Abstract
Certain anaerobic bacteria respire toxic selenium oxyanions and in doing so produce extracellular accumulations of elemental selenium [Se(0)]. We examined three physiologically and phylogenetically diverse species of selenate- and selenite-respiring bacteria, Sulfurospirillum barnesii, Bacillus selenitireducens, and Selenihalanaerobacter shriftii, for the occurrence of this phenomenon. When grown with selenium oxyanions as the electron acceptor, all of these organisms formed extracellular granules consisting of stable, uniform nanospheres (diameter, ∼300 nm) of Se(0) having monoclinic crystalline structures. Intracellular packets of Se(0) were also noted. The number of intracellular Se(0) packets could be reduced by first growing cells with nitrate as the electron acceptor and then adding selenite ions to washed suspensions of the nitrate-grown cells. This resulted in the formation of primarily extracellular Se nanospheres. After harvesting and cleansing of cellular debris, we observed large differences in the optical properties (UV-visible absorption and Raman spectra) of purified extracellular nanospheres produced in this manner by the three different bacterial species. The spectral properties in turn differed substantially from those of amorphous Se(0) formed by chemical oxidation of H2Se and of black, vitreous Se(0) formed chemically by reduction of selenite with ascorbate. The microbial synthesis of Se(0) nanospheres results in unique, complex, compacted nanostructural arrangements of Se atoms. These arrangements probably reflect a diversity of enzymes involved in the dissimilatory reduction that are subtly different in different microbes. Remarkably, these conditions cannot be achieved by current methods of chemical synthesis.
Selenium is a metalloid element that is chemically similar to sulfur and tellurium and in nature exists in four oxidation states, −2, 0, +4, and +6. The last two states occur in aqueous media as the soluble oxyanions selenite [SeO32− or Se(IV)] and selenate [SeO42− or Se(VI)]. Selenium also has unusual photo-optical and semiconducting physical properties and has industrial applications in devices such as photocopiers and microelectronic circuits. Recent interest in the field of nanotechnology has stimulated research into the chemical synthesis of selenium nanowires that are composed of elemental selenium [Se(0)] (1, 10, 11). However, the various allotropes of Se(0) are not well understood in terms of the internal structural arrangements of the selenium atoms, and hence selenium still is a complex element in terms of our understanding of its fundamental physical properties (3).
In biology, selenium is a key trace element that is found in representative species from all three domains of life (Bacteria, Archaea, and Eukaryota), as well as in viruses (34). At the molecular level, Se occurs in analogs of sulfur-containing amino acids (e.g., selenomethionine, selenocysteine) and is found in diverse enzymes (14, 37). Selenocysteine is an essential component of the geometry and corresponding electron density of certain enzymatic active sites, such as those of formate dehydrogenase (2). Ironically, although selenium is considered an essential dietary trace element, high concentrations of it are acutely toxic. Hydrologic changes in naturally seleniferous regions have caused serious environmental problems, notably in the western San Joaquin Valley of California (32). Selenium has a complete biogeochemical cycle in nature, with microbial redox reactions leading both to and from all of its oxidation states (7, 15). An important component of this cycle is the dissimilatory (respiratory) reduction of selenate and selenite to Se(0) in reactions that are catalyzed by anaerobic microorganisms inhabiting anoxic sediments (26, 27, 38).
To date, about 16 diverse species of Bacteria and Archaea have been described that grow anaerobically by linking the oxidation of organic substrates or H2 to the dissimilatory reduction of selenium oxyanions (30, 39). The end products of these reactions are the red, amorphous or monoclinic allotropes of Se(0), which accumulate in spent medium because the microorganisms reduce the 10 to 20 mM selenate or selenite provided to Se(0). Respiratory reductases for Se oxyanions contain molybdenum and are associated with the plasma membrane (18). In contrast, Se-resistant bacteria typically tolerate exposure to ≤1 mM selenite or ≤1 mM selenate, which they also reduce to Se(0) and excrete as distinct particles (8, 17, 23). Enzymes that confer Se resistance usually involve glutathione reductase (9).
