Abstract
Zwittermicin A represents a new chemical class of antibiotic and has diverse biological activities, including suppression of oomycete diseases of plants and potentiation of the insecticidal activity of Bacillus thuringiensis. To identify genes involved in zwittermicin A production, we generated 4,800 transposon mutants of B. cereus UW101C and screened them for zwittermicin A accumulation. Nine mutants did not produce detectable zwittermicin A, and one mutant produced eightfold more than the parent strain. The DNA flanking the transposon insertions in six of the nine nonproducing mutants contains significant sequence similarity to genes involved in peptide and polyketide antibiotic biosynthesis. The mutant that overproduced zwittermicin A contained a transposon insertion immediately upstream from a gene that encodes a deduced protein that is a member of the MarR family of transcriptional regulators. Three genes identified by the mutant analysis mapped to a region that was previously shown to carry the zwittermicin A self-resistance gene, zmaR, and a biosynthetic gene (E. A. Stohl, J. L. Milner, and J. Handelsman, Gene 237:403-411, 1999). Further sequencing of this region revealed genes proposed to encode zwittermicin A precursor biosynthetic enzymes, in particular, those involved in the formation of the aminomalonyl- and hydroxymalonyl-acyl carrier protein intermediates. Additionally, nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) homologs are present, suggesting that zwittermicin A is synthesized by a mixed NRPS/PKS pathway.
The virtuosity of bacteria as synthetic chemists is unparalleled in nature. Bacteria produce structurally diverse compounds with a wide spectrum of activities that have promoted their use by humans for numerous purposes. Antibiotics are key among the compounds of microbial origin, providing the basis for treatment of infectious diseases of humans, animals, and plants. The need for new antibiotics has intensified in recent years due to the emergence of new pathogens and development of resistance among old ones (18, 57), but the yield of new antibiotics from traditional searches has slowed, with a rediscovery rate exceeding 99% (61). The confluence of these trends has focused attention on new sources of microbial products and new approaches to synthesize antimicrobial agents. Therefore, two intriguing issues accompanied the discovery of the novel antibiotic zwittermicin A (48). First, it was surprising that decades of exhaustive screening of soil microorganisms for secondary metabolites had missed this antibiotic, which is produced by Bacillus cereus, an abundant, readily culturable member of most soil communities (38, 50). Second, zwittermicin A represents a new structural class of antibiotic, presenting the possibility that the enzymes involved in its synthesis could contribute to the chemical diversity produced by construction of hybrid biosynthetic pathways or protein domain shuffling (6, 24-26).
Zwittermicin A is a linear aminopolyol (Fig. 1) (22) that was first identified for its role in suppression of plant disease by B. cereus UW85 (48). It has a broad spectrum of activity, inhibiting certain gram-positive, gram-negative, and eukaryotic microorganisms (49). It also potentiates the insecticidal activity of the protein toxin produced by Bacillus thuringiensis, increasing mortality of insects that are typically recalcitrant to killing, such as gypsy moths reared on willow leaves (3, 4).
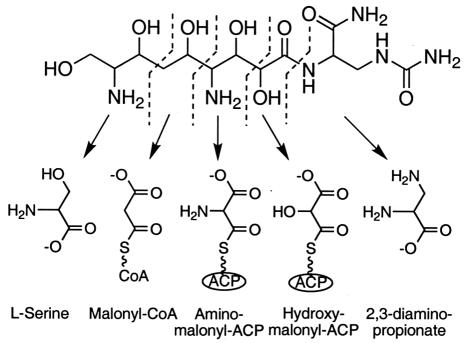
FIG. 1.
Chemical structure of zwittermicin A and the proposed precursors for its biosynthesis. The dashed lines delineate the individual precursors in the final molecule, while the arrows point to the corresponding precursor shown at the bottom. It is anticipated the 2,3-diaminopropionate component of zwittermicin A will be carbamoylated and amidated at some point after incorporation into the mixed nonribosomal peptide/polyketide backbone of the molecule.
The unusual structure and diverse biological activities of zwittermicin A make the pathway for its synthesis interesting and potentially useful. In an attempt to isolate the zwittermicin A biosynthetic genes, we previously identified the zwittermicin A self-resistance gene (zmaR) and some of the genes involved in biosynthesis that are adjacent to zmaR (33, 52). Here we report the use of an extensive mutant analysis to identify genes involved in zwittermicin A production, which appear to be related to genes involved in production of peptide and polyketide antibiotics. The portion of the zwittermicin A biosynthetic pathway presented here gives insights into aminomalonyl- and hydroxymalonyl-acyl carrier protein (ACP) formation, which expands the repertoire of precursors available for the biosynthesis of important natural products.
MATERIALS AND METHODS
Strains and plasmids.
Bacterial strains, plasmids, and phage used in this study are listed in Table 1.
TABLE 1.
Bacterial strains, plasmids, and phage used in this study
| Strain, plasmid, or phage | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | General purpose strain | 20 |
| E. coli GM2929 | dcm-6 dam-13::Tn9 | E. coli Genetic Stock Center |
| B. cereus UW85 | Wild type, ZmA+ | 21 |
| B. cereus UW101C | UW85 cured of pBC85 plasmid, ZmA+ | 47 |
| B. cereus UW11 | UW101C with Tn5401, ZmA− | This study |
| B. cereus UW32 | UW101C with Tn5401, ZmA− | This study |
| B. cereus UW52 | UW101C with Tn5401, ZmA− | This study |
| B. cereus UW56 | UW101C with Tn5401, ZmA+++ | This study |
| B. cereus UW64 | UW101C with Tn5401, ZmA− | This study |
| B. cereus UW78 | UW101C with Tn5401, ZmA− | This study |
| B. cereus UW89 | UW101C with Tn5401, ZmA− | This study |
| B. cereus UW96 | UW101C with Tn5401, ZmA− | This study |
| B. cereus UW119 | UW101C with Tn5401, ZmA− | This study |
| B. cereus UW120 | UW101C with Tn5401, ZmA− | This study |
| B. cereus UW85/pAD123 | UW85 with pAD123 | 13 |
| B. cereus UW85 ΔmarR | UW85 derivative containing an in-frame deletion in marR | This study |
| B. cereus UW101C ΔmarR | UW101C derivative containing an in-frame deletion in marR | This study |
| E. herbicola LS005 | Indicator strain for ZmA production | 48 |
| Plasmids | ||
| pEG922 | Carries Tn5401, temperature-sensitive shuttle vector, Cmr Tetr | 2 |
| pBeloBAC11 | BAC vector, Cmr | 27 |
| pDG1726 | Spectinomycin resistance cassette | 19 |
| pUC18 | General-use E. coli cloning vector | 59 |
| pSPUC | pUC18 containing Spr gene from pDG1726 cloned into the SmaI site | This study |
| pAD123 | B. cereus/E. coli shuttle vector, Cmr | 13 |
| Phage Pjer | B. cereus transducing phage | J. Dodsworth, J. Gross, and J. Handelsman, unpublished data |
ZmA+, strain produces zwittermicin A; ZmA−, strain produces no detectable zwittermicin A; ZmA+++, strain produces more zwittermicin A than the wild type. Cmr, chloramphenicol resistance; Tetr, tetracycline resistance; Spr, spectinomycin resistance.
Tn5401 mutants of B. cereus UW101C.
