable. Therefore, to study the total microbial community involved in organic matter decomposition, methods which include both culturable and nonculturable microorganisms are needed.
Molecular biological techniques offer new opportunities for analysis of the structure and species composition of a microbial community (20). In particular, sequence variation in rRNA genes has been exploited for inferring phylogenetic relationships among microorganisms (33) and may be used to estimate the genetic diversity of complex microbial communities in natural ecosystems (15, 17, 19, 22). Denaturing gradient gel electrophoresis (DGGE) allows one to directly determine the presence and the relative levels of different 16S rRNA amplicons and, thus, to profile the corresponding microbial populations in both a qualitative way and a semiquantitative way (4, 11, 12, 28, 32). The diversity can be estimated from the number of 16S rRNA gene sequence similarity groups (i.e., the number of DNA bands on the DGGE gel) (15). Each band is assumed to represent an operational taxonomic unit, which is called a species for simplicity (14). We tested the relationship between the number and intensity of DNA bands and the relationship between the number and relative levels of different bacterial isolates (species) in a preliminary experiment by adding known amounts of different bacteria to sterilized soil. This should have shown that DGGE can be used for semiquantitative comparison (20).
The main aim of this investigation was to determine the diversity of bacterial communities during litter decomposition in soil based on analysis of directly extracted DNA. Two types of litter were buried in comparable soil types exposed to different climates and types of vegetation in northern, central, and southern Germany. Using Odum's system-theoretical hypotheses (23, 24), we tested whether microbial diversity increases during the course of litter decomposition (hypothesis 1), whether refractory straw supports microbial communities with diversity lower than the diversity of microbial communities supported by readily available rye grass (hypothesis 2), and whether microbial diversity is different in the same litter at different locations (hypothesis 3). Hypothesis 2 was formulated because accelerated decomposition of readily available litter is correlated with a higher diversity of soil decomposer communities (10).
MATERIALS AND METHODS
Addition of bacteria to soil.
To determine if each bacterial genotype (species) produced one band and if the intensity of the band was proportional to the number of cells, strains of the following 10 bacteria isolated from soil (18) were used: Alcaligenes sp., Arthrobacter sp., Bacillus sp., Corynebacterium sp., Enterobacter cloacae, Flavobacterium sp., Nocardia sp., Pseudomonas cepacia, Pseudomonas fluorescens, and Rhodococcus. The strains were added to a sandy soil that was autoclaved twice for 30 min at 120°C, with 24 h between the two sterilization procedures. Suspensions of the separate bacterial strains at an optical density at 600 nm of 1.5 were added to sterilized soil; the amount added was 0.5 ml per 9.5 g of sterilized soil. Before addition of the bacteria the dry matter content of the soil was 92.1% (wt/wt).
To determine the relationship between band intensity and relative abundance, we performed an additional experiment with three strains which were added to sterilized soil in three different ratios. The ratios were based on optical densities. Cell numbers were measured by image analysis afterwards. An optical density at 600 nm of 1 corresponded to densities of 33.1 × 108, 5.80 × 108, and 1.34 × 108 cells ml−1 for P. fluorescens, Arthrobacter sp., and Bacillus sp., respectively. Thus, relative amounts were calculated, and DNA band intensities were measured. The mixture of species was added in 0.5 ml, but the optical density at 600 nm was 7.2. The numbers of cells in the suspensions were determined by automatic image analysis after filtration on black 0.2-μm-pore-size polycarbonate filters (5). The number of bacteria per milliliter was used to calculate the number of cells added per gram of soil. Thus, the following numbers of cells (relative abundance) were added: Alcaligenes sp., 2.6 × 107 cells g of soil−1 (22%); Bacillus sp., 0.25 × 107 cells g of soil−1 (2.1%); Arthrobacter sp., 1.1 × 107 cells g of soil−1 (9.2%); Corynebacterium sp., 4.9 × 107 cells g of soil−1 (40%); Nocardia sp., 0.01 × 107 cells g of soil−1 (0.08%); P. cepacia, 1.5 × 107 cells g of soil−1 (12%); P. fluorescens, 1.1× 107 cells g of soil−1 (8.8%); and Rhodococcus, 1.0 × 107 cells g of soil−1 (8.3%). The sum of the counts for the separate strains was 12 × 107 cells g−1. This corresponds well to the density of cells found in the mixture (14 × 107 cells g−1).
Litter experiments and sites.
