Abstract
Background
Chloroplasts and mitochondria evolved from the endosymbionts of once free-living eubacteria, and they transferred most of their genes to the host nuclear genome during evolution. The mechanisms used by plants to coordinate the expression of such transferred genes, as well as other genes in the host nuclear genome, are still poorly understood.
Results
In this paper, we use nuclear-encoded chloroplast (cpRPGs), as well as mitochondrial (mtRPGs) and cytoplasmic (euRPGs) ribosomal protein genes to study the coordination of gene expression between organelles and the host. Results show that the mtRPGs, but not the cpRPGs, exhibit strongly synchronized expression with euRPGs in all investigated land plants and that this phenomenon is linked to the presence of a telo-box DNA motif in the promoter regions of mtRPGs and euRPGs. This motif is also enriched in the promoter regions of genes involved in DNA replication. Sequence analysis further indicates that mtRPGs, in contrast to cpRPGs, acquired telo-box from the host nuclear genome.
Conclusions
Based on our results, we propose a model of plant nuclear genome evolution where coordination of activities in mitochondria and chloroplast and other cellular functions, including cell cycle, might have served as a strong selection pressure for the differential acquisition of telo-box between mtRPGs and cpRPGs. This research also highlights the significance of physiological needs in shaping transcriptional regulatory evolution.
Background
Mitochondria and chloroplasts evolved from the endosymbionts of once free-living α-proteobacteria and cyanobacteria, respectively [1]. Most of the genes in the endosymbionts were transferred to the nuclear genome during evolution, resulting in much smaller current organelle genomes than their ancient cousins [2-4]. Many transferred genes acquired promoters from their eukaryotic hosts [1], and most proteins expressed by these functional transferred genes were eventually translocated back to their organelle, guided by targeting peptides [4,5]. Intuitively, a tight coordination of gene expression between organelle genomes and the host nuclear genome should be important for cellular functions, as well as the overall fitness of the organism [6,7]. In fact, coordination between host and organelles through biochemical signaling has been extensively studied [8]. Genetically, the coordination between nuclear and mitochondrial genome expression [6], as well as the coordination between nuclear and chloroplast genome expression [7] has been investigated. However, the molecular mechanisms underlying the coordination between host and organelle functions are still far from understood. In this work, we aim to identify signals coordinating the expression of genes in mitochondrion, chloroplast and nucleus.
On the other hand, chloroplasts, mitochondria and the host cell all have protein translation machineries. In addition to the cytoplasmic ribosome of the host cell, mitochondria and chloroplasts each have their own respective ribosomes to translate proteins encoded in their genomes [9]. The existence of ribosomes in mitochondria and chloroplasts provides a certain freedom of independent biogenesis and/or development for these organelles [10,11]. Given that protein synthesis is required for organelle biogenesis, development and various biological processes in these different cellular compartments, investigation of the coordination of expression and regulation of ribosomal protein genes (RPGs) between host nucleus and each type of organelle can shed light on the coordination of organelle biogenesis, development and biological processes in these different compartments. Since most organelle RPGs were transferred to the nuclear genome during evolution (Figure S1 in Additional File 1), specialized trans-factors and cis-elements may have evolved to ensure the expression of these transferred RPGs in a coordinated manner. As such, identification of these factors or elements is critical to understanding the coordination of biogenesis and development between these different organelles. We thus chose to examine the expression and regulatory patterns of these transferred RPGs.
Our analysis show that the expression patterns of transferred mitochondrial (mtRPGs) and cytoplasmic (euRPGs) ribosomal protein genes are highly coordinated, while expressions of chloroplasts (cpRPGs) and euRPGs are not. This phenomenon appears in all investigated monocot and dicot plants, the expression datasets of which are available. By sequence analysis on the promoter regions of these RPGs, we identified a functional DNA motif, telo-box, which is linked to the observed differential coordination patterns. The telo-box is present in promoters of mtRPGs and euRPGs, but absent in promoters of cpRPGs across all examined land plants. The telo-box is also enriched in the promoter regions of genes encoding enzymes involved in DNA replication, indicating a potential role of telo-box in the cell cycle. Evidences from comparative genomics analysis indicated that the telo-box in mtRPGs was acquired from the host nuclear genome. Based on these results, we proposed a model of land plant nuclear genome evolution. In this model, after endosymbiosis, many genes in endosymbionts were transferred to the nuclear genome. The demand for a high-level coordination of energy supply might have been a strong selection pressure, which gradually led to coordinated expression of proteins in mitochondria with those involved in other cellular functions, including cell cycle.
Results
In this section, we first examined the expression patterns of cytoplasmic, mitochondrial and chloroplast ribosomal protein genes (RPGs) using expression profiles across a wide array of tissues of A. thaliana, and confirmed our findings in several other monocot and dicot plants. Such comprehensive tissue-specific expression datasets provide an unbiased sampling of gene expression. Based on the striking co-expression between euRPGs and mtRPGs, but not between euRPGs and cpRPGs, we further studied the promoter sequences of these three sets of RPGs. We found a DNA motif known as telo-box in promoters of euRPGs and mtRPGs but not cpRPGs in all studied land plants, which explains the observed differential coordination patterns. Functional implication of telo-box and regulatory evolution of mtRPGs and cpRPGs after gene transfer is then studied.
Co-expression between mtRPGs and euRPGs
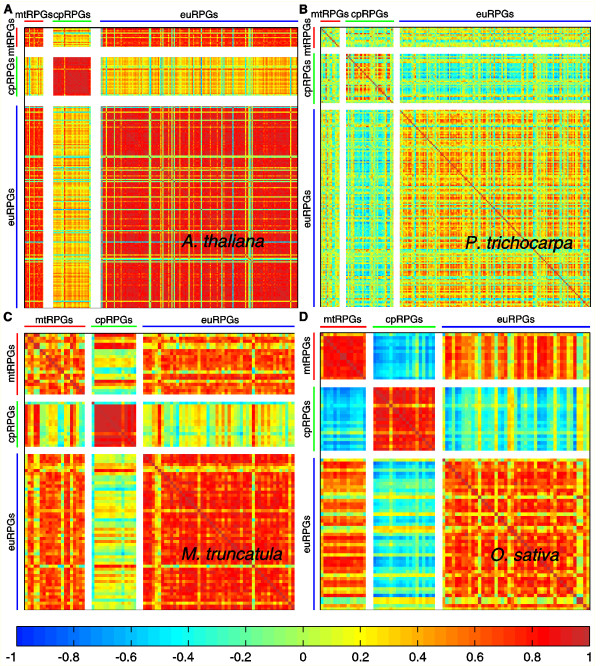
Intuitively, biogenesis of cellular organelles needs to be highly coordinated to ensure the optimal growth of a plant cell and hence the organism. Since production of proteins is the key step for organelle biogenesis, the study of protein translational machinery, i.e., ribosomes, may provide insights on the coordination between host and the organelles. We therefore first asked how the expression of mtRPGs, euRPGs and cpRPGs are coordinated across various tissues in plants. Here we chose to use tissue-specific expression data to minimize possibility of spurious findings due to sample bias. Correlation analysis of the expression of cpRPGs, mtRPGs and euRPGs in Arabidopsis thaliana showed that the expression of mtRPGs was strongly positively correlated with the expression of euRPGs (Pearson's Correlation Coefficient, PCC = 0.6260 ± 0.3220, p < 1.0E-10, t-test). On the other hand, although the expression of cpRPGs showed significantly positive correlation with mtRPGs and euRPGs (PCC = 0.1582 4; 0.1901, p < 1.0E-10), the magnitude was much lower (p < 1.0E-10, Figure 1A). Similar results were also obtained in Populus trichocarpa (Figure 1B), Medicago truncatula (Figure 1C), and Oryza sativa (Figure 1D). Furthermore, the above observed correlation patterns were also found using tissue-specific protein expression level data in A. thaliana (Figure S2 in Additional File 1), where the protein expression levels between mtRPGs and euRPGs were significantly positively correlated (PCC = 0.1844 ± 0.4031, p = 4.8E-12), while the protein expression levels of cpRPGs were significantly negatively correlated with those of both mtRPGs and euRPGs (PCC = -0.1461 ± 0.4115, p = 1.4E-31). These results indicated that some regulatory mechanisms might exist for the differential coordination of expression between mtRPGs and cpRPGs.