It has previously been noted that small spheres of Se(0) form on the cell surface of a gram-positive rod, Bacillus selenitireducens strain MLS10, after respiratory growth on selenite (41). Here we examined this phenomenon more closely to document exogenous Se(0) nanosphere formation by three physiologically and phylogenetically diverse species of Se-respiring bacteria: the haloalkaliphile B. selenitireducens isolated from Mono Lake in California, the halophile Selenihalanaerobacter shriftii (strain DSSE1) isolated from the Dead Sea (42), and the freshwater organism Sulfurospirillum barnesii (strain SES3) isolated from a drainage slough in Nevada (28, 40). We also harvested the Se(0) spheres from cultures of these bacteria and removed the cellular debris. We examined the spectroscopic characteristics (UV-visible and Raman) of the purified preparations of biologically formed Se(0) and compared them to the spectroscopic characteristics of abiotically formed Se(0). From this we gained insight into the unique internal structural arrangements of the selenium atoms.
MATERIALS AND METHODS
Preparation of biogenic and chemical Se(0) for physical characterization.
S. barnesii and S. shriftii were grown in anaerobic batch cultures with Se(VI) (10 to 20 mM) as the electron acceptor, while B. selenitireducens was grown with 10 mM Se(IV) (28, 41, 42). The compositions of the three media have been described previously and are not described in detail here; the media all contained lactate as the electron donor, and vitamins and yeast extract were added as growth supplements. However, the three media differed substantially in pH, salinity, and buffering systems, as follows: for S. barnesii the pH of the medium was 7.3, the salinity was 2 g/liter, and the buffer was phosphate plus bicarbonate-CO2; for S. shriftii the pH of the medium was 7.0, the salinity was 205 g/liter, and the buffer was piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES); and for B. selenitireducens the pH of the medium was 9.8, the salinity was 56 g/liter, and the buffer was carbonate-bicarbonate. For recovery of extracellular Se nanospheres, all three cultures were first grown with nitrate in lieu of Se oxyanions. Biogenic elemental selenium was formed by bacterial reduction of Se(IV) added to these cultures after harvesting and washing. Nitrate-grown cultures of B. selenitireducens strain MLS10, S. barnesii strain SES3, and S. shriftii strain DSSE1 were grown separately in 500 ml of medium by using the methods described above. Late-exponential-phase cells were washed and resuspended in anaerobic minimal salts medium devoid of any chemical reductant (e.g., cysteine-HCl). Na2SeO3 (3.0 mM) and 2.0 mM sodium lactate were added to each serum bottle containing 50 ml of a cell suspension. The bottles were placed in a 25°C shaking (140 rpm) incubator for 3 days, and during this time the cells reduced Se(IV) to Se(0), as indicated by the appearance of a bright red precipitate. Cellular material was removed from the Se(0) particles by using a modified procedure of Laddaga and MacLeod (21). The cell suspensions containing Se(0) were ultrasonicated at 100 W for 2 min and centrifuged at 1,500 × g for 30 min. The pellets were resuspended, ultrasonicated, and centrifuged (1,500 × g) sequentially in 0.5 M NaCl, 0.5 M sucrose, and finally a complete salts solution composed of 17.5 g of NaCl per liter, 0.74 g of KCl per liter, 12.3 g of MsSO4 · 7H2O per liter, and 0.15 g of Tris buffer per liter, and the pH was adjusted to 7.5. The cells were then lysed in 35 ml of the complete salts solution containing 0.020 g of egg white lysozyme which was incubated at 22°C for 18 h. The lysed cells were rinsed away from the Se(0) by resuspension, ultrasonication, incubation (3 to 4 h), and sequential centrifugation (8,000 to 10,000 × g) with the complete salts solution, 0.25 M NaOH, 0.1 M NaOH, 10 mM Na2HPO4 (adjusted to pH 7.3), and carbon-free, distilled, deionized water. The cleaned Se(0) was then resuspended in distilled, deionized water.
Two forms of elemental selenium were synthesized chemically. Colloidal amorphous red elemental selenium [Se(0)red] was prepared by oxidation of gaseous hydrogen selenide [H2Se(g)] in water. Hydrogen selenide (5,060 ppm) in N2 (Scientific Gas Products, Ashland Chemical Co., Columbus, Ohio) was sparged at a rate of ∼300 ml · min−1 into 800 ml of 0.1 μM NaOH (pH ∼10) simultaneously with O2 at a flow rate equal to that of H2Se. During the 6-h reaction the solution was vigorously stirred while colloidal, fine-grained, red Se(0) precipitated. After the H2Se source was removed, the solution was sparged with O2 for an additional 30 min to oxidize any remaining HSe−. The suspension was then bubbled with N2 for 1 h and pasteurized by heating it at 80°C for 1 h. During these procedures the Se(0) remained red and was easily deflocculated by agitation.