Litter bag experiments were carried out in northern, central, and southern Germany at Hohenschulen, Frankenhausen, and Scheyern. The site and soil characteristics are shown in Table 1. Two litter types, representing rapidly and slowly decomposing litter, were derived from Lolium perenne and Triticum aestivum cultivated at the northern and central German sites, respectively. The Lolium material was used as cattle fodder, and the Triticum material was used as straw in stables. The C/N ratios of the two types of material were 17 and 112 (wt/wt), respectively. Ten-gram portions of field-dried litter (2- to 20-cm fraction) were put in nylon bags (20 by 20 cm) with a mesh size of 2 mm. During early spring, 24 bags of each litter type were inserted vertically into the soil to an average depth of 20 cm in slit cuts made with a shovel (2). Depending on the mass remaining, four to eight bags were harvested on days 18, 58, 118, and 180, at different stages of decomposition. In the analyses we focused mainly on the central German site, which showed the most pronounced differences in the decomposition rate (data not shown). The diversity in the Lolium litter at the central site was compared to the diversity in the Lolium litter at the northern and southern sites during the initial stage of decomposition (day 18). At the northern, central, and southern sites the mass losses were 16, 29, and 33%, respectively, on day 18 and 37, 59, and 44%, respectively, on day 58. The fresh litter remaining from each bag was weighed, and aliquots that were cut into 5-mm pieces were used to estimate the water content (105°C), the pH, the loss after ignition, and complementary litter properties (results not shown). Duplicates were taken from two separate litter bags. The DNA profiles for all litter decomposition stages were compared on one gel to avoid variation between gels. Aliquots of the litter were stored at −21°C before analysis.
TABLE 1.
Site and soil characteristics of the northern, central, and southern German experimental sites
| Site | Location | Temp (°C)a | Precipi- tation (mm)b | Organic C concn (mg of C g of soil−1) | pH (CaCl2) | Texture | Crop in 2002 | Soil unitsc |
|---|---|---|---|---|---|---|---|---|
| Hohenschulen | 54°19′N, 10°00′E | 8.5 | 750 | 17 | 7.0 | sL-tL | Triticum | Luvisols, anthrosols |
| Frankenhausen | 51°24′N, 9°26′E | 9.7 | 622 | 12 | 7.3 | tU | Secale | Luvisols, anthrosols, pelsosols |
| Scheyern | 48°29′N, 11°26′E | 7.4 | 803 | 15 | 6.2 | suL | Medicago and Lolium | Luvisols, cambisols |
Mean annual temperature.
Mean annual precipitation.
Classification according to ISSS-ISRIC-FAO (1998).
Litter extraction.
Genomic DNA was extracted directly from litter with a FastDNA Spin kit for soil (Bio 101, Carlsbad, Calif.) (www.bio101.com). Litter (0.05 to 0.25 g [fresh weight], corresponding to approximately 0.05 g [dry weight]) was extracted in lysing matrix tubes containing a mixture of ceramic and silica particles. Sodium phosphate buffer (978 μl) and microtubule buffer (122 μl) were added from the kit. Samples were homogenized and DNA was solubilized by bead beating with a FastPrep instrument (Bio 101) for 30 s at level 5.5. The supernatant was transferred, and the DNA was precipitated with a protein precipitation solution (250 μl). The DNA was purified by the Geneclean procedure (Bio 101).
PCR.
The variable V3 region of 16S rRNA gene sequences from nucleotide 341 to nucleotide 534 (Escherichia coli numbering) was amplified by PCR by using eubacterial primers 2 and 3 and the hot-start touchdown protocol described by Muyzer et al. (20). DNA extracted from the litter was amplified with a PCR mixture (50 μl) containing 29.2 μl of sterilized MilliQ water, 5 μl of Mg-containing buffer, 2 μl of skim milk, 0.05 μl of T4 gen, 10 μl of a deoxynucleoside triphosphate mixture, 1 μl of primer 1, 1 μl of primer 2, 1 μl of the DNA solution, and 0.75 μl of Expand High Fidelity DNA polymerase (La Roche). The polymerase was added after a hot-start procedure (5 min at 94°C, followed by 5 min at 80°C). PCR was performed with a Perkin-Elmer 9600 thermocycler by using the following protocol: 1 min at 94°C (denaturation), 1 min at 65°C (annealing), and 3 min at 72°C (elongation) with a 1°C touchdown every second cycle during annealing for 20 cycles, followed by 10 cycles with an annealing temperature of 55°C and a final cycle consisting of 10 min at 72°C.
After gel electrophoresis (1.5% [wt/vol] agarose gel) of 4-μl subsamples of the PCR product, the amount of amplified DNA was quantified by comparing band intensities to standard curves obtained with a low DNA Mass ladder (GibcoBRL). Band intensities were measured with ONE-Dscan electrophoresis analysis software (Scanalytics, CSP Inc., Billerica, Mass.)
DGGE.