Figure 1.
Correlation between mRNA expression of mtRPGs, cpRPGs and euRPGs in four angiosperms. The expression profiles of RPGs of A. thaliana (A), P. trichocarpa (B), M. truncatula (C) and O. sativa (D). Each element of the matrix represents the Pearson's correlation coefficient between the expression profiles of two RPGs. Color code is illustrated at the bottom panel.
The correlation of expression among RPGs was further examined in detail by using A. thaliana microarray data at several developmental stages (Figure S3 in Additional File 1). We found that the expression of mtRPGs and euRPGs was positively correlated across all developmental stages. In contrast, the correlation between the expression of cpRPGs with either mtRPGs or euRPGs changed dramatically during developmental progression. In particular, cpRPGs were found to be negatively correlated with mtRPGs and euRPGs at early developmental stages (7th day: PCC = -0.2855 ± 0.3040, p < 1.0E-10 and 17th day: PCC = -0.3166 ± 0.3086, p < 1.0E-10; Figure S3A, S3B in Additional File 1), but cpRPGs became positively correlated with mtRPGs and euRPGs at later developmental stages (21st day: PCC = 0.3509 ± 0.2192, p < 1.0E-10; 8th week: PCC = 0.3488 ± 0.5719, p < 1.0E-10; Figure S3C, S3D in Additional File 1). Notably, cpRPGs showed a much higher level of coordination in their expression levels at the 8th week of seed development, as compared to the co-expression within either mtRPGs or euRPGs (within cpRPGs, PCC = 0.8681 ± 0.2332; within mtRPGs, PCC = 0.0361 ± 0.4613; within euRPGs, PCC = 0.3940 ± 0.3984; Figure S3D in Additional File 1).
Telo-boxes are Enriched in Promoters of mtRPGs and euRPGs
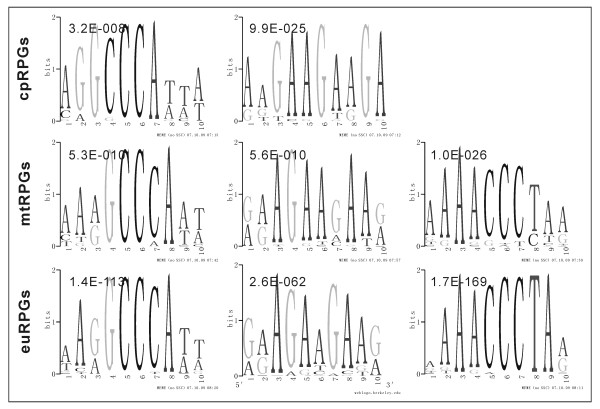
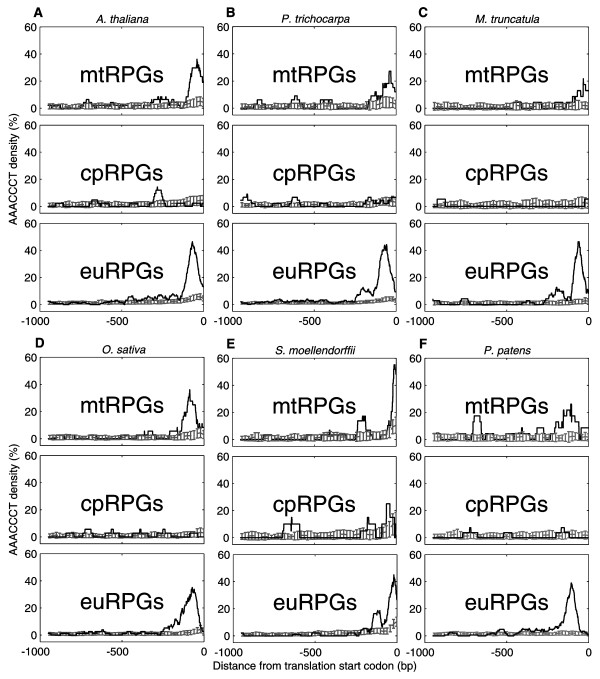
We next asked if the above observations could be explained by transcriptional regulatory elements. Analysis of the promoter sequences of nuclear-encoded cpRPGs, mtRPGs and euRPGs in A. thaliana using MEME [12] revealed conserved DNA motifs (Figure 2). The first motif, GCCCA, known as site II motif and highly enriched in promoters of all three classes of RPGs in A. thaliana (Figure 2), is a binding target of the transcription factor At-TCP20 [13]. The transcription factor corresponding to the second shared motif, GAAGAA, has not been identified. The third motif, AAACCCT, known as telo-box, is enriched in promoters of both euRPGs and mtRPGs, but not in promoters of cpRPGs in A. thaliana (Figure 2 and 3A). Similar results (Figure S5 in Additional File 1) were obtained using other DNA motif-finding tools e.g. AlignACE and DME [14,15]. Thus, the absence or presence of telo-box in RPG promoters was considered to be associated with the differential expression coordination patterns among cpRPGs, mtRPGs and euRPGs. In addition, mitochondrial and cytoplasmic RPGs with telo-box in their respective promoter regions showed significantly higher co-expression than those without telo-box (p < 0.001, Figure S6 in Additional File 1), further indicating the functional importance of telo-box in synchronizing the expression patterns of mtRPGs and euRPGs. In fact, such an association was also observed in three other angiosperm land plants, P. trichocarpa, M. truncatula and O. sativa (Figure 3B, 3C, 3D; though the appearance of telo-box in mtRPGs of Figure 3B and 3C is slightly less than that of Figure 3A and 3D, possibly due to noise in promoter annotation), for which both DNA sequence data and expression profiling data are available. In two recently sequenced land plant species, Selaginella moellendorffii and Physcomitrella patens, telo-boxes are also enriched in the promoter regions of corresponding mtRPGs and euRPGs, but not in cpRPGs (Figure 3E, 3F; the appearance of telo-box is slightly above background in cpRPGs of S. moellendorffii, possibly due to noise in promoter annotations). Furthermore, the chromosome location of telo-box is also consistently close to the translation start codon in all these examined plants (Figure 3), indicating that the positioning of telo-box may be functionally important for the proper expression of the regulated genes. The examined plant species covered a wide range of land plant species (including moss, spikemoss, monocot and dicot plants) (Figure S7 in Additional File 1); therefore, the association between the coordinated expression pattern of RPGs and the presence of telo-box pattern in the promoter regions of RPGs might be conserved in all land plants.
Figure 2.
Promoter motifs of cpRPGs, mtRPGs and euRPGs in A. thaliana. The first (GCCCA) and third (AAACCCT) motifs are known as site II motif and telo-box motif, respectively. The number on the upper left of each logo is the E-value of MEME prediction. Some motifs only enriched in certain classes of RPGs are shown in Figure S4 in Additional File 1.
Figure 3.
Positional distribution of the telo-box motif (AAACCCT) of RPGs in all studied land plants. Telo-box (dark solid line) is enriched in promoters of mtRPGs and euRPGs, but not in cpRPGs for A. thaliana (A), P. trichocarpa (B), M. truncatula (C), O. sativa (D), S. moellendorffii (E) and P. patens (F). Grey dashed line with error bar indicates motif density of background sequences. Window size is 50bp.
Telo-boxes are Enriched in Promoters of Non-RPGs Highly Co-expressed with mtRPGs or euRPGs
We next examined whether telo-boxes are enriched in the promoter regions of non-ribosomal protein genes (non-RPGs) which are highly co-expressed with either mtRPGs or euRPGs in A. thaliana. In the identified 243 non-RPGs with PCC ≥ 0.9 to mtRPGs or euRPGs, the telo-box motif is significantly enriched in their promoter regions (p = 8.7E-63, Chi-square test, Figure 4). In contrast, in 341 non-RPGs which are highly co-expressed (PCC ≥ 0.9) with cpRPGs no significant enrichment of telo-box in their promoters is observed (Figure 4). This result further supports telo-box as the molecular mechanism underlying differential coordination patterns among cpRPGs, mtRPGs and euRPGs.
Figure 4.