We transformed plasmid pEG922 into B. cereus UW101C by the method of Silo-Suh et al. (48), and transformants were selected on 0.5× tryptic soy agar (TSA) containing 10 μg of tetracycline/ml. pEG922 carries a version of Tn5401 that has been tagged with a gene for tetracycline resistance (2). Mutants of UW101C, which is cured of pBC85, were generated instead of UW85 because of the tendency of Tn5401 to preferentially insert into plasmids (2). To construct Tn5401 mutants in UW101C, individual colonies of UW101C carrying pEG922 were inoculated into 0.5× tryptic soy broth (TSB) containing 10 μg of tetracycline/ml. The cultures were grown with shaking overnight at 28°C and subcultured (1:100 dilution) into 0.5× TSB and grown for 6 to 8 h with shaking at 42°C. These cultures were then diluted 100-fold in water, and 100-μl samples of the dilute culture were plated on 0.5× TSA containing 10 μg of tetracycline/ml. The plates were incubated overnight at 43°C. Large and small colonies were present after the overnight incubation. Individual colonies were patched onto 0.5× TSA containing 10 μg of tetracycline/ml and onto 0.5× TSA containing 5 μg of chloramphenicol/ml. Small colonies were often chloramphenicol resistant, and large ones were often chloramphenicol sensitive. Southern analysis confirmed that chloramphenicol-sensitive, tetracycline-resistant colonies contained Tn5401 insertions. From each culture subjected to this regimen, 24 or 48 Tn5401 mutants were generated. Southern analysis of 18 mutants taken from the same culture indicated that each mutant contained only one insertion and the insertion in each mutant was in a different genomic location (data not shown). Using this protocol, we generated approximately 4,800 Tn5401 mutants in UW101C.
Screening and phenotypic characterization of mutants.
Each mutant was screened for the ability to inhibit Erwinia herbicola, which is an indicator of zwittermicin A production (48, 50). We scored zones of inhibition after 2 days incubation at 28°C and retested mutants that failed to inhibit E. herbicola. To verify that the size of the zone of inhibition was predictive of zwittermicin A production, we purified zwittermicin A from B. cereus cultures, using Sep-Pak cartridges as described previously (32), and quantified it by endpoint dilution using high-voltage paper electrophoresis (48). The limit of detection of zwittermicin A was 1.5 μg/ml of culture. This detection limit is higher than in previous studies that used this method (32, 48). Since the previous studies were conducted, we have refined zwittermicin A purification and have discovered that previous preparations of known amounts of zwittermicin A contained a significant amount of salt, likely accounting for the discrepancies in detection limit.
Cloning and sequencing of DNA regions involved in zwittermicin A production.
We prepared genomic DNA of each mutant according to the protocol recommended in the Easy-DNA kit (Invitrogen, Carlsbad, Calif.) except for the addition of vortexing the cells for 4 min in the presence of 0.1-mm diameter silica beads before addition of solution A. Genomic DNA was digested with PstI, which does not cut within the transposon sequence, cloned into pUC18 in Escherichia coli DH5α, and plated on Luria-Bertani agar containing 5 μg of tetracycline/ml. Cycle sequencing reactions were performed with the Big Dye Ready Reaction mix (Perkin-Elmer Corp., Foster City, Calif.) by using primers P1 and P2, which were designed based on the sequence of each end of the transposon (Table 2). Sequencing gels were run at the University of Wisconsin—Madison Biotechnology Center, on an ABI 377 automated DNA sequencer.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) | Use |
|---|---|---|
| P1 | GGTCTTCTGAATCGAAGAACC | Sequencing out of the ends of Tn5401 |
| P2 | CCCAGAAGAAGTAAAAGATGGG | |
| P3 | GGAGTAACCTTTTGATGCC | Sequencing into the unique HpaI site in Tn5401 |
| P4 | CCACCTGCGAGTACAAACTGG | |
| UW11 PF | GGTATTTGGAGTATTGGCG | Probe for hybridization |
| UW11 PR | CAGTCGCTCCAGTTAGATTACC | |
| UW32 PF | GGATTACCTGTTATTCCAACGC | Probe for hybridization |
| UW32 PR | CATAGACGCTGCTATCATTGG | |
| UW52 PF | GGTATCAGTAGATTTTGGGG | Probe for hybridization |
| UW52 PR | CCTGTTTGTTTGATGTCCGG | |
| UW56 PF | GGTAAAGATGGTCTTCACTCCC | Probe for hybridization |
| UW56 PR | GGGCAATATTTGTACCTTGTCG | |
| UW64 PF | GACATCGTGTATACCAGTCGG | Probe for hybridization |
| UW64 PR | CTTCATCAGCTACATCTGCACC | |
| UW78 PF UW78 PF | CATGGAGATCGTTGAGGGG GATAACAGCAACCTCATCTGGC | Probe for hybridization; PCR amplification and sequencing of region flanking Tn5401 in UW78 |
| UW89 PF | GAAAGATCAGGTTCTTCCGCC | Probe for hybridization |
| UW89 PR | GTGCATTTGTCCACCAATACC | |
| UW96 PF | GCTTATCCTCATACCTTAACCG | Probe for hybridization |
| UW96 PR | GTAAGCGCTAATTCCAAGACC | |
| UW119 PF | GGCTAGGCAACAATTAGGAGC | Probe for hybridization |
| UW119 PR | GTCCTCATCCATTACTTCACC | |
| UW120 PF | GGATTACCTGTAGTTCCAGAGG | Probe for hybridization |
| UW120 PR | CTCCTGTTATGCGATTTGGCC | |
| marRup-F marRup-R | CAGTGGATCCTACCTGATGAGTAGAAGTG CATTGCATGCTGCTGTTGTTCGGTCTAC | Construction of ΔmarR mutant |
| marRdown-F | CTGAGCATGCCTTCTTCAAAGAGTGAGG | Construction of ΔmarR mutant |
| marRdown-R | CCACAAGCTTCGTCACATCGCTATCAAGC | |
| orf2-a | CTCCTATGATGCAACCTGC | PCR amplification and sequencing of region flanking Tn5401 in UW78 |
Inverse PCR primers were designed to amplify out from the ends of Tn5401 and into the unique HpaI site in Tn5401 (Table 2). Primers were synthesized by the University of Wisconsin Biotechnology Center. Genomic DNA from each mutant was used as template for PCR with Taq Plus Long polymerase (Stratagene, La Jolla, Calif.). HpaI-digested DNA was extracted with phenol-chloroform, precipitated with ethanol, self-ligated, and used as template for inverse PCR. PCR conditions consisted of an initial hot melt at 95°C for 3 min followed by 30 cycles of 30 s at 95°C, 30 s at 55°C, and 5 min at 72°C and a final extension at 72° for 7 min. PCR products were partially purified by using the Qiaquick PCR purification kit (Qiagen Inc., Valencia, Calif.).
A 3-kb region adjacent to the transposon in UW78 was PCR amplified from UW85, using primers based on sequences obtained by inverse PCR (UW78 PR/orf2-a). The PCR product was purified using the Qiaquick PCR Purification Kit and sequenced by primer walking. Further sequence downstream from the 3-kb region was obtained by sequencing zmaR-containing clones HM11 and HG90 from a bacterial artificial chromosome (BAC) library (42, 52) by primer walking. BAC DNA was isolated by using the Qiagen Large-Construct Kit. The contig was assembled and open reading frame (ORF) analysis was performed using DNASTAR software (Madison, Wis.).
Reconstruction of transposon mutants via transduction.