Black vitreous elemental selenium [Se(0)black] was formed by reduction of Se(IV) with ascorbic acid. Twenty-five milliliters of 5% (vol/vol) ascorbic acid was added to 30 ml of 167 mM Na2SeO3, which initiated rapid formation of a red Se(0) precipitate. The suspension was stirred for 1 h and then centrifuged at 11,000 × g for 70 min. After decanting, the Se(0) precipitate was rinsed with 50 ml of 50% (vol/vol) ethanol-water and sonicated at 100 W for 3 min to disperse the precipitate. This procedure was repeated with 10 mM Na2HPO4 and distilled deionized water. The suspension was then bubbled with N2 for 1 h and pasteurized by heating it at 80°C for 1 h. During these procedures the Se(0) gradually darkened from a red form to a dense gray-black form that exhibited vitreous morphology in scanning electron micrographs (data not shown).
Transmission electron microscopy (TEM) and scanning electron microscopy (SEM).
Stained thin sections (used for micrographs) and unstained thin sections (used for collecting energy dispersive X-ray spectroscopy [EDS] spectra) of cell suspensions were prepared as described previously (12) by using Spurr low-viscosity resin (Marivac, St. Laurent, Quebec, Canada). Thin sections and untreated whole mounts of bacteria were placed on carbon-Formvar-coated 200-mesh copper grids, and images were obtained with a Philips EM400T at 100 kV under standard operating conditions with the liquid nitrogen anticontaminator in place. EDS was also performed at 100 kV by using a spot size of 200 nm, a current of 25 mA, and a (live) counting time of 100 s. For selected area electron diffraction we used the EM400T in the diffraction mode with a camera length of 575 mm and an exposure time of 20 s. SEM images were obtained as described previously (36).
UV-visible and Raman spectroscopy.
Samples of Se(0) particles suspended in water were placed in a quartz cell (path length, 1 cm) and analyzed to obtain UV-visible absorption spectra with a Varian Cary 500 Scan UV-VIS-NIR spectrophotometer. Raman spectra were obtained with a Renishaw Ramascope Raman spectrometer equipped with an integral Leica DMLM microscope; 514.5-nm radiation was produced from a 20-mW air-cooled Ar+ laser (Spectra-Physics model 263C). The silicon Raman band at 520 nm was used to calibrate the spectrometer, and the spectral resolution was approximately 1.5 cm−1.
RESULTS
SEM and TEM of Se-respiring bacteria.
Cells of selenite-grown B. selenitireducens produced abundant Se(0) nanospheres on the exterior of the cell envelope, as shown by the SEM image in Fig. 1A. TEM of thin sections also revealed the common presence of intracellular Se(0) granules when cells were grown on Se(IV) (Fig. 1B). When nitrate-grown washed cells were fed Se(IV), external Se(0) granules were the predominant form of Se(0) present, as shown by the wider-field image in Fig. 2A. The EDS spectra derived from a nanosphere indicated that it was composed entirely of selenium (Fig. 2B). The Cu peaks were associated with the TEM grid, the Na and Cl peaks reflected the high salt content of the medium, and the C and O peaks most likely were associated with cellular exudate. The lack of any other metal peaks in the spectrum indicated that the selenium occurred in the elemental state [Se(0)] rather than as a metal selenide [Se2−]. A higher magnification revealed the attachment of the external Se nanospheres on the cell surface (Fig. 2C). The smaller, dark, fine-grained precipitates on the cell surface were composed of Se as well, but they also produced EDS peaks for Na, Cl, K, and P (data not shown), suggesting that they were mixed precipitates from the highly saline medium. The cell envelope and the external Se nanospheres also appeared to be encapsulated with an exopolymer. Selected area electron diffraction of nanospheres revealed a monoclinic crystal structure. Although patterns were difficult to generate (due to decay of the selenium nanospheres when they were exposed to the electron beam), we measured d-spacings of 0.303, 0.284, 0.266, 0.224, 0.202, and 0.185 nm (selected area electron diffraction transform data not shown). Owing to the small size of the nanospheres, this diffraction pattern is attributable to only the strongest reflections from the phases present.
FIG. 1.