Positional distribution of telo-box (AAACCCT) and Site II motif (GCCCA) of non-RPGs. Telo-box (solid line) is enriched in genes involved in both DNA replication and non-RPGs (non-ribosomal protein genes) that are highly correlated with (PPC ≥ 0.9) mtRPGs or euRPGs, but not in non-RPGs that are highly correlated with (PPC ≥ 0.9) cpRPGs. Site II motif (dash-dot line) is enriched in all three groups.
In the 243 non-RPGs highly co-expressed with mtRPGs or euRPGs (PCC ≥ 0.9), gene ontology analysis using DAVID [16] (on 127 genes with "biological process" annotation) indicates that most of these gene products are significantly related to "RNA processing and metabolism", "cell organization and biogenesis", and "protein translation and location" (Table 1). In addition, 140 genes with "molecular function" annotation are mostly related to "RNA, nucleotide, nucleic acid or protein binding", and "translation initiation factor activity" (Table 1).
Table 1.
GO enrichment of non-RPGs highly correlated with mtRPGs or euRPGs
| NO.* | GO term | Count § | FDR † |
|---|---|---|---|
| BP1 | RNA processing | 24 | 8.2E-9 |
| BP 2 | ncRNA metabolic process | 19 | 2.3E-8 |
| BP 3 | protein folding | 17 | 1.2E-6 |
| BP 4 | ribonucleoprotein complex biogenesis | 16 | 8.9E-6 |
| BP 5 | mitochondrion organization | 8 | 1.5E-5 |
| BP 6 | rRNA processing | 12 | 1.8E-5 |
| BP 7 | ribosome biogenesis | 15 | 4.9E-5 |
| BP 8 | ncRNA processing | 13 | 1.6E-4 |
| BP 9 | chromatin organization | 13 | 8.8E-4 |
| BP 10 | translational initiation | 8 | 6.0E-3 |
| BP 11 | protein targeting to mitochondrion | 5 | 2.4E-2 |
| MF1 | RNA binding | 43 | 1.6E-9 |
| MF2 | nucleotide binding | 68 | 2.5E-8 |
| MF3 | translation factor activity, nucleic acid binding | 14 | 1.7E-6 |
| MF4 | translation initiation factor activity | 12 | 3.5E-6 |
| MF5 | unfolded protein binding | 12 | 7.8E-6 |
* BP: Biological process, MF: Molecular function. § The total numbers of annotated genes in GO biological process and molecular function are 127 and 140, respectively. † P-value is corrected by False Discovery Rate (FDR) using DAVID [16].
Since it has been shown in Drosophila [17] that euRPGs are regulated by a transcription factor, DREF, which participates in DNA replication, and since the expression of ribosomal protein genes is related to cell proliferation [18], we next studied whether DNA replication genes (e.g., origin recognition, replicative helicases, helicase loading factors) in A. thaliana had coordinated expression with mtRPGs/euRPGs and showed enriched telo-boxes in their promoters. Interestingly, our analysis revealed that expression of these DNA replication genes was also highly positively correlated with that of mtRPGs/euRPGs (PCC = 0.5643 ± 0.2998). In addition, these genes have significantly enriched telo-boxes in their promoter regions (p = 3.8E-6, Chi-square test, Figure 4). Therefore, the shared regulation between ribosomal protein genes and DNA replication genes are conserved between insects and plants.
Conservation of Transcription Factor Purα that Binds to Telo-box
Given the coordinated expression pattern between mtRPGs and euRPGs, and the common telo-box in promoters of mtRPGs and euRPGs, it will be interesting to ask whether the trans-factor of telo-box is conserved among the studied species. We therefore studied the conservation pattern of transcription factor Purα, which is known to recognize telo-box [19-21]. First, the homolog of Purα was found to be present in all the examined land plants. Secondly, multiple sequence alignment for the Purα protein (Figure S8 in Additional File 1) reveals that several domains are highly conserved in all studied land plants, including the DNA-binding domain [21]. This observation is consistent with the highly conserved sequence of telo-boxes in land plants (Figure 3), and provides further support that telo-box might be a controlling mechanism of the coordinated expression between mtRPGs and euRPGs. In addition, this result indicates that Purα may participate in regulating the biogenesis and development of mitochondria.
Evolutionary Origin of Telo-box in Promoters of Transferred mtRPGs
Mitochondrial RPGs may have acquired telo-boxes for their coordinated expression with euRPGs in one of two ways: (1) they acquired telo-boxes after transferring into the nuclear genome or (2) they possessed telo-boxes in the endosymbionts, and the regulatory regions were carried on during transfer. To study these hypotheses, we searched genomic sequences of mitochondrion and chloroplast ancestors, respectively. Our result indicated that neither proto-mitochondrial ancestor (Rickettsia prowazekii str. Madrid E [22,23]) nor proto-chloroplast ancestor (Synechocystis sp. PCC6803 [4,24]) contained telo-boxes (data not shown). In addition, to account for the possibility that telo-boxes may have been lost during the evolution of R. prowazekii and Synechocystis, we also searched all available 132 chloroplast genomes and 25 plant mitochondria genomes, and telo-box was still not found (data not shown). These results indicated that mtRPGs acquired telo-box after endosymbiosis, whereas cpRPGs did not either successfully acquire or keep the telo-box after endosymbiosis.
Given the above results, it is interesting to ask if these transferred mtRPGs happened to be inserted into nuclear genomic regions where telo-box was enriched. To test this hypothesis, we first studied the appearances of telo-boxes in the vicinity of mtRPGs and cpRPGs (between upstream 40kb and downstream 40kb of the translation start codon). As can be seen in Figure S9A in Additional File 1, no significant difference of telo-box enrichment in flanking sequences between mtRPGs and cpRPGs was found. This result indicates that biased insertion during gene transfer between mtRPGs and cpRPGs is unlikely. To further confirm this conclusion, we studied the distances from each mtRPG/cpRPG to the closest non-ribosomal nuclear gene in the same chromosome which has telo-box in its promoter region, as these non-ribosomal nuclear genes might provide source of telo-boxes for transferred mtRPGs. As seen in Figure S9B in Additional File 1, no significant difference was observed between mtRPGs and cpRPGs in their distances to their respective closest non-RPG neighbors regulated by telo-boxes (p > 0.1, t-test for the mean distances of mtRPGs and cpRPGs to the closest non-RPG neighbors with telo-box). Taken together, these results indicate that selective pressures, rather than preferential insertion regions, may be the reason for mtRPGs and cpRPGs to acquire different regulatory mechanisms to coordinate their biogenesis with host after gene transfer.
The Coordination between mtRPGs and euRPGs is Land Plant-Specific
To check whether the coordinated pattern between mtRPGs and euRPGs is unique in land plants, we studied whether this pattern also existed in algae. We chose to study the brown algae Ectocarpus siliculosus, which is phylogenetically distant from land plants [25] (Figure S7 in Additional File 1) and genome sequence and gene expression profiling data of which are available [25,26]. Analysis on RPGs of E. siliculosus indicated that telo-boxes were not enriched in promoters of any types of RPGs (data not shown) and that the expression of mtRPGs is clearly independent from that of euRPGs (PCC = -0.0365 ± 0.2127, p > 0.01; Figure S10A in Additional File 1). This result indicated that the coordination of mtRPGs and euRPGs might be land plant-specific. To further confirm this conclusion, we used the unicellular green algae Chlamydomonas reinhardtii which diverged from land plants over a billion years ago [27]. We separately measured the expression levels of 6, 7 and 10 highly reliable mtRPGs, cpRPGs and euRPGs of C. reinhardtii under four conditions, including continuous light, continuous dark, high and low nitrogen treatments (see Methods). Results indicate that the expression levels of mtRPGs and euRPGs are not coordinated (Figure S10B in Additional File 1); furthermore, RPGs in C. reinhardtii lack telo-box motifs (data not shown). Taken together, these results indicate that the differential transcriptional modulation of cpRPGs and mtRPGs by telo-box is land plant-specific.