Phage Pjer was propagated on each zwittermicin A mutant on 0.1× TSA containing 10 μg of tetracycline/ml. The phage lysate was mixed with an overnight culture of UW85/pAD123 at multiplicities of infection of 0.1 and 1 and plated on 0.1× TSA containing 10 μg of tetracycline/ml and 7.5 μg of chloramphenicol/ml. Resulting Tetr Cmr colonies were analyzed by Southern blotting to determine whether they were authentic transductants.
DNA sequence analysis.
All sequence similarity searches were performed by using the BLASTX or BLASTP algorithms on the NCBI Web site (www.ncbi.nlm.nih.gov/BLAST/) (1). We obtained sequence data for DNA flanking each end of the transposon for four of the five mutant regions cloned into pUC18 (UW11, UW32, UW56, and UW64); sequence from only one side of the transposon in UW89 was obtained. For the inverse PCR sequences, we submitted sequence flanking one end of the transposon for UW52 and UW78 and for UW96, UW119, and UW120 we submitted sequence that contained DNA adjacent to both ends of the transposon. The sequences submitted to BLASTX contained no transposon or pUC18 sequences and, where applicable, one-half of the sequence was converted to the opposite strand so that the DNA submitted to BLASTX matched a genomic sequence in B. cereus UW101C.
Mapping genes to a bacterial artificial chromosome library of B. cereus UW85.
Using the sequences surrounding Tn5401 for each of the 10 mutants, we designed PCR primers to amplify small DNA regions to make DNA probes for each of the mutant sequences (Table 2). Primers were synthesized by the University of Wisconsin Biotechnology Center. PCR conditions were the same as described above except that annealing was 45 s and extension was 3 min for each cycle and the final extension was 5 min. The template was 0.5 μl of B. cereus UW85 genomic DNA. PCR products were partially purified as described above, and the DNA was labeled with digoxigenin according to the manufacturer's protocol (Roche Molecular Biochemicals, Indianapolis, Ind.).
A bacterial artificial chromosome library of B. cereus UW85 genomic DNA was previously constructed in pBeloBAC11, with 323 clones having an average insert size of 98 kb, resulting in a calculated 5.75-fold coverage of the genome (42). For mapping, we used a subset of the 216 largest clones, which had an average insert size of 117 kb, resulting in a calculated 4.6-fold coverage of the genome. DNA from each BAC clone was prepared by using a modified miniprep procedure (42), spotted onto a nylon membrane, and UV-cross-linked to it. Digoxigenin-labeled probes based on each of the 10 mutant sequences were hybridized to the UW85 BAC library subset by using a standard protocol (Roche Molecular Biochemicals). Probes that did not hybridize to the BAC library subset were tested against B. cereus UW85 genomic DNA as a positive control for hybridization.
Construction of a ΔmarR mutant.
EcoRI-digested DNA from BAC139 was subcloned into pAD123, and the ligation mixture was electroporated into UW85 and UW101C as described previously (48). Pools of transformants were screened for the presence of the marR locus by using primers UW56 PF and UW56 PR. Sequence encompassing the entire marR gene was obtained from a positive clone by primer walking. To construct an allele of marR containing an in-frame deletion, we PCR-amplified two ∼800-bp fragments containing either the beginning or end of the marR gene, along with flanking DNA (primer sets marRup-F/marRup-R and marRdown-F/marRdown-R, respectively). The primers contained restriction sites (BamHI/SphI and SphI/HindIII, respectively) used to directionally clone the fragments into the suicide plasmid pSPUC. The resulting construct contained an allele of marR that had an in-frame deletion spanning 63 amino acids. A single amino acid substitution was introduced at the engineered SphI site. The deletion was verified by sequencing the pSPUC::ΔmarR construct across the deletion. We performed a two-step allelic exchange in UW85 and UW101C to replace the wild-type copy of marR with the copy containing the deletion. pSPUC::ΔmarR was electroporated into UW85 and UW101C, and plasmid integrants were selected on 0.5× TSA containing 100 μg of spectinomycin/ml. Putative integrants were grown in 0.5× TSB without antibiotics for 2 days with regular subculturing to allow for the excision and loss of the plasmid. Cultures were plated on nonselective media and screened for spectinomycin sensitivity. Sps clones were screened by PCR to determine whether they contained the mutant allele, and putative deletion mutants were verified by Southern blot analysis.
RESULTS
Identification of transposon mutants affected in zwittermicin A production.
A screen of 4,800 Tn5401 mutants of B. cereus UW101C identified 10 mutants affected in zwittermicin A production. In the initial screen for inhibition of E. herbicola, nine mutants produced no zone of inhibition and one produced a larger zone than the wild type. The mutants that did not inhibit E. herbicola did not produce detectable zwittermicin A, and mutant UW56, which inhibited E. herbicola more than the wild type, produced eightfold more zwittermicin A than UW101C (Table 3). Zwittermicin A purified from cultures of UW56 had the same Rf value in paper electrophoresis as authentic zwittermicin A (data not shown). Southern blot analysis of the 10 mutants affected in zwittermicin A production indicated that each mutant contained one insertion and the insertions were in different genomic locations (data not shown). All of the mutants that did not produce detectable zwittermicin A were significantly reduced in the ability to suppress damping off of tomato (16).
TABLE 3.
Characteristics of loci carrying insertions that affect zwittermicin A production in B. cereusa
| Strain | Amt of zwittermicin A produced (μg/ml of culture) | BAC clone carrying DNA homologous to site of insertion identified by hybridization | Estimated insert size in BAC clone (kb) |
|---|---|---|---|
| UW11 | <1.5a | 363 | 110 |
| UW32 | <1.5 | 209 | 80 |
| UW52 | <1.5 | NHb | NA |
| UW56 | 25 | 139 | 120 |
| UW64 | <1.5 | Multiplec | 100-210 |
| UW78 | <1.5 | 209 | 80 |
| UW89 | <1.5 | NH | NA |
| UW96 | <1.5 | 209 | 80 |
| UW119 | <1.5 | 225 and 295 | 85 and 125 |
| UW120 | <1.5 | NH | NA |
| UW85 | 12.5 | NAd | NA |
| UW101C | 3.1 | NA | NA |
Data shown are from one experiment and are representative of two experiments.
NH, no hybridization.
BAC clones that hybridized are 142, 150, 151, 152, 153, 159, 236, 302, 304, 309, and 321.
NA, not applicable.
Reconstruction of the transposon mutants via transduction.
To determine whether the Tn5401 insertions were causal for the zwittermicin A phenotypes, we attempted to transduce the transposons from the original mutants into B. cereus UW85/pAD123 by using phage Pjer. We reconstructed 6 of the 10 mutants (UW11, UW32, UW52, UW64, UW89, and UW120) by this method, and all of the reconstructed mutants were phenotypically indistinguishable from the original mutants and produced no detectable zwittermicin A (data not shown).
DNA sequence analysis of transposon mutants.