Profiles of the amplified 16S rRNA gene sequences were produced by DGGE as described by Muyzer et al. (20) by using the Ingeny U-Phor system (Ingeny, Goes, The Netherlands). The PCR products were loaded onto a polyacrylamide gel (8% [wt/vol] acrylamide in 0.5× TAE buffer [4.84 g of Tris base per liter, 11.42 ml of acetic acid per liter, 20 ml of 0.5 M EDTA per liter; pH 8.0]) with a 45 to 75% denaturant gradient (100% denaturant was 7 M urea and 40% [vol/vol] deionized formamide). The wells were loaded with equal amounts of DNA, and electrophoresis was carried out in 0.5× TAE buffer at 75 V for 16 h at 60°C. The DNA fragments were stained for 20 min in 0.5× TAE buffer with ethidium bromide (final concentration, 0.5 μg/liter). The gel was destained in distilled water for 5 min. Images of the gels were obtained by using a UV 300 transilluminator (Fotodyne, Hartland, Wis.) and an Image Point cooled charge-coupled device video camera (Photometrics Ltd., Tucson, Ariz.). The video images were acquired with a Quantimet 570 image analysis system (Leica, Cambridge, United Kingdom) and were stored as TIFF files. Band patterns were analyzed by using GelCompar II software (Applied Maths, Sint-Martens-Latem, Belgium). The background intensity was subtracted (10%), the DNA bands were identified interactively, and the position and mass (intensity) of each band were determined. The data were used for principal-component analysis (PCA) with GelCompar to evaluate differences between the DNA profiles. Qualitative PCA in which the presence but not the intensity of bands was used gave the best separation between different samples.
Statistics.
The data were used to calculate the Shannon-Weaver diversity index (H̄), the evenness (e), and the equitability (J) (1), as follows: H̄ = (C/N)(N · log N − Σni · log ni), where C is 2.3, N is the total mass of all DNA bands, and ni is the mass of the ith DNA band; e = H̄/log S, where S is the number of DNA bands; and J = H̄/Hmax, where Hmax is the theoretical maximal Shannon-Weaver diversity index for the population examined, assuming that each species has only one member. The Shannon-Weaver diversity index is a general diversity index which increases with the number of species and which is higher when the mass is distributed more evenly over the species. The evenness is independent of the number of species. Evenness is lower if a small number of bands are dominant and highest if the relative abundance of all bands is the same. The equitability correspondingly indicates whether there are dominant bands.
RESULTS
Number of DNA bands and number of bacterial species.
Eight of ten bacterial strains isolated from soil yielded a single band. Two strains yielded more than one band. Flavobacterium sp. yielded a strong band and a weak band, and E. cloacae clearly produced five bands whose intensities were similar.
The eight strains that produced a single band were added to autoclaved soil both separately and in a mixture. The autoclaved soil did not produce any DNA band. No Nocardia and Rhodococcus DNA bands were detected (Fig. 1). For Rhodococcus the reason is not clear. The densities of Nocardia (105 cells g−1) were probably below the detection limit of our DGGE protocol. This protocol is normally used for bacterial communities in field soils with total densities of about 109 bacteria g−1. The other six isolates each yielded a clear DNA band at a density of about 107 cells g−1. The individual isolates were clearly reflected in the DNA band pattern of the mixture. When the mixture was diluted 10-fold, the pattern was weaker but still visible. This indicates that the detection limit of our DGGE protocol is about 106 cells g of soil−1. The Arthrobacter and Corynebacterium bands were so close that they appeared to be one band in the mixture.
FIG. 1.
DGGE DNA bands of single bacterial isolates and of mixtures added to autoclaved soil. The mixture was added undiluted and diluted 10-fold. Most isolates are reflected in the mixture.
DNA band intensity and relative abundance of bacteria.
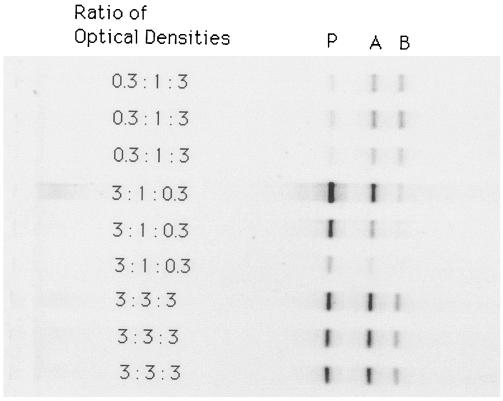
The ratio of the optical densities was reflected by the ratio of the intensities of the three DNA bands in each lane (Fig. 2). The relative abundance calculated from the intensity of the DNA bands correlated with the relative abundance calculated from the optical densities (r = 0.73; P < 0.0001; n = 27) and with the relative abundance calculated from the cell numbers (r = 0.67; P < 0.0001; n = 27).