Regulatory Changes are Common for RPGs in other Species
In addition to the mitochondrion shared by all eukaryotes, plants, as compared to the animal species, have chloroplasts. Therefore, the transcriptional evolution of plant organelle ribosomal proteins may have been more complicated than other species. As demonstrated in this work, the acquisition of telo-boxes of transferred genes after endosymbiosis is different between mtRPGs and cpRPGs. In fact, such a dramatic change in the transcriptional regulation of ribosomal protein genes has already been seen in other organisms. For example, the cis-elements for cytoplasmic ribosomal protein genes are found to be significantly different among fungi, insects and mammals [17]. It was also reported that the ribosomal regulation is highly evolvable in yeast through the use of an intermediate redundant regulatory program [28]. Most strikingly, it was shown that the loss of a cis-element AATTTT in promoters of mtRPGs following whole-genome duplication is linked to rapid anaerobic growth of S. cerevisiae [29]. However, unlike the above-mentioned discoveries, our results highlight the differential acquisition of cis-elements after gene transfer, possibly due to the different physiological needs between mitochondria and chloroplast. Taken together, these discoveries indicate that the gene expression regulatory programs are highly evolvable for ribosomal protein genes, which are one of the most conserved gene families among all kingdoms. Therefore it will not be surprise to see dramatic changes in regulatory programs in other less conserved gene families which are specific to certain species. In fact, our discovery adds more support for the view that speciation primarily arise from changes in gene regulatory regions [30].
Discussion
The co-expression pattern of RPGs in different cellular compartments and the regulatory elements controlling this pattern
Analysis of the expression pattern of genes involved in ribosomal proteins in different compartments showed that mtRPGs exhibited a high level of co-expression with euRPGs, while cpRPGs did not show such a high level of co-expression with euRPGs (Figure 1). This pattern of expression was conserved across different land plant species examined in this study (Figure 1, Figure S2 in Additional File 1). These results indicated that the strong coordination of expression of mtRPGs and euRPGs possibly increased plant fitness, although the detailed mechanisms have not yet been elucidated. Interestingly, although the expression of cpRPGs was not well correlated with euRPGs when data from different developmental stages were pooled together in the analysis, cpRPG expression showed negative correlation with expression of mtRPGs and euRPGs in the early developmental stage in A. thaliana (Figure S3A, S3B in Additional File 1). In contrast, at the later developmental stages of A. thaliana, cpRPGs showed a positive correlation with expression of euRPGs and mtRPGs (Figure S3C, S3D in Additional File 1). Thus, while the expression levels of mtRPGs showed a strongly positive correlation with euRPGs under different developmental stages, the relationship between the expression of cpRPGs and euRPGs was developmental stage-dependent. Since expression data at developmental stage resolution are only available for Arabidopsis, it will be interesting to see if similar observations can be made in more plant species. Nonetheless, since the plants species we studied give a very good representation of the land plants (Figure S7 in Additional File 1), it is highly likely that the differential modulation of the transcriptional regulation of mtRPGs, cpRPGs, and euRPGs is a ubiquitous phenomenon for all land plants. However, the physiological significance of these patterns awaits further investigation.
Analyses of the promoter regions of RPGs help identify the cis-regulatory motifs potentially responsible for the different patterns of co-expression between RPGs in different cellular compartments (Figure 1). Three distinct cis-regulatory motifs were identified, two shared by mtRPGs, cpRPGs and euRPGs and one only existing in mtRPGs and euRPGs (Figure 2). The first motif, GCCCA, is called a site II motif, which is the binding site of a transcription factor known as At-TCP20 [31]. At-TCP20 is expressed in many different tissues in A. thaliana and can influence cell division and growth coordination [31]. The third motif, AAACCCT, called telo-box, is the binding site of the Purα transcription factor, which has been suggested to be a partner of the At-TCP20 [20]. Our analysis provided additional evidence for this viewpoint, since the telo-box is in close proximity to the site II motif in promoter regions of the RPGs (Figure S11 in Additional File 1). Therefore, both cis-elements could work as a module to coordinate gene expression, or they could also participate in controlling the cell cycle. The function of the second motif, GAAGAA, is not clear at this point, but it is preferentially enriched in chromosomal locations close to the other motifs (Figure S11 in Additional File 1), which indicates that its function might also be related to cell cycle control. Interestingly, the identified promoter motifs do not seem to be shared with either mammals or insects [17,32]. This result indicates that the regulatory mechanisms of ribosomal protein genes, one of the most conserved gene families, are highly evolvable and highlights the contribution of regulatory network changes in evolution, in addition to the contribution of gene sequences.
The special role of telo-box in coordinating DNA, protein synthesis, energy production and cell cycle
Telo-box, which is the binding site of the Purα transcription factor, clearly does not exist in chloroplasts (Figure 2 and 3). This motif (AAACCCT or AACCCTA) is homologous to a telomere repeat (AAACCCT)n of land plants, which is enriched in the ends of chromosomes [33]. Telo-box was first observed in promoters of translation elongation factor eEF1A [34-37] and subsequently found within the promoters of PCNA (proliferating cell nuclear antigen) and RNR (ribonucleotide reductase), both of which are over-expressed in cycling cells [19]. Our analysis further showed that telo-box was enriched in genes involved in nucleotide (DNA or RNA), protein binding and in processes ranging from "cell organization and biogenesis", "RNA processing and metabolism", to "protein translation and location" (Table 1). These results indicated that the telo-box motif likely functioned at the top of the hierarchy coordinating host and mitochondrion in these different processes.
Furthermore, this study indicated that the telo-box might be a major regulator of cell cycle activity, which is supported by the following evidences: 1) The telo-box motif is enriched in mtRPGs, euRPGs and genes of DNA replication machinery; 2) DNA replication, protein synthesis, and energy production by mitochondria are all required for normal cell cycle [38]; 3) cells need to synthesize large amounts of DNA and protein in order to increase cell size before mitosis [39,40]; 4) the transcription factor, At-Purα, which is a trans-factor for the telo-box, controls both gene transcription and DNA replication [41].
A hypothetical model for the differential acquisition of telo-box during organelle evolution
Both mitochondria and chloroplasts are the descendents of endosymbionts; however, mtRPGs showed a high level of co-expression with euRPGs, while cpRPGs did not (Figure 1). We have linked this phenomenon to a lack of telo-box in the promoter regions of cpRPGs (Figure 3), which indicates the possibility that telo-box might be the critical binding motif contributing to the difference in the expression of RPGs in different cellular compartments. This, in turn, has raised a number of important questions.
First, what is the origin of telo-box? The fact that the genomic sequence of ancestors of mitochondrion and chloroplast did not have telo-box indicated that mtRPGs acquired and successfully maintained the telo-box after endosymbiosis, while cpRPGs either did not acquire or failed to maintain the telo-box during the evolutionary process after endosymbiosis. Although cpRPGs have a relatively short evolutionary span (1.2~1.5 Ga) compared to mtRPGs (>1.5 Ga) [1], it is unlikely that the cpRPGs never acquired telo-box. In fact, after endosymbiosis, most of the genes in the endosymbionts' genome were transferred to the host nuclear genome [4,42,43] and formed a unique metabolic network of the current chloroplast [44,45]. Most of these genes have acquired new cis-regulatory motifs in their promoter sequences (Figure S12 in Additional File 1).
Secondly, why did cpRPGs fail to maintain the telo-box during evolution? Although telo-box could have been integrated into the promoter regions of cpRPGs, these regulatory elements were clearly selectively purged out after gene transfer. This indicates that a strong negative selection pressure may have resulted from simultaneous expression of cpRPGs with those of mtRPGs and euRPGs. One possible mechanism of this negative selection pressure is that photosynthesis generates oxygen, which can potentially generate reactive oxygen species under high light [46]. Reactive oxygen species, such as superoxide, not only cause direct damage to DNA [47], but also influence structure and, correspondingly, the function of proteins [48,49], including proteins involved in DNA replication and protein synthesis. As a result, it is disadvantageous to have photosynthesis occur simultaneously with the DNA replication and protein synthesis, which are required for normal cell cycle. Indeed, DNA replication usually occurs at midnight, quite possibly to avoid damaging DNA by UV radiation [50,51]. Therefore, the potential damage caused by concurrence of photosynthesis and processes related to cell cycle might have generated a strong negative selection pressure, which purged the telo-box from the cpRPGs.