The DNA regions flanking the transposon in six of the zwittermicin A-deficient mutants contained sequences similar to genes involved in either peptide or polyketide antibiotic biosynthesis (Table 4). Sequences flanking the transposon insertions in UW52, UW78, UW96, and UW120 exhibit significant similarity to known nonribosomal peptide synthetases (NRPSs) in Bacillus species, while flanking regions from two mutations (in UW64 and UW89) contain sequences with similarity to type I polyketide synthases (PKSs) in Nostoc sp. strain PCC7120. Mutant UW56, which overproduced zwittermicin A, contained a transposon 5 bp upstream from a gene that encodes a deduced protein that is 88% identical to a Bacillus anthracis transcriptional regulator in the MarR family, which is comprised of both negative and positive regulators (14, 53). The prototypic MarR protein was identified in E. coli, in which MarR represses expression of the multiple antibiotic resistance operon (43). Sequence flanking the transposon in UW11 has similarity to NprR, which is reported to activate transcription of neutral protease NprB (bacillolysin) in B. thuringiensis (GenBank accession no. AAK00601). Directly downstream of nprR in UW11 is a region with similarity to bacillolysin, an arrangement that is also found in the B. cereus ATCC 14579 and B. thuringiensis ATCC 35646 genomes (Integrated Genomics Inc.). The transposon in UW119 inserted within a region with 93% amino acid similarity to an ABC transporter permease protein in B. thuringiensis ATCC 35646. Immediately downstream of the permease-encoding gene in the B. thuringiensis genome is a gene that encodes an ABC transporter ATP-binding protein. The B. thuringiensis permease and ATP-binding protein are highly similar to YclI and YclH, respectively, in Bacillus subtilis, which together compose an ABC transporter that is a member of a subfamily of exporters in B. subtilis that is related to a transporter involved in resistance to rapamycin in Streptomyces hygroscopicus (37).
TABLE 4.
Sequence similarity of DNA flanking Tn5401 in mutants affected in zwittermicin A productiona
| Mutant | Homology of gene product | Organism | E value | % Identity (over no. of aab) |
|---|---|---|---|---|
| UW11 | Transcriptional activator NprR | B. thuringiensis | 1e-05 | 61 (46) |
| UW32 | Alkanesulfonate monooxygenase | B. subtilis | 7e-06 | 36 (97) |
| UW52 | Surfactin synthetase subunit 2 | B. subtilis | 9.2 | 51 (27) |
| UW56 | MarR, MarR family | B. anthracis | 3e-41 | 85 (111) |
| UW64 | PKS type I | Nostoc sp. strain PCC 7120 | 2e-22 | 29 (156) |
| UW78 | Tyrocidine synthetase | Brevibacillus brevis | 2e-13 | 59 (59) |
| UW89 | PKS type I | Nostoc sp. strain PCC 7120 | 5e-24 | 60 (65) |
| UW96 | Mycosubtilin synthetase subunit B (MycB) | B. subtilis | 9e-42 | 39 (159) |
| UW119 | Similar to transporter | Listeria innocua | 3e-21 | 27 (218) |
| UW120 | Bacitracin synthetase 1 | Bacillus licheniformis | 8e-51 | 36 (193) |
Comparisons are with GenBank entries.
aa, amino acids.
Mapping BAC clones carrying sequences involved in zwittermicin A production.
We mapped the 10 mutant loci to clones in a BAC library of B. cereus UW85 genomic DNA (Table 3). The two loci identified in mutants UW11 and UW56 each hybridized to a different BAC clone; one probe (generated from UW119) hybridized to two BAC clones, one probe (generated from UW64) hybridized to multiple BAC clones, and three probes (generated from UW52, UW89, and UW120) failed to hybridize to any BAC clones. The loci identified in mutants UW32, UW78, and UW96 hybridized to BAC clone 209, which also contains zmaR (E. Stohl, personal communication). These BAC clones carried inserts from 80 to 210 kb.
Construction of a ΔmarR mutant.
Because the transposon in UW56, the one mutant that overproduced zwittermicin A, inserted immediately upstream from an ORF with similarity to the MarR family of transcriptional regulators, we tested the possibility that this gene was responsible for the increase in zwittermicin A production. We replaced the wild-type copy of marR with a copy containing an in-frame deletion in both UW85 and UW101C. Both ΔmarR constructs produced levels similar to their respective parental strains (data not shown), suggesting that a lack of the MarR protein is not responsible for the zwittermicin A overproduction phenotype in UW56.
Zwittermicin A biosynthetic cluster flanking zmaR.
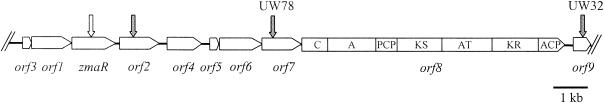
Sequence analysis of DNA flanking Tn5401 in UW78 revealed that the transposon had inserted near zmaR, the zwittermicin A self-resistance gene. Previous study of zmaR and adjacent DNA identified three ORFs (orfs1 to -3) whose gene products exhibit similarity to proteins involved in polyketide and peptide antibiotic biosynthesis (33, 52). We obtained approximately 12 kb of sequence downstream from the 4 kb previously characterized by sequencing a PCR product and zmaR-containing BAC clones from a genomic library of UW85 (42) (GenBank Accession no. AF155831). (We identified a G at nucleotide position 3933 which is reported as a T in our previous publication [52].) The overall G+C content of the entire 16-kb region is 35%, which is characteristic of B. cereus (23). Sequence analysis revealed five additional ORFs (orf4 to orf8) and one partial ORF (orf9), all with the same transcriptional orientation as orf1 to -3 and zmaR (Fig. 2 and Table 5). Several pairs of adjacent genes exhibit overlapping stop and start codons (orf3 and orf1, orf1 and zmaR, orf5 and orf6, and orf6 and orf7). Furthermore, orf7 and orf8 overlap by 26 bp. This overlapping arrangement of genes is common among PKS and NRPS operons and suggests the genes are translationally coupled (11, 34, 41). The proteins predicted from orf5 and orf6 exhibit high similarity to those from orf3 and orf1, respectively, suggesting that one group of genes may have arisen through a duplication of the other.
FIG. 2.
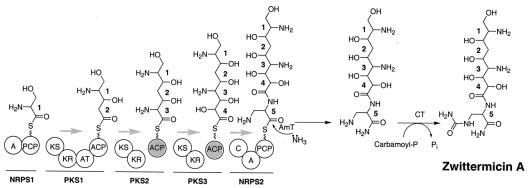
Organization of genes identified in the zwittermicin A biosynthetic cluster of B. cereus UW85. Vertical arrows represent mutations within the designated gene. An open arrow indicates that the mutant produced zwittermicin A (52); a shaded arrow indicates that the mutant did not produce detectable levels of zwittermicin A (52; this work). The domain organization of orf8 is indicated (see Results for details). Abbreviations: C, condensation; A, adenylation; PCP, peptidyl carrier protein; KS, ketoacyl synthase; AT, acyltransferase; KR, ketoreductase; ACP, acyl carrier protein.
TABLE 5.