FIG. 2.
DGGE DNA bands of P. fluorescens (P), Arthrobacter sp. (A), and Bacillus sp. (B) added to autoclaved soil in different ratios.
Bacterial DNA content in plant litter.
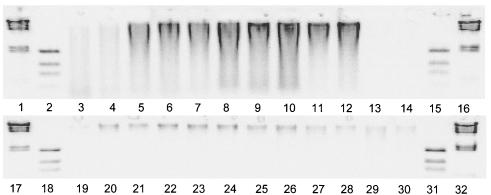
The original litter material contained a small amount of DNA, and the amount increased rapidly after the litter bags were inserted into the soil (Fig. 3). The DNA content of the crude extract was generally higher for the rye grass than for the wheat straw. The DNA content of the rye litter was higher at the central German site than at the northern and southern sites on day 18. The target gene for the bacterial communities could successfully be amplified in all extracts with two exceptions: one rye litter replicate on day 180 and one fresh wheat straw replicate on day zero. These samples were not used for the statistical analyses. Equal quantities of amplified DNA were loaded into the slots of the different lanes on the gel for semiquantitative comparison of the diversity of litter-decomposing bacteria by DGGE.
FIG. 3.
DNA extracted from rye grass and wheat straw on agarose gels. Lanes 1, 16, 17, and 32, 23-kbp marker; lanes 2, 15, 18, and 31, DNA Mass ladder (Life Technologies); lanes 3 to 12, five decomposition stages (days 0, 18, 58, 118, and 180, duplicate samples) for rye grass; lanes 13, 14, and 19 to 26, five decomposition stages (days 0, 18, 58, 118, and 180, duplicate samples) for wheat straw; lanes 27 to 30, rye grass (duplicate samples) from northern and southern Germany obtained on day 18.
DGGE.
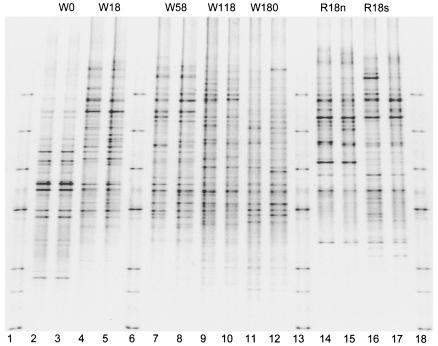
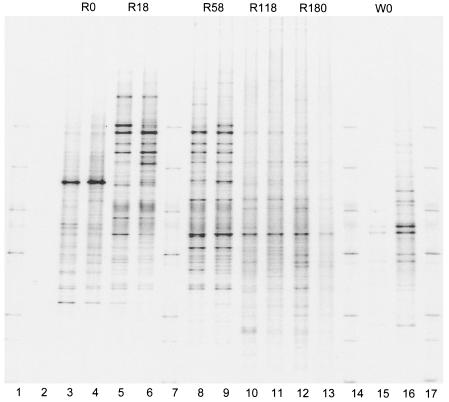
The melting profiles revealed different DNA fragments that were the same length but had different base sequences (operational taxonomic units or species) over the 180-day decomposition period (Fig. 4 and 5). Between 28 and 52 bands were distinguished in the rye litter and the wheat litter (Table 2). The lowest number was in the fresh rye litter, and the highest number was obtained after 118 days of decomposition in the wheat straw. Dominating bands were present particularly during the first part of the decomposition period on days 0, 18, and 58, in contrast to the later stages of decomposition on days 118 and 180, when the bands were more similar.
FIG. 4.
DGGE patterns of 16S rRNA gene sequences during litter decomposition in soil in central Germany. Lanes 1, 6, 13, and 18, marker; lanes 2 to 12, five decomposition stages for wheat straw on days 0, 18, 58, 118, and 180 (W0, W18, W58, W118, and W180, respectively) (duplicate samples); lanes 14 to 17, rye grass on day 18 in soil in northern and southern Germany (R18n and R18s, respectively) (duplicate samples). The two replicates obtained on day 0 were derived from the second extract since DGGE for the first extract was not successful (see Fig. 6).
FIG. 5.