Thirdly, what is the reason for the strong coordinated expression of mtRPGs and euRPGs? In all the examined land plant species, the expressions of mtRPGs and euRPGs are highly co-expressed (Figure 1). This co-expression does not seem to be dependent on developmental stage or growth conditions (Figure S3 in Additional File 1). This fact indicates that there is a strong positive selection for co-expression of mtRPGs and euRPGs. Again, the clue may come from the cell cycle. Two of the three conserved binding sites, Site II and telo-box, are related to cell cycle control [13,20,31]. Furthermore, the genes involved in DNA replication are highly positively correlated with mtRPGs and euRPGs and also harbor enriched Site II and telo-box motif in their promoters (Figure 4). In addition, the genes highly co-expressed with euRPGs or mtRPGs are enriched in nucleotide (DNA or RNA) and protein binding function (Table 1). This indicates that cell cycle might be coordinated with mitochondrial function. To enable a cell go through cell cycle, cells need to have large amounts of protein synthesized and have DNA replicated in order to reach a certain cell size [40]. Protein synthesis and DNA replication requires energy, which will be supplied by mitochondria. Therefore, a highly coordinated function of mitochondria and cell cycle might have created a positive selection pressure, which facilitated the maintenance of telo-box after mtRPGs gained it from the host nuclear genome.
Following the above reasoning, we proposed a model to explain the evolution of the promoter structure in mitochondria and chloroplast (Figure 5). In this model, after endosymbiosis, endosymbionts transferred most of their genes into the host nuclear genome. The transferred genes, in turn, acquired regulatory elements, including the telo-box, from the nuclear genome. During the evolutionary process, DNA damage by the reactive oxygen species from aberrant cpRPG expression created a negative selection pressure and purged telo-box from the cpRPGs, while, on the other hand, the high level of coordination between mitochondria function and cell cycle created a positive selection force, thus maintaining the telo-box in mtRPGs. Therefore, the selective removal or maintenance of telo-box in RPGs by possibly different mechanisms, one being negative selection force and another being positive selection force, created the dramatic differences we found in the expression pattern of these two organelles. Furthermore, these negative and positive selection forces might have been major forces in shaping the evolution of promoter structure in these two important organelles in plant cell.
Figure 5.
The model of plant nuclear genome evolution. (A) α-proteobacteria and cyanobacteria with RPGs not containing telo-box motif (purple curve) were engulfed in host cell, which possesses euRPGs with telo-boxes. (B) RPGs of endosymbionts (chloroplast and mitochondrion) are transferred into host nuclear genome. (CI) mtRPGs, but not cpRPGs, acquired telo-boxes from host nuclear genome. (CII) both mtRPGs and cpRPGs acquired telo-boxes from host nuclear genome. (DII) under negative selection pressures, telo-boxes of cpRPGs are purged. As a result, mtRPGs and euRPGs share telo-box and exhibit synchronized expression (thick red arrow). On the other hand, cpRPGs do not have telo-boxes in their promoter regions, and the expression coordination between cpRPGs and euRPGs is weak (dashed green arrow).
Conclusions
This study showed that mtRPGs, but not cpRPGs, displayed strongly correlated expression with euRPGs in land plants. This phenomenon is linked to a highly conserved cis-regulatory element, AAACCCT, known as the telo-box motif, which is present in promoters of cytoplasmic and mitochondrial RPGs, but not in cpRPGs. Considering the fact that the telo-box is also enriched in promoters of genes involved in DNA replication, it seems likely that coordination of mitochondria function (mainly ATP production) with other cellular functions might have been a strong positive selection pressure in shaping the genome structure of land plants. Similarly, the potential damage caused by the concurrence of photosynthesis and cell cycle might have created a strong negative selection pressure which purged telo-box from the promoters of cpRPGs. This study indicated that the gain and loss of a single cis-element, possibly by different reasons, could result in dramatic differences in transcriptional regulation between chloroplast and mitochondria in land plants (Figure 5).
Methods
Species
Five plant species with both gene expression data and genome sequence data available are included in this study: Arabidopsis thaliana (mouse-ear cress), Populus trichocarpa (black cottonwood), Medicago truncatula (barrel medic), Oryza sativa (rice) and Ectocarpus siliculosus (brown algae). Two other recently sequenced species, Selaginella moellendorffii (spikemoss) and Physcomitrella patens (moss), are also included in our analysis to demonstrate the conservation of discovered DNA motifs. In addition, we also include the green algae Chlamydomonas reinhardtii, which has complete genome sequence and gene expression data, although, unfortunately, its micro-array did not include mtRPGs. We thus measured the expression levels of its ribosomal protein genes by RT-PCR experiments as described in the section subtitled "RT-PCR experiment for RPGs in C. reinhardtii". The evolutionary relationship of the studied species is shown in Figure S7 in Additional File 1[25,52,53].
Catalogs of cytoplasmic and transferred organelle ribosomal protein genes
Sequence information for A. thaliana, M. truncatula and E. siliculosus was downloaded from http://www.ncbi.nlm.nih.gov/http://www.medicago.org/index.php and http://bioinformatics.psb.ugent.be/webtools/bogas/[25], respectively. Sequences for P. trichocarpa, O. sativa, S. moellendorffii, P. patens and C. reinhardtii were obtained from the U.S. Department of Energy Joint Genome Institute (http://genome.jgi-psf.org/). To collect a full catalog of cytoplasmic and transferred organelle ribosomal protein genes, BLASTP was used to search the protein sequences of all studied species, using E-value of 10-5 as the significance cutoff. To obtain nuclear-encoded cpRPGs and mtRPGs, ribosomal proteins encoded in Syn (Synechocystis sp. PCC6803, current-day cyanobacteria as proto-chloroplast ancestor [22,23]) and Rpr (Rickettsia prowazekii str. Madrid E, current-day α-proteobacteria as proto-mitochondrial ancestor [4,24]) were used as query sequences, respectively. However, some genes in Syn and Rpr may have been lost since the endosymbiosis events. Therefore, we further collected ribosomal protein genes still present in any of the currently available plant chloroplast (132 plant species) and mitochondrial (25 plant species) genomes to account for the above concern. In A. thaliana, cytoplasmic ribosomal protein genes (euRPGs) were obtained from NCBI. These well annotated euRPGs were further used to annotate euRPGs in other species (provided as Additional File 2).
We then used TargetP [54] to predict the cellular localization of the above collected proteins in each species. Since proteins were predicted to be targeted to a specific organelle with reliability class ≥ 3 are considered to be highly reliable [54], here only proteins with E-value < 10-5 by BLASTP and reliability class ≥ 3 by TargetP were selected as RPGs for corresponding organelles, respectively. For C. reinhardtii, euRPGs were obtained from the Ribosomal Protein Gene database [55], and cpRPGs were identified using the experimental results [56,57]. The promoter sequences and expression datasets of ribosomal protein genes for each class in each species are summarized in Additional File 2 and Additional File 3, respectively. The catalog of cytoplasmic and transferred organelle ribosomal protein genes is also provided in Table S1 in Additional File 1.
Gene expression data
To study the expression characteristics of ribosomal protein genes, large-scale microarray expression datasets were collected. The gene expression profile focusing on A. thaliana development [58], which includes various tissue samples across 4 developmental stages, i.e., 7th, 17th, 21st day and 8th week, was downloaded from http://www.weigelworld.org. The gene expression data for O. sativa and M. truncatula were downloaded from http://www.plexdb.org/. The gene expression data with platform number GPL5921 and GPL963 were downloaded from NCBI GEO for P. trichocarpa and the green algae C. reinhardtii, respectively. The gene expression data for E. siliculosus were downloaded from http://www.ebi.ac.uk/microarray-as/ae/ (accession numbers: E-TABM-578 [26]). Each array was first standardized to have a mean value of 0 and a standard deviation of 1. Probe sets corresponding to the same gene were collapsed to a single number by taking the mean value. Expression data for ribosomal protein genes were then extracted for further expression analysis (provided as Additional File 3). The gene expression data for special developmental stages in A. thaliana were extracted from whole expression data according to developmental stages of samples, as described [58]. In addition, tissue-specific protein expression level data for A. thaliana were obtained from http://fgcz-atproteome.unizh.ch/[59]. We used Pearson's Correlation Coefficient (PCC) to measure the co-expression between gene/protein pairs.