Characteristics of genes identified in the zwittermicin A biosynthetic cluster of B. cereus UW85
| Gene | Nucleotide position | No. of amino acids | Homology of deduced protein |
|---|---|---|---|
| orf3 | 78-341 | 87 | ACP |
| orf1 | 338-1486 | 382 | Acyl-CoA dehydrogenase |
| zmaR | 1483-2610 | 375 | Acetyltransferase (acetylates zwittermicin A) |
| orf2 | 2630-3847 | 405 | Malonyl-CoA-ACP transacylase |
| orf4 | 3888-4736 | 282 | 3-Hydroxybutyryl-CoA dehydrogenase |
| orf5 | 4767-5012 | 81 | ACP |
| orf6 | 5012-6205 | 397 | Acyl-CoA dehydrogenase |
| orf7 | 6202-7779 | 525 | Mycosubtilin synthetase subunit C (MycC) |
| orf8 | 7754-15442 | 2,562 | NRPSs/PKSs |
| orf9 (partial) | 15461-15879 | 139 | Alkanesulfonate monooxygenase |
The newly identified ORFs encode putative proteins with similarities to known proteins that are involved in the biosynthesis of polyketides and nonribosomal peptides (Fig. 2 and Table 5). The deduced proteins encoded by orf4 to -6 are similar to enzymes in a myxobacterium and two actinomycetes that are proposed to be involved in the synthesis of an unusual polyketide extender unit, methoxymalonyl-acyl carrier protein (ACP) (29, 58, 60). orf4 and orf6 encode putative proteins that are similar to SorD and SorE from the myxobacterium Sorangium cellulosum, which encode a beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase and an acyl-CoA dehydrogenase, respectively, speculated to be involved in the synthesis of methoxymalonyl-ACP which is incorporated into the polyketide soraphen A (29). Orf5 is similar to two ACPs: Asm14 in Actinosynnema pretiosum ssp. auranticum (60) and FkbJ in S. hygroscopicus (58), which are thought to participate in the formation of methoxymalonyl-ACP, used in the biosynthesis of the antitumor agent ansamitocin and the macrolide FK520, respectively.
The protein encoded by orf2, previously shown by gene disruption to be necessary for zwittermicin A production (52), has high similarity to FenF and ItuD, which are putative malonyl-CoA transacylases involved in the synthesis of the lipopeptide antibiotics mycosubtilin and iturin A, respectively, in B. subtilis (12, 56). FenF is predicted to catalyze the attachment of malonyl-CoA to an ACP domain of MycA, a hybrid peptide synthetase, amino transferase, and fatty acid synthase (12).
The transposon in UW78 inserted into orf7, which is highly similar to genes encoding NRPSs. The best matches to Orf7 are the serine adenylation domains in the B. subtilis NRPS enzymes MycC and ItuC, which are involved in the synthesis of the peptide antibiotics mycosubtilin and iturin A, respectively (12, 56). Orf7 contains all 10 highly conserved core motifs found in the adenylation domains of peptide synthetases (30). To predict the amino acid specificity of the Orf7 adenylation domain, we used a recently designed algorithm that identifies eight or nine residues that line the substrate-binding pocket of the adenylation domain and compares them to a database of known NRPS adenylation domains for which the substrate has been determined experimentally (7, 51; http://raynam.chm.jhu.edu/∼nrps/). Analysis of the Orf7 adenylation domain indicates that it is most similar to the serine activation domain of mycosubtilin synthetase C (MycC). Notably, Orf7 lacks both a condensation and an apparent peptidyl carrier protein (also called thiolation) domain.
orf8 encodes a putative protein of 2,562 amino acids that appears to be a multifunctional hybrid of a NRPS and PKS. The amino-terminal portion of the protein has strong homology to NRPSs in Bacillus species, containing a single amino acid-activating module that includes the condensation, adenylation, and peptidyl carrier protein (or thiolation) domains that are typical of NRPSs. The first 1,000 amino acids of Orf8 exhibit the highest similarity to iturin A synthetase C (ItuC) from B. subtilis. The adenylation domain of Orf8 contains all 10 of the conserved amino acid motifs characteristic of peptide synthetases (30). Substrate-binding pocket analysis suggests that the adenylation domain of Orf8 activates threonine.
The carboxy-terminal portion of Orf8 contains regions with a high degree of similarity to domains typical of type I PKSs, which are large multifunctional proteins that catalyze the sequential condensation of short-chain carboxylic acids into the growing polyketide structure. The last 1,500 amino acids of Orf8 contain regions with strong similarity to ketoacyl synthase, acyltransferase, ketoreductase, and ACP domains from type I PKS enzymes. When compared with proteins of known function, the C-terminal region of Orf8 exhibits the highest similarity to type I PKS enzymes from Nostoc sp. and to the PKS portion of hybrid NRPS-PKS enzymes from a variety of cyanobacteria and myxobacteria (8, 9, 17, 44, 46, 55).
A partial ORF starts 19 nucleotides downstream from the end of orf8 and is the site of insertion of the transposon in mutant UW32. The deduced protein sequence resembles alkanesulfonate monooxygenases from a variety of bacteria, with the highest similarity to one in Pseudomonas putida. SsuD, the alkanesulfonate monooxygenase in E. coli, catalyzes the conversion of alkanesulfonates to sulfite and the corresponding aldehydes (15).
DISCUSSION
We identified new loci involved in the production of zwittermicin A by B. cereus UW85 through a broad mutant search. Most of the genes identified have significant sequence similarity to genes involved in polyketide and peptide antibiotic biosynthesis in other organisms. Three of the loci identified by the mutant analysis mapped to a BAC clone (BAC209) shown previously to contain the zwittermicin A self-resistance gene, zmaR, and extensive sequence revealed a large cluster of genes with homology to genes involved in synthesis of peptide and polyketide antibiotics. One large gene in this region appears to encode a hybrid peptide synthetase and polyketide synthase. Only two of the three insertions that mapped to BAC209 mapped to the sequenced 16-kb cluster, suggesting that the complete cluster contains additional ORFs.
The six zwittermicin A-nonproducing mutants we reconstructed by transduction of Tn5401 from the original mutant into a clean genetic background failed to produce detectable zwittermicin A, confirming the role of these loci in zwittermicin A biosynthesis. While we were unable to reconstruct the remaining four mutations from UW56, UW78, UW96, and UW119 by transduction, two of the four (UW78 and UW96) mapped to BAC209, which was previously shown to be important in zwittermicin A biosynthesis, suggesting that the transposons were likely causal for the phenotypes of the mutants.
The transposon in UW56, the only mutant which overproduced zwittermicin A, inserted directly upstream of a gene whose deduced protein exhibits homology to the MarR family of transcriptional regulators, which is comprised largely of negative regulators but also includes a number of positive regulators (14, 53). The in-frame deletion in the marR locus did not affect zwittermicin A levels, suggesting that either the overproduction phenotype in UW56 is due to a secondary mutation elsewhere in the genome or the transposon causes an increase in expression of marR or a downstream gene, which in turn causes an overproduction of zwittermicin A. The ORF immediately downstream (about 50 nucleotides) of marR, which we designate orfX, has homology to acetyltransferase genes from a variety of bacteria. OrfX is most similar to acetyltransferases of the GNAT family, to which zmaR also has some homology (52). Our earlier work showed that a zmaR knockout mutant produced near-wild-type levels of zwittermicin A but was not compromised in growth compared to the parent strain, suggesting that another mechanism of self-resistance is present in B. cereus. Multiple mechanisms of self-resistance are common in antibiotic-producing organisms (5, 45). Moreover, Crameri and Davies (10) reported that the introduction of a plasmid carrying an antibiotic resistance gene (for an acetyltransferase) into aminoglycoside-producing bacteria resulted in increased antibiotic production. Based on this finding, we speculate that OrfX is an acetyltransferase that serves as an alternative zwittermicin A self-resistance determinant and that orfX is overexpressed in UW56 due to a promoter at the end of the transposon, increasing zwittermicin A production.