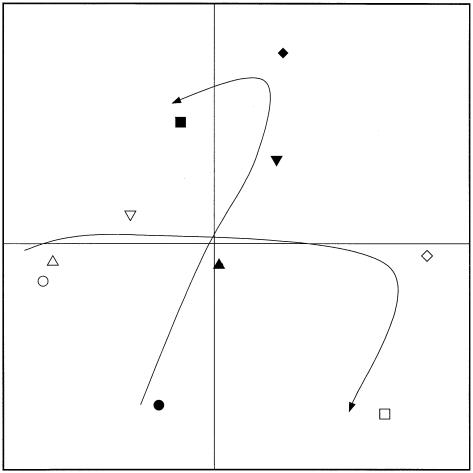
Qualitative PCA of 16S rRNA gene sequences during rye litter decomposition (A, X = 29.8%; Y = 25.2%; Z = 14.0%; Σ = 69.1%) and wheat litter decomposition (B, X = 36.2%; Y = 18.8%; Z = 15.5%; Σ = 70.5%). Symbols: •, day 0; ▴, day 18; ▾, day 58; ♦, day 118; ▪, day 180.
TABLE 2.
Bacterial diversity as indicated by the number of DNA bands, the Shannon-Weaver diversity index, the evenness, and the equitability of 16S rRNA gene sequences during rye and wheat litter decomposition in agricultural soils in northern (54°N), central (51°N), and southern (48°N) Germanyalegend
| Material | Site | Day | No. of DNA bands | Shannon-Weaver diversity index | Evenness | Equitability |
|---|---|---|---|---|---|---|
| Rye grass | 51°N | 0 | 27.5 ± 1.5 | 2.77 ± 0.01 | 1.92 ± 0.02 | 0.84 ± 0.01 |
| 51°N | 18 | 36.5 ± 1.5 | 3.09 ± 0.08 | 1.98 ± 0.03 | 0.86 ± 0.01 | |
| 51°N | 58 | 40.0 ± 2.0 | 3.19 ± 0.03 | 1.99 ± 0.01 | 0.87 ± 0.00 | |
| 51°N | 118 | 37.0 ± 2.0 | 3.21 ± 0.05 | 2.05 ± 0.00 | 0.89 ± 0.00 | |
| 51°N | 180 | 39.0 | 3.34 | 2.10 | 0.91 | |
| Wheat straw | 51°N | 0 | 35.5 | 3.01 | 1.94 | 0.84 |
| 51°N | 18 | 39.5 ± 0.5 | 3.33 ± 0.02 | 2.08 ± 0.00 | 0.91 ± 0.00 | |
| 51°N | 58 | 44.5 ± 4.5 | 3.30 ± 0.10 | 2.01 ± 0.01 | 0.87 ± 0.00 | |
| 51°N | 118 | 52.0 ± 1.0 | 3.61 ± 0.01 | 2.10 ± 0.00 | 0.91 ± 0.00 | |
| 51°N | 180 | 49.0 ± 1.0 | 3.50 ± 0.01 | 2.07 ± 0.02 | 0.90 ± 0.01 | |
| Rye grass | 54°N | 18 | 29.5 ± 0.5 | 2.91 ± 0.01 | 1.98 ± 0.02 | 0.86 ± 0.01 |
| 48°N | 18 | 37.0 ± 1.0 | 3.13 ± 0.04 | 2.00 ± 0.01 | 0.87 ± 0.00 |
aThe values are means ± standard deviations for two replicates.
In lane 13 of Fig. 6, the main DNA bands are hardly visible even though the DNA content in this sample was similar to the DNA content in the replicate lane, lane 12 (Fig. 3). However, the difference between lanes 12 and 13 indicates that amplification of the target sequences was inhibited for unknown reasons. Therefore, lane 13 was not used for further analysis.
FIG. 6.
DGGE pattern of 16S rRNA gene sequences during litter decomposition in soil in central Germany. Lanes 1, 7, 14, and 17, marker; lane 2, negative control; lanes 3 to 13, five decomposition stages for rye grass on days 0, 18, 58, 118, and 180 (R0, R18, R58, R118, and R180, respectively) (duplicate samples); lanes 15 and 16, wheat straw on day 0 (W0).
Diversity.
The Shannon-Weaver diversity index, evenness, and equitability showed that generally microbial diversity increased as decomposition proceeded (Table 2). When evenness and equitability were examined, this trend was not significant after 18 days, particularly for wheat litter. The average number of bacterial DNA bands increased significantly from 32 on day 0 to 45 on day 180 (P = 0.0031, as determined by analysis of variance). The average diversity in the wheat straw (44 DNA bands) was significantly higher than that in the rye litter (36 DNA bands) (P = 0.0004). The highest diversity was found in the wheat straw on day 118. The increasing evenness and equitability of the bacterial communities during the course of litter decomposition indicated that the species were more evenly represented during the later stages of decomposition.