Promoter sequences of RPGs
To analyze the transcriptional regulatory mechanism of cpRPGs, mtRPGs and euRPGs, we retrieved the promoter sequences (upstream 1kb relative to the translation start codon) for each ribosomal protein gene in each species (also provided as Additional File 2). These sequences were further used to search for potential cis-regulatory motifs using the MEME software with parameters of 5-10 in width for motif discovery [12]. To avoid possible bias in motif discovery, other tools including AlignACE [14] and DME [15] were also used. Motif width was set to be 10 for both software, and the background sequences for DME were the promoters of non-RPGs. Background sequences used in Figure 3 were randomly selected from promoters (upstream 1kb) of genes except RPGs in each species. We bootstrapped fifty datasets, each consisting of the same number as the set of cpRPGs, mtRPGs and euRPGs, and then took the mean value and standard deviation as the motif density and corresponding errors of background sequences, respectively. The distances between RPGs and non-RPGs with telo-box were calculated according to their translation start codon. The non-RPGs located in the same chromosome and transcribed at the same orientation with RPGs were choose for calculating the shortest distances.
RT-PCR experiment for RPGs in C. reinhardtii
C. reinhardtii strain CC-503 cw92 mt+ [60] was cultured in liquid TAP medium [61] at 25°C under continuous light or dark. NH4Cl concentration in TAP medium (7 mM) was increased five-fold for the high nitrogen treatment and reduced ten-fold for the low nitrogen treatment. There were at least three replicates for each condition. Cells were collected at their mid-exponential phase of growth by centrifugation (4000 × g for 3 min). Isolation of total RNA was performed with the Triazol reagent (Invitrogen) according to instructions of the manufacturer. After DNase treatment, single-stranded cDNA was synthesized from total RNA according to the manual of PrimeScript II 1st Strand cDNA Synthesis Kit (TaKaRa) and used as templates for real-time PCR reactions. Real-time PCR was performed on the LightCycler® instrument (Bio-Rad CFX96 Real-time PCR Detection System) using SYBR Green as a fluorescent dye (iQ SYBR Green Supermix, Bio-Rad; 2x mixture contains 100 mM KCl, 40 mM Tris-HCl, pH 8.4, 0.4 mM each dNTP, 50 U/ml iTaq DNA polymerase, 6 mM MgCl2, SYBR Green I and 20 Nm fluorescein). Each individual reaction contains 1.0 pmole of the indicated primers (provided in Additional File 4) and 1 μl of 5-fold diluted single-stranded cDNA. The final volume of each reaction was 20 μl. PCR conditions were as follows: 10 min at 95°C for activation of the hot start Taq polymerase and 40 cycles for the melting (30s at 95°C), annealing (30s at 60°C) and extension (30s at 72°C). The fluorescence measurement was made at the end of the annealing step. The Ubiquitin ligase (Chlamydomonas GenBank ESTs: BU648530, BE237749, BE237718, BU648531) was used as the housekeeping gene ([62], the primer sequences are provided as Additional File 4). Expression of this gene was previously shown to be constitutive under the different conditions used [63]. For each condition and gene, we first filtered the undetected values, calculated the mean value of CT (cycle threshold), and then normalize the expression value with formula: 2[Mean value (CT) - Control value]/Control value. The resulted value was used as expression level for the analysis of expression correlation by calculating Pearson's Correlation Coefficient (PCC) of each gene pair (provided as Additional File 3).
List of Abbreviations
RPGs: Ribosomal Protein Genes; mtRPGs: mitochondrial RPGs; cpRPGs: chloroplast RPGs; euRPGs: cytoplasmic RPGs; non-RPGs: non-Ribosomal Protein Genes; PCC: Pearson's Correlation Coefficient.
Authors' contributions
JW and XM collected data. YW and XZ performed experiments on C. reinhardtii. JW, YW, ZW, LL, XZ and XM analyzed the data and wrote the manuscript. XZ proposed the evolutionary model to explain the data. XM conceived of the study. All authors read and approved the final manuscript.
Supplementary Material
Table S1 and Figures S1-S12. Table S1: Catalogs of cytoplasmic and transferred organelle ribosomal protein genes. Figure S1: Occurrence frequency of RPGs in 132 plant chloroplast and 25 plant mitochondrial genomes. (A) More than half of chloroplast RPGs are absent in most chloroplast genomes of 132 plants surveyed (green dash line), providing a raw estimate of the lower bound of transfer frequency of cpRPGs. (B) Similarly, there are over 50 RPGs in Rickettsia prowazekii str. Madrid E (α-proteobacteria, an ancient cousin of mitochondrion), whereas only 18 mtRPGs can be found in the union of all 25 plant mitochondrial genomes and most mtRPGs are absent in over half studied species (red dash line). Genes with zero occurrence frequency are present in corresponding ancient cousin but absent in chloroplast/mitochondria of all studied plants. Figure S2: Protein expression correlation of mtRPGs, cpRPGs and euRPGs in A. thaliana. Each element of the matrix represents the Pearson's correlation coefficient between the expression profiles of two ribosomal proteins. Figure S3: RNA expression correlation of mtRPGs, cpRPGs and euRPGs in different tissues of several developmental stages in A. thaliana. 7th day (A), 17th day (B), 21st day (C) and 8th week (D) corresponds to stage of seedling, leaf, flower and seed of A. thaliana, respectively. Each element of the matrix represents the Pearson's correlation coefficient of the expression profiles of each two RPGs. Color code is illustrated in bottom panel. Figure S4: Putative promoter motifs only enriched in one of cpRPGs, mtRPGs, and euRPGs in A. thaliana. The number on the left of each logo is E-value of MEME prediction. Figure S5: Putative promoter motifs predicted by AlignACE and DME. The number on the left of each logo is score of AlignACE and DME prediction. Figure S6: Expression correlation between RPGs with telo-box and those without telo-box. Telo-box here indicates the sequence AAACCCT or AACCCTA. The error-bar is the standard deviation of expression correlation. A symbol of the three stars (***) indicates the p-value is less than 0.001. Figure S7: The phylogenetic relationship of all investigated species. The phylogenetic relationship of angiospermae is derived according to the 16S rRNA of chloroplast genome in each species, using the neighbor-joining method. Both gene expression data and genome sequence data are available for O. sativa, A. thaliana, P. trichocarpa, M. truncatula and E. siliculosus, whereas only genome sequence data are available for P. patens and S. moellendorffii. The current gene expression data for C. reinhardtii do not include mtRPGs. Gray fonts indicate species that are not sequenced yet. Branch lengths are not scaled to time. Figure S8: Alignment of Purα proteins in all studied land plants. Some species have two Purα proteins. Here three highly conserved domains of Purα are shown. The tree on the left is drawn based on the sequences of Purα proteins with neighbor-joining method. The sequence labeled with the cyan line is involved in DNA-binding. Asterisks, colons, and dots indicate identical (red), strongly similar (green) and weakly similar (blue) residues, respectively. Figure S9: The occurrence of telo-boxes (A) and non-RPGs with telo-box (B) relative to mtRPGs or cpRPGs: (A) The star symbol indicates the significance of the occurrence within -1kb for mtRPGs. (B) The insets indicates mean distances of mtRPGs (cyan) and cpRPGs (magenta) relative to their closest upstream (left) or downstream (right) non-ribosomal protein genes (non-RPGs) with telo-box, respectively. The error-bar in the insets is the standard deviation of the distances. Figure S10: RNA Expression correlation of mtRPGs, cpRPGs and euRPGs in brown (A) and green (B) algae. Each element of the matrix represents the Pearson's correlation coefficient between the expression profiles of two RPGs. RNA expression level is measured by RT-PCR experiment for green algae C. reinhardtii (see Methods). Figure S11: Positional distribution of Site II, GAAGAA and telo-box of RPGs in A. thaliana. Site II motif (GCCCA, magenta line), GAAGAA (dark green line) and telo-box motif (AAACCCT, cyan line) are close to each other in promoter regions of RPGs. Figure S12: Promoter motifs of nuclear-encoded chloroplast genes in A. thaliana. (A) The logos of the motifs are predicted with MEME. The upper-left number of each logo is E-value. (B) The motif density of nuclear-encoded chloroplast genes (NCGs) in A. thaliana (Ath) and corresponding genes in Synechocystis sp. PCC6803 (Syn). The nuclear-encoded chloroplast genes in A. thaliana are identified by Martin et al. (2002).
The promoter sequences of RPGs for studied species
The expression levels of RPGs for studied species
The primers of RPGs for RT-PCR experiment in C. reinhardtii
Contributor Information
Jie Wang, Email: wjwmss@gmail.com.