Elemental selenium spheres formed by B. selenitireducens. (A) SEM of cells growing on selenite and forming chains of Se(0) spheres. (B) TEM (thin section) of a cell grown on selenite that formed internalized Se(0).
FIG. 2.
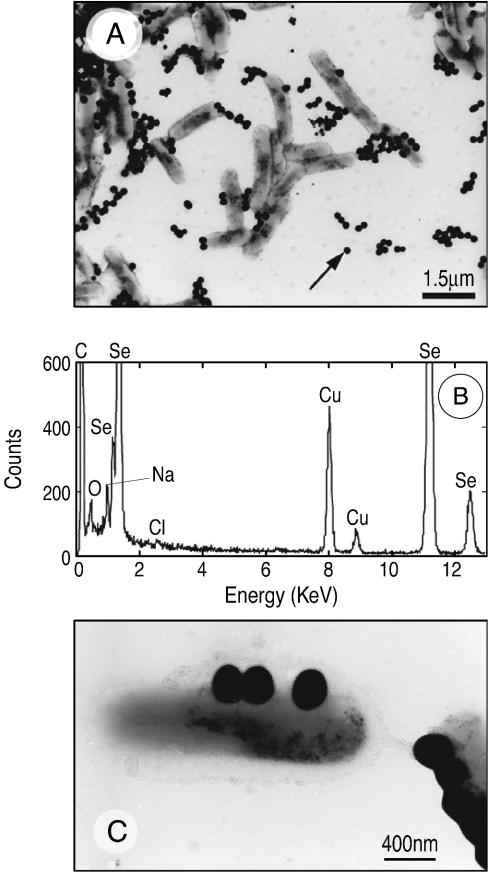
(A) TEM (whole mount) of nitrate-grown washed cells of B. selenitreducens that were fed selenite, showing a large number of external Se(0) spheres. (B) EDS of the particle in panel A indicated by an arrow. (C) Higher-magnification TEM (whole mount) of nitrate-grown cells.
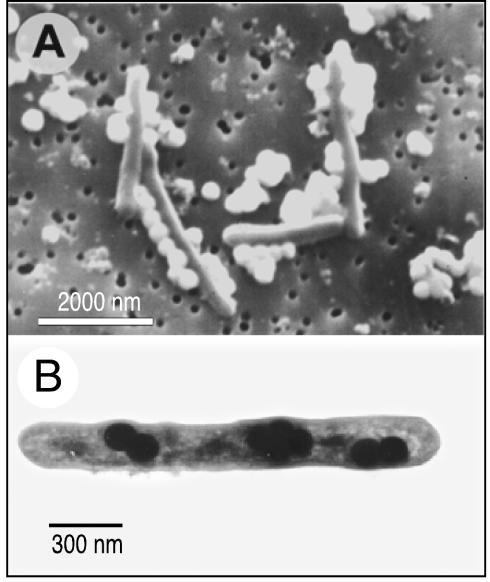
We also observed similar external and internal accumulations of Se nanospheres in selenate-grown cells of the gram-negative species S. shriftii and S. barnesii. For Se(VI)-grown S. barnesii, TEM images typically showed that there were extensive accumulations of Se granules on the exterior of the cells, which apparently sloughed off the cell surfaces and formed even larger aggregates composed of many individual granules (Fig. 3A and B). The presence of internal Se nanospheres in this organism was also confirmed by examining thin sections, which documented the presence of nanospheres after growth with Se(VI) (Fig. 3C) and with nitrate-grown washed cells that were fed Se(IV) (Fig. 3D). S. shriftii produced both internal and external Se nanospheres when it was grown on nitrate, washed, and subsequently fed Se(VI) (Fig. 4A) or Se(IV) (Fig. 4B).
FIG. 3.
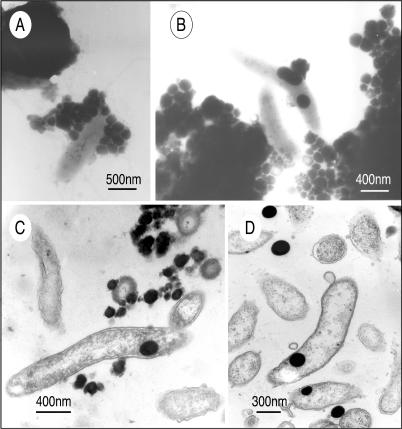
TEM of S. barnesii, showing external and internal accumulations of Se granules. (A) Whole mount of cells grown on selenate, showing ∼30 external granules attached to a cell. (B) Whole mount of two cells close to very large external aggregations of Se granules. EDS spectra indicated that all the dark granules in panels A and B are elemental selenium, similar to the spectrum shown in Fig. 2 (data not shown). (C) Thin section of selenate-grown cells, showing external and internal Se granules. (D) Thin section of cells grown on nitrate, washed, and given selenite, showing internal and external Se granules.