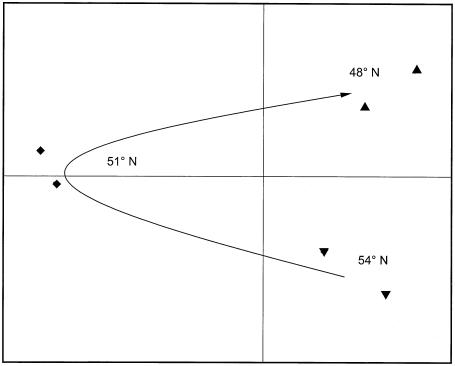
On day 18, the number of DNA bands and the Shannon-Weaver diversity index were significantly higher at the central and southern German sites (P = 0.023). The evenness and equitability values were similar at the three sites.
Similarity and succession.
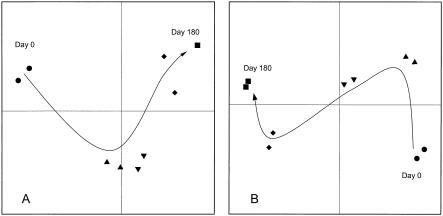
PCA showed that the (qualitative) differences in DNA profiles among the five decomposition stages were much greater than the differences between the duplicates (Fig. 5). The rye litter and straw litter showed divergent development of bacterial communities (Fig. 7) which was associated with significantly different diversity indices. Also, the differences among the bacterial DNA profiles at the sites in northern, central, and southern Germany were much greater than the differences between the duplicates (Fig. 8). This indicates that site-specific bacterial communities developed in the decomposing rye litter that had the same origin.
FIG. 7.
Qualitative PCA of 16S rRNA gene sequences during decomposition of rye litter (open symbols) and wheat straw (solid symbols) on day 0 (○ and •), day 18 (▵ and ▴), day 58 (▿ and ▾), day 118 (⋄ and ♦), and day 180 (□ and ▪) (X = 19.3%; Y = 16.9%; Z = 14.2%; Σ = 50.4%).
FIG. 8.
Qualitative PCA of 16S rRNA gene sequences for rye litter after 18 days of decomposition in northern ▴, central ♦, and southern ▾ Germany (X = 29.8%; Y = 25.2%; Z = 14.0%; Σ = 69.1%).
DISCUSSION
The DNA band pattern obtained by DGGE is an attractive way to study complex communities in environmental samples because most of the bacterial genotypes (species) gave one band in our experiments with species added to a sterilized soil. The intensities of the DNA bands reflected the relative levels of the bacterial strains, and the bands for most individual strains were also present in the DNA band pattern of the mixture. However, both under- and overestimation of the real number of genotypes could occur. Although we occasionally observed missing and overlapping bands which may have complicated the interpretation of band patterns, bacterial DNA profiles obtained by DGGE can be used as a semiquantitative measure of bacterial diversity, and we therefore used this method to study bacterial diversity during litter decomposition in agricultural soils.
The DNA band patterns obtained from amplified 16S rRNA gene sequences and DGGE indicated that the structure and diversity of bacterial communities changed significantly during 180 days of litter decomposition in an agricultural soil. The number of bacterial DNA bands increased from 28 to 40 in rye litter and from 36 to 52 in wheat litter (Table 2). The numbers of bands in the litter were in the range found in rhizosphere and bulk soil by Duineveld et al. (8) but were lower than the numbers of bands in mineral soils in northern Germany (more than 50 bands [unpublished data]) and in The Netherlands (about 50 bands [4]). Compared to the DGGE patterns for rhizosphere soil, the DGGE patterns for bulk soil are generally complex, with many distinct bands (30).
The number of DNA bands increased as litter decomposition proceeded, whereas the microbial biomass and activity decreased (6). This can be regarded as due to bacterial adaptation to more heterogeneous environmental conditions and the complex composition of the remaining organic matter. Soil microbial communities have extreme phenotypic and genotypic diversity (28). The increase in diversity as the activity decreased appeared to reflect the conversion of litter to soil organic matter and the concomitant development of diverse microbial communities adapted to lower availability of nutrients. The disappearance of dominating bands and the subsequent development of a more uniform band pattern as decomposition proceeded can be interpreted as follows: r strategists (opportunists) that prevailed on fresh litter were replaced by a variety of K strategists (persisters) related to resistant organic matter and humic substances (1). The development of a higher level of diversity is probably related to an increased importance of biotic interactions within the community (25, 26).
Only approximately 30% of the wheat straw had been decomposed after 180 days, whereas more than 80% of the rye litter had disappeared (data not shown). The bacterial communities in the slowly decomposing wheat straw appeared to be more diverse than those in the rapidly decomposing rye litter. Apparently, more species (or genotypes) were required for decomposition or were able to grow when the litter quality was low. The low nutritional quality of the wheat straw was reflected by the content of bacterial DNA, which was much lower for wheat straw than for rye litter (Fig. 3). In the rapidly decomposing rye grass litter with higher nutritional value, the amount of bacterial DNA increased rapidly and the bacterial DNA was dominated by fewer organisms, as indicated by the lower number of bands and lower diversity.