The results we present here support our hypothesis that zmaR is part of the zwittermicin A biosynthetic cluster (52). In this work we extend the sequence of the cluster to 16 kb and confirm the importance of this region in biosynthesis of zwittermicin A. While four of the loci mapped to several other BAC clones, based on sequence similarity they likely have an indirect role in zwittermicin A expression; they may be involved in regulation, export, or resistance to zwittermicin A. The three loci that failed to map to the BAC library exhibit similarity to NRPS and PKS enzymes. It is possible that these loci are clustered together and may even be closely linked to the biosynthetic region characterized here. Since the genes for antibiotic biosynthetic pathways are typically clustered (28, 31, 35, 36), it seems unlikely that these unmapped loci with NRPS and PKS homology are scattered throughout the chromosome. Genome sequencing of B. cereus UW85 should elucidate the placement of these loci with respect to the 16-kb core cluster.
Recently, the genomes of the B. cereus type strain (ATCC 14579), and closely related species from the B. cereus group, B. anthracis and B. thuringiensis, have been published (23, 39, 40; Integrated Genomics Inc. [http://www.integratedgenomics.com/]). Although many of the gene products in the zwittermicin A core cluster exhibit similarity to proteins in the B. cereus group, none appear to be true homologs, as the identities and similarities range from 27 to 41% and 51 to 65%, respectively. Moreover, the related genes in the B. cereus group genomes do not appear to be clustered. However, there is an entry in GenBank from B. thuringiensis serovar kurstaki (accession no. AF235003; unpublished data) spanning orfs1 to -5 and zmaR that is nearly identical at the nucleotide level to the corresponding sequence presented in our previous work and the current study.
The core gene cluster offers considerable insight into the mechanism of synthesis of zwittermicin A, which was not apparent from the structure because zwittermicin A represents a novel class of antibiotic (Fig. 1). The structural analysis indicates motifs resembling peptides and polyketides, which coincides with the genetic analysis that identified numerous genes that have similarity to genes involved in peptide and polyketide antibiotic biosynthesis, including three of the four (orfs1 to -3 and zmaR) we identified in earlier work (52). All of the ORFs in the core cluster are oriented in the same direction and exhibit an overlapping gene structure, suggesting they are part of an operon.
Although the deduced gene products in the core cluster share significant sequence similarity with proteins of known function, some display notable features that give insights into zwittermicin A precursor biosynthesis. In particular, the similarities of Orf1 and Orf3 to -6 to enzymes proposed to be involved in methoxymalonyl-ACP formation suggests hypotheses about how both aminomalonyl- and hydroxymalonyl-ACP are generated by B. cereus for zwittermicin A production.
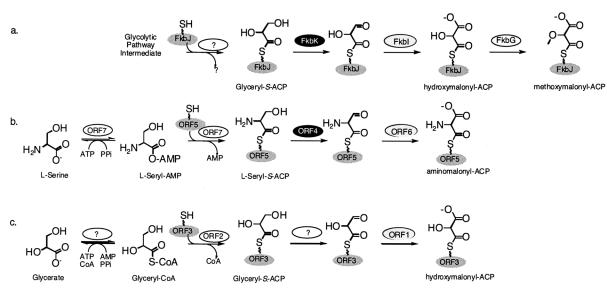
The proposed mechanism for methoxymalonyl-ACP formation during FK520 biosynthesis (58) is a model for how aminomalonyl- and hydroxymalonyl-ACP formation might proceed in B. cereus. Briefly, a proposed glycolytic pathway intermediate is converted to a glyceryl-ACP intermediate (Fig. 3a). While tethered to the ACP (FkbJ), the 3-hydroxyl of the glyceryl moiety is converted to an acid by the concerted actions of enzymes similar to 3-hydroxybutyryl-CoA dehydrogenases (FkbK) and acyl-CoA dehydrogenases (FkbI). The O-methylation of the 2-hydroxyl by FkbG produces methoxymalonyl-ACP, which is then incorporated into the growing FK520 polyketide (58).
FIG. 3.
Schematic representations of methoxymalonyl-ACP (a), aminomalonyl-ACP (b), and hydroxymalonyl-ACP (c) biosynthesis. The oval above each arrow indicates the enzyme proposed to catalyze the step being shown. Black and grey ovals are color coded to highlight homologous enzymes. The squiggled lines above FkbJ, ORF5, and ORF3 represent the 4′-phosphopentheinyl prosthetic group of each ACP. Question marks indicate enzymes or products yet to be identified.
We predict that Orf4, -5, and -6, which are homologs to FkbK, FkbJ, and FkbI, respectively, play analogous roles in aminomalonyl-ACP biosynthesis (Fig. 3b). However, the precursor for the pathway is not a glycolytic intermediate, but is predicted to be l-serine, which is activated and tethered to Orf5 by the action of Orf7, which is a homolog of the adenylation domains of NRPSs, and has an amino acid specificity code for l-serine. Thus, the conversion of l-serine to aminomalonyl-ACP is hypothesized to proceed as shown in Fig. 3b. If this hypothesis is correct, it will be the first example of aminomalonyl-ACP as an extender unit in polyketide biosynthesis.
We hypothesize that Orf1 to -3 are a portion of the hydroxymalonyl-ACP pathway. The presence of Orf2, a malonyl-CoA-ACP transacylase homolog, suggests that glyceryl-CoA is the substrate for the acylation of Orf3 (ACP) by Orf2 (Fig. 3c). Thus, we hypothesize that the zwittermicin A pathway encodes an acyl-CoA ligase to generate glyceryl-CoA. Once glyceryl-ACP is formed, the same steps for 3-hydroxyl conversion to an acid, as discussed above for methoxymalonyl- and aminomalonyl-ACP synthesis, would be followed. While a second homolog of FkbI is encoded in the sequenced portion of the gene cluster (Orf1), a second homolog of FkbK has yet to be identified. This suggests that either Orf4 can function in both pathways or that the gene encoding the second 3-hydroxybutyryl-CoA dehydrogenase homolog has yet to be located. It should be noted that a homolog for FkbG (O-methyltransferase) is not expected in the zwittermicin A pathway because of the absence of a methoxymalonyl group in the chemical structure of the antibiotic (Fig. 1).
Further sequencing of the zwittermicin A biosynthetic gene cluster is needed to propose hypotheses for the roles of Orf8 and Orf9 in antibiotic biosynthesis. Orf8 is of interest due to its homology to NRPSs and PKSs, suggesting that it is involved in zwittermicin A assembly once all the necessary precursors have been biosynthesized. In addition, as we sequence more of the biosynthetic gene cluster, we expect to find more NRPS/PKS homologs along with the enzymes necessary for 2,3-diaminopropionate biosynthesis and modification (Fig. 1). The biosynthetic enzymes for the two other precursors, l-serine and malonyl-CoA, are most likely available from primary metabolism. Thus, the enzymes catalyzing their production are not likely to be encoded by the zwittermicin A biosynthetic gene cluster.
The sequence and bioinformatic analysis presented here has given considerable insights into the probable mechanism of zwittermicin A biosynthesis. In particular, the prediction that two of the PKS extender units are hydroxymalonyl-ACP and aminomalonyl-ACP allows for a reasonable hypothesis to be formulated for how zwittermicin A is assembled. As shown in Fig. 4, we predict that a five-module NRPS-PKS hybrid “megasynthase” condenses the five components of the zwittermicin A core. The release of an intermediate from the megasynthase is predicted to be catalyzed by an amidotransferase, and downstream modification by a carbamoyltransferase will complete the zwittermicin A biosynthetic pathway.
FIG. 4.