Yu Wang, Email: wangyu@picb.ac.cn.
Zhuo Wang, Email: zhuowang@sjtu.edu.cn.
Lei Liu, Email: leiliu@sibs.ac.cn.
Xin-Guang Zhu, Email: zhuxinguang@picb.ac.cn.
Xiaotu Ma, Email: xiaotuma@utdallas.edu.
Acknowledgements
We thank Keyan Zhao and Guangyong Zheng for suggestions of gene expression data and protein expression analysis. XZ and YW were supported by Chinese National Science Foundation general program 30970213, Shanghai Institutes for Biological Sciences (grant number: 2009GS-YJ-04) and Shanghai government Pujiang program; XM was partly supported by NIH grant HG001696 to Michael Q. Zhang. ZW was supported by grants from the National Natural Science Foundation of China (30800199). The authors are grateful for the helpful suggestions from two anonymous reviewers. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- Glockner G, Rosenthal A, Valentin K. The structure and gene repertoire of an ancient red algal plastid genome. J Mol Evol. 2000;51:382–390. doi: 10.1007/s002390010101. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Heazlewood JL, Kristensen B, Bruan HP, Moller IM. The Plant Mitochondrial Proteome. Trends Plant Sci. 2005;10:36–43. doi: 10.1016/j.tplants.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Giege P, Sweetlove LJ, Cognat V, Leaver CJ. Coordination of Nuclear and Mitochondrial Genome Expression during Mitochondrial Biogenesis in Arabidopsis. Plant Cell. 2005;17:1497–1512. doi: 10.1105/tpc.104.030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean D, Jerome CA, Brown APC, Gary JC. Co-regulation of nuclear genes encoding plastid ribosomal proteins by light and plastid signals during seedling development in tobacco and Arabidopsis. Plant Mol Biol. 2008;66:475–490. doi: 10.1007/s11103-007-9279-z. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. Plastidial retrograde signalling - a true "plastid factor" or just metabolite signatures? Trends Plant Sci. 2010;15:427–435. doi: 10.1016/j.tplants.2010.05.009. [DOI] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raf M, Roberts K, Walter P. The molecular biology of the cell. 4. Garland Science, Abingdon, New York; 2002. p. 1268 pp. [Google Scholar]
- Byrne ME. A role for the ribosome in development. Trends Plant Sci. 2009;14:512–519. doi: 10.1016/j.tplants.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Schippers JHM, Mueller-Roeber B. Ribosomal composition and control of leaf development. Plant Sci. 2010;179:307–315. doi: 10.1016/j.plantsci.2010.06.012. [DOI] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Li C, Potuschak T, Colon-Carmona A, Gutierrez RA, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA. 2005;102:12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. Journal of Molecular Biology. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Zhang MQ. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc Natl Acad Sci USA. 2005;102:1560–1565. doi: 10.1073/pnas.0406123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang K, Li X. Evolution of Drosophila ribosomal protein gene core promoters. Gene. 2009;432:54–59. doi: 10.1016/j.gene.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FW, Ioannou YA. Ribosomal proteins in cell proliferation and apoptosis. Int Rev Immunol. 1999;18:429–448. doi: 10.3109/08830189909088492. [DOI] [PubMed] [Google Scholar]
- Manevski A, Bertoni G, Bardet C, Tremousaygue D, Lescure B. In synergy with various cis-acting elements, plant insterstitial telomere motifs regulate gene expression in Arabidopsis root meristems. FEBS Lett. 2000;483:43–46. doi: 10.1016/S0014-5793(00)02056-1. [DOI] [PubMed] [Google Scholar]
- Tremousaygue D, Garnier L, Bardet C, Dabos P, Herve C, Lescure B. Internal telomeric repeats and 'TCP domain' protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J. 2003;33:957–966. doi: 10.1046/j.1365-313X.2003.01682.x. [DOI] [PubMed] [Google Scholar]
- Tremousaygue D, Manevski A, Bardet C, Lescure N, Lescure B. Plant interstitial telomere motifs participate in the control of gene expression in root meristems. Plant J. 1999;20:553–561. doi: 10.1046/j.1365-313X.1999.00627.x. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR. Mitochondrial origins. Proc Natl Acad Sci USA. 1985;82:4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA, Allen JF. Genomics and chloroplast evolution: what did cyanobacteria do for plants? Genome Biol. 2003;4:209. doi: 10.1186/gb-2003-4-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouze P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, Beszteri B, Billiau K, Bonnet E, Bothwell JH, Bowler C, Boyen C, Brownlee C, Carrano CJ, Charrier B, Cho GY, Coelho SM, Collen J, Corre E, Silva CD, Delage L, Delaroque N, Dittami SM, Doulbeau S, Elias M, Farnham G, Gachon CMM, Gschloessl B, Heesch S, Jabbari K, Jubin C, Kawai H, Kimura K, Kloareg B, Kupper FC, Lang D, Bail AL, Leblanc C, Lerouge P, Lohr M, Lopez PJ, Martens C, Maumus F, Michel G, Miranda-Saavedra D, Morales J, Moreau H, Motomura T, Nagasato C, Napoli CA, Nelson DR, Nyvall-Collen P, Peters AF, Pommier C, Potin P, Poulain J, Quesneville H, Read B, Rensing SA, Ritter A, Rousvoal S, Samanta M, Samson G, Schroeder DC, Segurens B, Strittmatter M, Tonon T, Tregear JW, Valentin K, von Dassow P, Yamagishi T, Van de Peer Y, Wincker P. The Ectocarpus genome and the independent evolution of multicellularity in the brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Dittami SM, Scornet D, Petit JL, Segurens B, Silva CD, Corre E, Dondrup M, Glatting KH, Konig R, Sterck L, Rouze P, Van de Peer Y, Cock JM, Boyen C, Tonon T. Global expression analysis of the brown alga Ectocarpus siliculosus (Phaeophyceae) reveals large-scale reprogramming of the transcriptome in response to abiotic stress. Genome Biol. 2009;10:R66. doi: 10.1186/gb-2009-10-6-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J. Chlamydomonas Annotation Team; JGI Annotation Team. Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–251. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir A. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci USA. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Li W, Saunders MA. The chimpanzee and us. Nature. 2005;437:50–51. doi: 10.1038/437050a. [DOI] [PubMed] [Google Scholar]
- Herve C, Dabos P, Bardet C, Jauneau A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 2009;149:1462–1477. doi: 10.1104/pp.108.126136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP. The architecture of mammalian ribosomal protein promoters. BMC Evol Biol. 2005;5:15. doi: 10.1186/1471-2148-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- Axelos M, Bardet C, Liboz T, Le Van Thai A, Curie C, Lescure B. The gene family encoding the Arabidopsis thaliana translation elongation factor EF-1α: molecular cloning, characterization and expression. Mol Gen Genet. 1989;219:106–112. doi: 10.1007/BF00261164. [DOI] [PubMed] [Google Scholar]
- Curie C, Liboz T, Bardet C, Gander E, Medale C, Axelos M, Lescure B. Cis and trans-acting elements involved in the activation of the Arabidopsis thaliana A1 gene encoding the translation elongation factor EF-1α. Nucleic Acids Res. 1991;19:1305–1310. doi: 10.1093/nar/19.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liboz T, Bardet C, Le Van Thai A, Axelos M, Lescure B. The four members of the gene family encoding the Arabidopsis thaliana translation elongation factor EF-1α are actively transcribed. Plant Mol Biol. 1989;14:107–110. doi: 10.1007/BF00015660. [DOI] [PubMed] [Google Scholar]
- Shewmaker CK, Ridge NP, Pokalsky AR, Rose RE, Hiatt WR. Nucleotide sequence of an EF-1 alpha genomic clone from tomato. Nucleic Acids Res. 1990;18:4276. doi: 10.1093/nar/18.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaloe O, Kjeligard NO. Control of macromolecular synthesis. WA Benjamin, Inc, New York; 1966. [Google Scholar]
- Beemster GTS, Fiorani F, Inz D. Cell cycle: the key to plant growth control? Trends Plant Sci. 2003;8:154–158. doi: 10.1016/S1360-1385(03)00046-3. [DOI] [PubMed] [Google Scholar]
- Tyson JJ, Chen K, Novak B. Network dynamics and cell physiology. Nat Rev Mol Cell Biol. 2001;2:908–916. doi: 10.1038/35103078. [DOI] [PubMed] [Google Scholar]