Previous studies on decomposition of various litter types showed that there were litter-specific biomass pools (21), decomposition-stage-dependent changes in biomass, respiration, and enzyme activity (6), and also specific community compositions of culturable bacteria and fungi (7). This study showed that the total communities, including both culturable and nonculturable bacteria, became more diverse during litter decomposition in agricultural soils, as revealed by DGGE of specifically amplified PCR products. More information concerning the identities of the dominant members of the bacterial community could be obtained by excision of DNA bands from the gels, followed by cloning and sequencing of PCR-amplified gene fragments, as has been done for ammonia-oxidizing communities by Laverman et al. (16). Identification of species was beyond the scope of this study, in which we focused on quantification of diversity during litter decomposition.
Based on band pattern and bacterial diversity indices in the same litter investigated in soils in northern, central, and southern Germany, site-specific differences for the same litter were found, and such differences had to be expected. However, we acknowledge that common soil components may form complexes with proteins and may inhibit the PCR by interaction with Taq DNA polymerase (31).
In conclusion, microbial diversity increased during the course of litter decomposition (hypothesis 1 was confirmed), in accordance with the system-theoretical hypotheses of Odum (23, 24). Compared to the microbial communities in readily available rye grass, refractory straw enabled development of microbial communities with greater diversity (hypothesis 2 was rejected). Bacterial diversity differed in the same litter buried in similar soils at different locations (hypothesis 3 was confirmed). Thus, not only the origin of the litter but also the surrounding soil affected the development of bacteria in the litter.
Acknowledgments
We are grateful for the excellent technical assistance of Sabine Splitzer and Sandra Wulff during the litter bag experiment.
Financial support was provided by the German Research Foundation (project MU 831/12-1) and the state of Bavaria and Schleswig-Holstein.
REFERENCES
- 1.Atlas, R. M., and R. Bartha. 1998. Microbial ecology: fundamentals and applications. Addison-Wesley Publishing Company, Reading, Pa.
- 2.Beare, M. 1997. Fungal and bacterial pathways of organic matter decomposition and nitrogen mineralization in arable soils, p. 37-70. In L. Brussard and R. Ferrera-Cerrato (ed.), Soil ecology in sustainable agricultural systems. CRC Lewis Publishers, Boca Raton, Fla.
- 3.Bengtsson, J. 2002. Disturbance and resilience in soil animal communities. Eur. J. Soil Biol. 38:119-125. [Google Scholar]
- 4.Bloem, J., and A. M. Breure. 2003. Microbial indicators, p. 259-282. In B. A. Markert, A. M. Breure, and H. G. Zechmeister (ed.), Bioindicators/biomonitors—principles, assessment, concepts. Elsevier, Amsterdam, The Netherlands.
- 5.Bloem, J., M. Veninga, and J. Shepherd. 1995. Fully automatic determination of soil bacterium numbers, cell volumes, and frequencies of dividing cells by confocal laser scanning microscopy and image analysis. Appl. Environ. Microbiol. 61:926-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dilly, O., and J.-C. Munch. 1996. Microbial biomass content, basal respiration and enzyme activities during the course of decomposition of leaf litter in a black alder (Alnus glutinosa (L.) Gaertn.) forest. Soil Biol. Biochem. 28:1073-1081. [Google Scholar]
- 7.Dilly, O., S. Bartsch, P. Rosenbrock, F. Buscot, and J.-C. Munch. 2001. Shifts in physiological capabilities of the microbiota during the decomposition of leaf litter in a black alder (Alnus glutinosa (Gaertn.) L.) forest. Soil Biol. Biochem. 33:921-930. [Google Scholar]
- 8.Duineveld, B. M., G. A. Kowalchuk, A. Keijzer, J. D. van Elsas, and J. A. van Veen. 2001. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekschmitt, K., A. Klein, B. Pieper, and V. Wolters. 2001. Biodiversity and functioning of ecological communities—why is diversity important in some cases and unimportant in others? J. Plant Nutr. Soil Sci. 164:239-246. [Google Scholar]
- 10.Giller, K. E., and G. Gadisch. 1997. Driven by nature: a sense of arrival or departure, p. 393-399. In G. Cadisch and K. E. Giller (ed.), Driven by nature. Plant litter quality and decomposition. CAB International, Wallingford, United Kingdom.