Proposal for the minimum biosynthetic machinery needed for zwittermicin A assembly. The monomers incorporated into zwittermicin A are as follows: 1, serine; 2, malonyl; 3, aminomalonyl; 4, hydroxymalonyl; 5, 2,3-diaminopropionate. Each circle represents an individual domain of the NRPS/PKS megasynthase. The grey circles identify the ACP components of the aminomalonyl- or hydroxymalonyl-ACP precursors. The grey arrows indicate the direction of zwittermicin A assembly. The bars mark the borders between NRPS and PKS portions of the putative megasynthase. Abbreviations: A, adenylation; PCP, peptidyl carrier protein; KS, ketosynthase; KR, ketoreductase; AT, acyltransferase; C, condensation; AmT, amidotransferase; CT, carbamoyltransferase.
The NRPS portions of the megasynthase are predicted to recognize and activate serine (residue 1) or 2,3-diaminopropionate (residue 5). While the source of serine is likely the cellular pool of this proteinogenic amino acid, 2,3-diaminopropionate is predicted to be biosynthesized specifically for zwittermicin A production. Recently, a mechanism for 2,3-diaminopropionate biosynthesis was predicted for the antibiotic viomycin (54). Thus, it is reasonable to assume that the zwittermicin A biosynthetic gene cluster encodes enzymes that are orthologs of VioB and VioK, the two enzymes predicted to generate 2,3-diaminopropionate.
Using Fig. 4 as a guide, the predicted zwittermicin A megasynthase would function as follows. NRPS1 would activate and tether serine to its peptidyl carrier protein domain. PKS1 would tether a malonyl moiety from malonyl-CoA to its ACP domain and subsequently condense the upstream serine with the covalently bound malonyl moiety. The ketoreductase domain of PKS1 would reduce the carbonyl from serine, generating the intermediate shown on PKS1 (Fig. 4). The two subsequent condensations and reductions with aminomalonyl and hydroxymalonyl moieties would be catalyzed in an analogous manner to generate the intermediate shown on PKS3 (Fig. 4). We point out that the catalytic domains shown in Fig. 4 are the minimum number of functional domains needed. We cannot eliminate the possibility that the aminomalonyl and hydroxymalonyl moieties are transferred to ACP domains embedded within the megasynthase, as previously seen for methoxymalonyl incorporation into soraphen (29), FK520 (58), and ansamitocin (60). The final module of the megasynthase, NRPS2, would activate, tether, and condense 2,3-diaminopropionate with the PKS3-tethered intermediate to generate the five-residue intermediate on NRPS2. The release and subsequent modification of the zwittermicin A backbone is predicted to occur as shown in Fig. 4.
To determine whether these predictions are correct, the completion of the sequencing of the zwittermicin A biosynthetic gene cluster and the biochemical and genetic testing of these hypotheses will be required. We are currently working towards these goals. The results presented in the current study suggest two new mechanisms for PKS extender unit biosynthesis and broaden our understanding of the metabolic capabilities of NRPS and PKS enzymology.
The work presented here increases our understanding of the biology of B. cereus and the genetics of zwittermicin A production. Moreover, the genes identified in this analysis may have utility in construction of novel antibiotic biosynthetic pathways or in discovery of other members of the zwittermicin A class of antibiotics.
Acknowledgments
We are grateful to James Baum for providing the transposon vector pEG922 and to Jeremy Dodsworth for providing transducing phage Pjer. We thank Jeremy Dodsworth and Jessica Gross for reconstructing some of the zwittermicin A mutants.
This work was funded by the USDA, The Howard Hughes Medical Institute, and Hatch Project 4038 from the College of Agricultural and Life Sciences of the University of Wisconsin—Madison.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum, J. A. 1994. Tn5401, a new class II transposable element from Bacillus thuringiensis. J. Bacteriol. 176:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broderick, N. A., R. M. Goodman, J. Handelsman, and K. F. Raffa. 2003. Effect of host diet and insect source on synergy of gypsy moth (Lepidoptera: Lymantriidae) mortality to Bacillus thuringiensis subsp. kurstaki by zwittermicin A. Environ. Entomol. 32:387-391. [Google Scholar]
- 4.Broderick, N. A., R. M. Goodman, K. F. Raffa, and J. Handelsman. 2000. Synergy between zwittermicin A and Bacillus thuringiensis subsp. Kurstaki against gypsy moth (Lepidoptera: Lymantriidae). Environ. Entomol. 29:101-107. [Google Scholar]
- 5.Calcutt, M. J., and F. J. Schmidt. 1994. Gene organization in the bleomycin-resistance region of the producer organism Streptomyces verticillus. Gene 151:17-21. [DOI] [PubMed] [Google Scholar]
- 6.Cane, D. E., C. T. Walsh, and C. Khosla. 1998. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science 282:63-68. [DOI] [PubMed] [Google Scholar]
- 7.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Z., P. Flatt, W. H. Gerwick, V. A. Nguyen, C. L. Willis, and D. H. Sherman. 2002. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene 296:235-247. [DOI] [PubMed] [Google Scholar]
- 9.Christiansen, G., J. Fastner, M. Erhard, T. Borner, and E. Dittmann. 2003. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J. Bacteriol. 185:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crameri, R., and J. E. Davies. 1986. Increased production of aminoglycosides associated with amplified antibiotic resistance genes. J. Antibiot. (Tokyo) 39:128-135. [DOI] [PubMed] [Google Scholar]
- 11.Du, L., C. Sanchez, M. Chen, D. J. Edwards, and B. Shen. 2000. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol. 7:623-642. [DOI] [PubMed] [Google Scholar]
- 12.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn, A. K., and J. Handelsman. 1999. A vector for promoter trapping in Bacillus cereus. Gene 226:297-305. [DOI] [PubMed] [Google Scholar]
- 14.Egland, P. G., and C. S. Harwood. 1999. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an Fnr family member. J. Bacteriol. 181:2101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichhorn, E., J. R. van der Ploeg, and T. Leisinger. 1999. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 274:26639-26646. [DOI] [PubMed] [Google Scholar]
- 16.Emmert, E. A. B. 1999. Chemical biology of Bacillus cereus. Ph. D. thesis. University of Wisconsin—Madison, Madison.