- Bergemann AD, Johnson EM. The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol. 1992;12:1257–1265. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nature. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhu XG, Chang X, Chen YZ, Li YX, Liu L. Though with constraints imposed by endosymbiosis, preferential attachment is still a plausible mechanism responsible for the evolution of the chloroplast metabolic network. J Evol Biol. 2009;22:71–79. doi: 10.1111/j.1420-9101.2008.01621.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhu XG, Chen YZ, Li YY, Hou J, Li Y, Liu L. Exploring photosynthesis evolution by comparative analysis of metabolic networks between chloroplasts and photosynthetic bacteria. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss I, Koelmann M, Wojtera J, Holtgrefe S, Kitzmann C, Backhausen JE, Scheibe R. Knockout of major leaf ferredoxin reveals new redox-regulatory adaptations in Arabidopsis thaliana. Physiol Plant. 2008;133:584–598. doi: 10.1111/j.1399-3054.2008.01112.x. [DOI] [PubMed] [Google Scholar]
- Arai T, Kelly VP, Minowa O, Noda T, Nishimura S. High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis. 2002;23:2005–2010. doi: 10.1093/carcin/23.12.2005. [DOI] [PubMed] [Google Scholar]
- Ledford HK, Niyogi KK. Singlet oxygen and photo-oxidative stress management in plants and algae. Plant Cell and Environ. 2005;28:1037–1045. doi: 10.1111/j.1365-3040.2005.01374.x. [DOI] [Google Scholar]
- Niyogi KK. Photoprotection revisited: Genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- Pregueiro AM, Liu QY, Baker CL, Dunlap JC, Loros JJ. The Neurospora checkpoint kinase 2: A regulatory link between the circadian and cell cycles. Science. 2006;313:644–649. doi: 10.1126/science.1121716. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol and Cell Biol. 2005;25:3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M. Genomes, phylogeny, and evolutionary systems biology. Proc Natl Acad Sci USA. 2005;102:6630–6635. doi: 10.1073/pnas.0501984102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nakao A, Yoshihama M, Kenmochi N. RPG: the ribosomal protein gene database. Nucleic Acids Res. 2004;32:D168–D170. doi: 10.1093/nar/gkh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Beligni MV, Prieto S, Haynes PA, McDonald WH, Yates JR III, Mayfield SP. Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome: identification of proteins unique to the 70S ribosome. J Biol Chem. 2003;278:33774–33785. doi: 10.1074/jbc.M301934200. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Prieto S, Beligni MV, Haynes PA, McDonald WH, Yates JR III, Mayfield SP. Proteomic characterization of the small subunit of Chlamydomonas reinhardtii chloroplast ribosome: identification of a novel S1 domain-containing protein and unusually large orthologs of bacterial S2, S3, and S5. The Plant Cell. 2002;14:2957–2974. doi: 10.1105/tpc.004341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- Hyams J, Davies DR. The induction and characterization of cell wall mutants of Chlamydomonas reinhardi. Mutat Res. 1972;14:381–389. [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila M, Couso I, Leon R. Carotenoid content in mutants of the chlorophyte Chlamydomonas reinhardtii with low expression levels of phytoene desaturase. Proc Biochem. 2008;43:1147–1152. doi: 10.1016/j.procbio.2008.06.014. [DOI] [Google Scholar]
- Gonzalez-Ballester D, Camargo A, Fernandez E. Ammonium transported genes in Chlamydomonas: the nitrate-specific regulatory gene Nit2 is involved in Amt1;1 expression. Plant Mol Biol. 2004;56:863–878. doi: 10.1007/s11103-004-5292-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 and Figures S1-S12. Table S1: Catalogs of cytoplasmic and transferred organelle ribosomal protein genes. Figure S1: Occurrence frequency of RPGs in 132 plant chloroplast and 25 plant mitochondrial genomes. (A) More than half of chloroplast RPGs are absent in most chloroplast genomes of 132 plants surveyed (green dash line), providing a raw estimate of the lower bound of transfer frequency of cpRPGs. (B) Similarly, there are over 50 RPGs in Rickettsia prowazekii str. Madrid E (α-proteobacteria, an ancient cousin of mitochondrion), whereas only 18 mtRPGs can be found in the union of all 25 plant mitochondrial genomes and most mtRPGs are absent in over half studied species (red dash line). Genes with zero occurrence frequency are present in corresponding ancient cousin but absent in chloroplast/mitochondria of all studied plants. Figure S2: Protein expression correlation of mtRPGs, cpRPGs and euRPGs in A. thaliana. Each element of the matrix represents the Pearson's correlation coefficient between the expression profiles of two ribosomal proteins. Figure S3: RNA expression correlation of mtRPGs, cpRPGs and euRPGs in different tissues of several developmental stages in A. thaliana. 7th day (A), 17th day (B), 21st day (C) and 8th week (D) corresponds to stage of seedling, leaf, flower and seed of A. thaliana, respectively. Each element of the matrix represents the Pearson's correlation coefficient of the expression profiles of each two RPGs. Color code is illustrated in bottom panel. Figure S4: Putative promoter motifs only enriched in one of cpRPGs, mtRPGs, and euRPGs in A. thaliana. The number on the left of each logo is E-value of MEME prediction. Figure S5: Putative promoter motifs predicted by AlignACE and DME. The number on the left of each logo is score of AlignACE and DME prediction. Figure S6: Expression correlation between RPGs with telo-box and those without telo-box. Telo-box here indicates the sequence AAACCCT or AACCCTA. The error-bar is the standard deviation of expression correlation. A symbol of the three stars (***) indicates the p-value is less than 0.001. Figure S7: The phylogenetic relationship of all investigated species. The phylogenetic relationship of angiospermae is derived according to the 16S rRNA of chloroplast genome in each species, using the neighbor-joining method. Both gene expression data and genome sequence data are available for O. sativa, A. thaliana, P. trichocarpa, M. truncatula and E. siliculosus, whereas only genome sequence data are available for P. patens and S. moellendorffii. The current gene expression data for C. reinhardtii do not include mtRPGs. Gray fonts indicate species that are not sequenced yet. Branch lengths are not scaled to time. Figure S8: Alignment of Purα proteins in all studied land plants. Some species have two Purα proteins. Here three highly conserved domains of Purα are shown. The tree on the left is drawn based on the sequences of Purα proteins with neighbor-joining method. The sequence labeled with the cyan line is involved in DNA-binding. Asterisks, colons, and dots indicate identical (red), strongly similar (green) and weakly similar (blue) residues, respectively. Figure S9: The occurrence of telo-boxes (A) and non-RPGs with telo-box (B) relative to mtRPGs or cpRPGs: (A) The star symbol indicates the significance of the occurrence within -1kb for mtRPGs. (B) The insets indicates mean distances of mtRPGs (cyan) and cpRPGs (magenta) relative to their closest upstream (left) or downstream (right) non-ribosomal protein genes (non-RPGs) with telo-box, respectively. The error-bar in the insets is the standard deviation of the distances. Figure S10: RNA Expression correlation of mtRPGs, cpRPGs and euRPGs in brown (A) and green (B) algae. Each element of the matrix represents the Pearson's correlation coefficient between the expression profiles of two RPGs. RNA expression level is measured by RT-PCR experiment for green algae C. reinhardtii (see Methods). Figure S11: Positional distribution of Site II, GAAGAA and telo-box of RPGs in A. thaliana. Site II motif (GCCCA, magenta line), GAAGAA (dark green line) and telo-box motif (AAACCCT, cyan line) are close to each other in promoter regions of RPGs. Figure S12: Promoter motifs of nuclear-encoded chloroplast genes in A. thaliana. (A) The logos of the motifs are predicted with MEME. The upper-left number of each logo is E-value. (B) The motif density of nuclear-encoded chloroplast genes (NCGs) in A. thaliana (Ath) and corresponding genes in Synechocystis sp. PCC6803 (Syn). The nuclear-encoded chloroplast genes in A. thaliana are identified by Martin et al. (2002).
The promoter sequences of RPGs for studied species
The expression levels of RPGs for studied species
The primers of RPGs for RT-PCR experiment in C. reinhardtii