- 11.Griffiths, B. S., K. Ritz, R. D. Bardgett, R. Cook, S. Christensen, F. Ekelund, S. Sørensen, E. Bååth, J. Bloem, P. de Ruiter, J. Dolfing, and B. Nicolardot. 2000. Ecosystem response of pasture soil communities to fumigation-induced microbial diversity reductions: an examination of the biodiversity-ecosystem function relationship. Oikos 90:279-294. [Google Scholar]
- 12.Griffiths, B. S., K. Ritz, R. Wheatly, H. L. Kuan, B. Boag, S. Christensen, F. Ekelund, S. J. Sørensen, S. Muller, and J. Bloem. 2001. An examination of the biodiversity-ecosystem function relationship in arable soil microbial communities. Soil Biol. Biochem. 33:1713-1722. [Google Scholar]
- 13.Heal, O. W., J. M. Anderson, and M. J. Swift. 1997. Plant litter quality and decomposition: an historical overview, p. 3-10. In G. Cadisch and K. E. Giller (ed.), Driven by nature. Plant litter quality and decomposition. CAB International, Wallingford, United Kingdom.
- 14.Hughes, J. R., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim-Jong, S., M. Sakai, A. Hosoda, T. Matsuguchi, and J. S. Kim. 1999. Application of DGGE analysis to the study of bacterial community structure in plant roots and in nonrhizosphere soil. Soil Sci. Plant Nutr. 45:493-497. [Google Scholar]
- 16.Laverman, A. M., A. G. C. L. Speksnijder, M. Braster, G. A. Kowalchuk, H. A. Verhoef, H. W. van Verseveld, and H. W. van Verseveld. 2001. Spatiotemporal stability of an ammonia-oxidizing community in a nitrogen-saturated forest soil. Microb. Ecol. 42:35-45. [DOI] [PubMed] [Google Scholar]
- 17.McCaig, A. E., L. Glover, and J. I. Posser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grassland pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel, P. H., and J. Bloem. 1993. Conversion factors for estimation of cell production rates of soil bacteria from thymidine and leucine incorporation. Soil Biol. Biochem. 25:943-950. [Google Scholar]
- 19.Muyzer, G. 1999. DGGE/TGGE: a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 20.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified gene coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neely, C. L., M. H. Beare, W. L. Hargrove, and D. C. Coleman. 1991. Relationships between fungal and bacterial substrate-induced respiration, biomass and plant residue decomposition. Soil Biol. Biochem. 23:947-954. [Google Scholar]
- 22.Nübel, U., F. Garcia-Pichel, M. Kühl, and G. Muyzer. 1999. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotinoids of oxygenic phototrophs in microbial mats. Appl. Environ. Microbiol. 65:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odum, E. P. 1969. The strategy of ecosystem development. Science 164:262-270. [DOI] [PubMed] [Google Scholar]
- 24.Odum, H. T. 1956. Efficiencies, size of organisms, and community structure. Ecology 37:592-597. [Google Scholar]
- 25.Pielou, E. C. 1983. Population and community ecology. Principles and methods. Gordon and Breach Science Publishers, New York, N.Y.
- 26.Putman, R. J. 1994. Community ecology. Chapman & Hall, London, United Kingdom.
- 27.Rosenbrock, P., F. Buscot, and J.-C. Munch. 1995. Fungal succession and changes in the fungal degradation potential during the early stages of litter decomposition in black alder forest (Alnus glutinosa (Gaertn.) L.). Eur. J. Soil Biol. 31:1-11. [Google Scholar]
- 28.Sessitsch, A., A. Weilharter, M. H. Gerzabek, H. Kirschmann, and E. Kandeler. 2001. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl. Environ. Microbiol. 67:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slater, J. H. and D Lovatt. 1984. Biodegradation and the significance of microbial communities, p. 439-485. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, Inc., New York, N.Y.
- 30.Thirup, L., Johansen, A., and A. Winding. 2003. Microbial succession in the rhizosphere of live and decomposing barley roots as affected by the antagonistic strain Pseudomonas fluorescens DR54-BN14 or the fungicide Imazalil. FEMS Microbiol. Ecol. 43:383-392. [DOI] [PubMed] [Google Scholar]
- 31.Watson, R.-J., and B. Blackwell. 2000. Purification and characterization of a common soil component which inhibits the polymerase chain reaction Can. J. Microbiol. 46:633-642. [DOI] [PubMed] [Google Scholar]
- 32.Westergaard, K., A. K. Müller, S. Christensen, J. Bloem, and S. J. Sørensen. 2001. Effects of tylosin as a disturbance on the soil microbial community. Soil Biol. Biochem. 33:2061-2071. [Google Scholar]
- 33.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zvyagintsev, D. G. 1994. Vertical distribution of microbial communities in soils, p. 29-37. In K. Ritz, J. Dighton, and K. E. Giller (ed.), Beyond the biomass. Blackwell Scientific Publications, Oxford, United Kingdom.