- 17.Gaitatzis, N., B. Silakowski, B. Kunze, G. Nordsiek, H. Blocker, G. Hofle, and R. Muller 2002. The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J. Biol. Chem. 277:13082-13090. [DOI] [PubMed] [Google Scholar]
- 18.Gold, H. S., and R. C. Moellering, Jr. 1996. Antimicrobial-drug resistance. N. Engl. J. Med. 335:1445-1453. [DOI] [PubMed] [Google Scholar]
- 19.Guerout-Fleury, A.-M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 21.Handelsman, J., S. Raffel, E. H. Mester, L. Wunderlich, and C. R. Grau. 1990. Biological control of damping-off of alfalfa seedlings with Bacillus cereus UW85. Appl. Environ. Microbiol. 56:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, H., L. A. Silo-Suh, J. Clardy, and J. Handelsman. 1994. Zwittermicin A, an antifungal and plant protection agent from Bacillus cereus. Tetrahedron Lett. 35:2499-2502. [Google Scholar]
- 23.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Milhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 24.Khosla, C. 2000. Natural product biosynthesis: a new interface between enzymology and medicine. J. Org. Chem. 65:8127-8133. [DOI] [PubMed] [Google Scholar]
- 25.Khosla, C., R. McDaniel, S. Ebert-Khosla, R. Torres, D. H. Sherman, M. J. Bibb, and D. A. Hopwood. 1993. Genetic construction and functional analysis of hybrid polyketide synthases containing heterologous acyl carrier proteins. J. Bacteriol. 175:2197-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khosla, C., and R. J. Zawada. 1996. Generation of polyketide libraries via combinatorial biosynthesis. Trends Biotechnol. 14:335-341. [DOI] [PubMed] [Google Scholar]
- 27.Kim, U.-J., B. W. Birren, T. Slepak, V. Mancino, C. Boysen, H-L. Kang, M. I. Simon, and H. Shizuya. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213-218. [DOI] [PubMed] [Google Scholar]
- 28.Lacalle, R. A., J. A. Tercero, and A. Jimenez. 1992. Cloning of the complete biosynthetic gene cluster for an aminonucleoside antibiotic, puromycin, and its regulated expression in heterologous hosts. EMBO J. 11:785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ligon, J., S. Hill, J. Beck, R. Zirkle, I. Molnar, J. Zawodny, S. Money, and T. Schupp. 2002. Characterization of the biosynthetic gene cluster for the antifungal polyketide soraphen A from Sorangium cellulosum So ce26. Gene 285:257-267. [DOI] [PubMed] [Google Scholar]
- 30.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2674. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Bueno, M., E. Valdivia, A. Galvez, J. Coyette, and M. Maqueda. 1998. Analysis of the gene cluster involved in production and immunity of the peptide antibiotic AS-48 in Enterococcus faecalis. Mol. Microbiol. 27:347-358. [DOI] [PubMed] [Google Scholar]
- 32.Milner, J. L., S. J. Raffel, B. J. Lethbridge, and J. Handelsman. 1995. Culture conditions that influence accumulation of zwittermicin A by Bacillus cereus UW85. Appl. Microbiol. Biotechnol. 43:685-691. [DOI] [PubMed] [Google Scholar]
- 33.Milner, J. L., E. A. Stohl, and J. Handelsman. 1996. Zwittermicin A resistance gene from Bacillus cereus. J. Bacteriol. 178:4266-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molnar, I., J. F. Aparicio, S. F. Haydock, L. E. Khaw, T. Schwecke, A. Konig, J. Staunton, and P. F. Leadlay. 1996. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene 169:1-7. [DOI] [PubMed] [Google Scholar]
- 35.Mootz, H. D., and M. A. Marahiel. 1997. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 179:6843-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Motamedi, H., and C. R. Hutchinson. 1987. Cloning and heterologous expression of a gene cluster for the biosynthesis of tetracenomycin C, the anthracycline antitumor antibiotic of Streptomyces glaucescens. Proc. Natl. Acad. Sci. USA 84:4445-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quentin, Y., G. Fichant, and F. Denizot. 1999. Inventory, assembly and anlysis of Bacillus subtilis ABC transport system. J. Mol. Biol. 287:467-484. [DOI] [PubMed] [Google Scholar]
- 38.Raffel, S. J., E. V. Stabb, J. L. Milner, and J. Handelsman. 1996. Genotypic and phenotypic analysis of zwittermicin A-producing strains of Bacillus cereus. Microbiology 142:3425-3436. [DOI] [PubMed] [Google Scholar]
- 39.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 40.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A.-B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 41.Recktenwald, J., R. Shawky, O. Puk, F. Pfennig, U. Keller, W. Wohlleben, and S. Pelzer. 2002. Nonribosomal biosynthesis of vancomycin-type antibiotics: a heptapeptide backbone and eight peptide synthetase modules. Microbiology 148:1105-1118. [DOI] [PubMed] [Google Scholar]
- 42.Rondon, M. R., S. J. Raffel, R. M. Goodman, and J. Handelsman. 1999. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc. Natl. Acad. Sci. 96:6451-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seoane, A. S., and S. B. Levy. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shalev-Alon, G., A. Sukenik, O. Livnah, R. Schwarz, and A. Kaplan. 2002. A novel gene encoding amidinotransferase in the cylindrospermopsin producing cyanobacterium Aphanizomenon ovalisporum. FEMS Microbiol. Lett. 209:87-91. [DOI] [PubMed] [Google Scholar]
- 45.Sheldon, P. J., Y. Mao, M. He, and D. H. Sherman. 1999. Mitomycin resistance in Streptomyces lavendulae includes a novel drug-binding-protein-dependent export system. J. Bacteriol. 181:2507-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silakowski, B., H. U. Schairer, H. Ehret, B. Kunze, S. Weinig, G. Nordsiek, P. Brandt, H. Blocker, G. Hofle, S. Beyer, and R. Muller. 1999. New lessons for combinatorial biosynthesis from myxobacteria. The myxothiazol biosynthetic gene cluster of Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 274:37391-37399. [DOI] [PubMed] [Google Scholar]
- 47.Silo-Suh, L. A. 1994. Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85. Ph. D. thesis. University of Wisconsin—Madison, Madison. [DOI] [PMC free article] [PubMed]
- 48.Silo-Suh, L. A., B. J. Lethbridge, S. J. Raffel, H. He, J. Clardy, and J. Handelsman. 1994. Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85. Appl. Environ. Microbiol. 60:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silo-Suh, L. A., E. V. Stabb, S. J. Raffel, and J. Handelsman. 1998. Target range of zwittermicin A, an aminopolyol antibiotic from Bacillus cereus. Curr. Microbiol. 37:6-11. [DOI] [PubMed] [Google Scholar]
- 50.Stabb, E. V., L. M. Jacobson, and J. Handelsman. 1994. Zwittermicin A-producing strains of Bacillus cereus from diverse soils. Appl. Environ. Microbiol. 60:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 52.Stohl, E. A., J. L. Milner, and J. Handelsman. 1999. Zwittermicin A biosynthetic cluster. Gene 237:403-411. [DOI] [PubMed] [Google Scholar]
- 53.Sulavik, M. C., L. F. Gambino, and P. F. Miller. 1995. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1:436-446. [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas, M. G., Y. A. Chan, and S. G. Ozanick. 2003. Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster. Antimicrob. Agents Chemother. 47:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tillett, D., E. Dittmann, M. Erhard, H. von Dohren, T. Borner, and B. A. Neilan. 2000. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 7:753-764. [DOI] [PubMed] [Google Scholar]
- 56.Tsuge, K., T. Akiyama, and M. Shoda. 2001. Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 183:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Virk, A., and J. M. Steckelberg. 2000. Clinical aspects of antimicrobial resistance. Mayo Clin. Proc. 75:200-214. [DOI] [PubMed] [Google Scholar]
- 58.Wu, K., L. Chung, W. P. Revill, L. Katz, and C. D. Reeves. 2000. The FK520 gene cluster of Streptomyces hygroscopicus var. ascomyceticus (ATCC14891) contains genes for biosynthesis of unusual polyketide extender units. Gene 251:81-90. [DOI] [PubMed] [Google Scholar]
- 59.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 60.Yu, T.-W., L. Bai, D. Clade, D. Hoffmann, S. Toelzer, K. Q. Trinh, J. Xu, S. J. Moss, E. Leistner, and H. G. Floss. 2002. The biosynthetic gene cluster of the maytansinoid antitumor agent ansamitocin from Actinosynnema pretiosum. Proc. Natl. Acad. Sci. USA 99:7968-7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaehner, H., and F. P. Fiedler. 1995. The need for new antibiotics: possible ways forward, p. 67-84. In P. A. Hunter, G. K. Darby, and N. J. Russell (ed.), Fifty years of antimicrobials. Proceedings of the 53rd Symposium of the Society for General Microbiology. Cambridge University Press, Cambridge, United Kingdom.