FIG. 4.
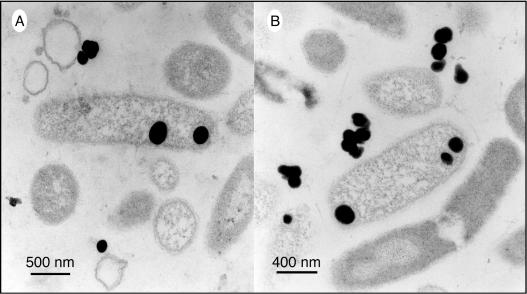
TEM (thin sections) of S. shriftii grown on nitrate, washed, and resuspended with selenate (A) and selenite (B). The dark spheres are composed of elemental selenium, as determined from EDS spectra (data not shown).
SEM images of purified Se nanospheres.
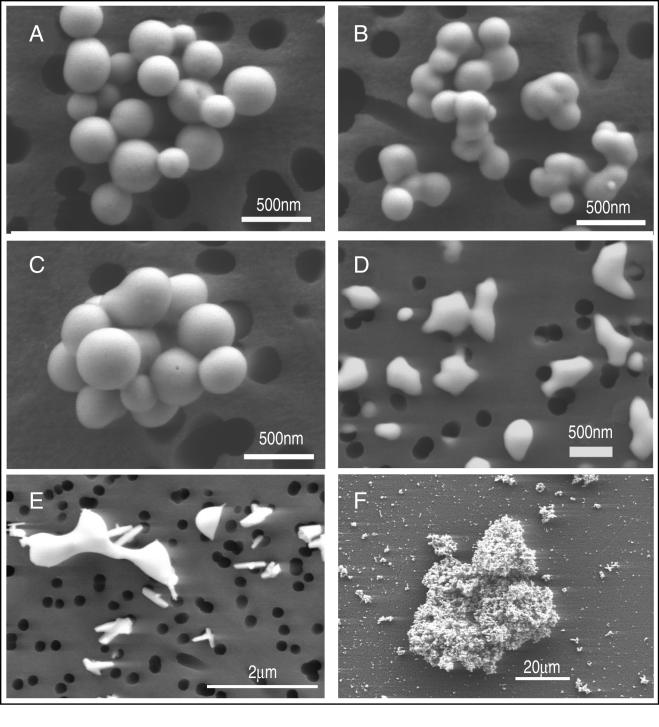
The external Se(0) particles from all three cultures, after harvesting and cleansing, consisted of nanospheres that ranged in diameter from 200 to 400 nm, and the most common diameter was ∼300 nm, as observed in SEM images (Fig. 5A to C). The Se(0) nanospheres were stable, as these structures persisted at least for several months when they were kept as solid suspensions in distilled water. In contrast, Se(0) chemically formed by auto-oxidation of H2Se gas with O2 produced only unstructured, amorphous aggregates that ranged in diameter from 200 to 800 nm (Fig. 5D). Elemental Se formed by chemical reduction of Se(IV) with ascorbate produced a vitreous, black allotrope consisting of unstructured aggregates having various dimensions. The particle size distribution for this allotrope was extremely variable, and the range was particles as small as 10 nm (Fig. 5E) to aggregations as large as 50 μm (Fig. 5F) because of the sticky nature of this material.
FIG. 5.
SEM of cleansed elemental selenium formed by bacteria (A to C) or synthesized chemically (D to F). (A to C) Selenium spheres formed by B. selenitireducens MLS10 (A), S. barnesii SES3 (B), and S. shriftii DSSE1 (C). (D) Elemental selenium formed by oxidation of H2Se with O2 in alkaline (pH 10.0) water. (E and F) High- and low-magnification images, respectively, of vitreous black elemental selenium formed by reduction of selenite with ascorbate.
Spectral properties and Se atom arrangements of the purified Se nanospheres.
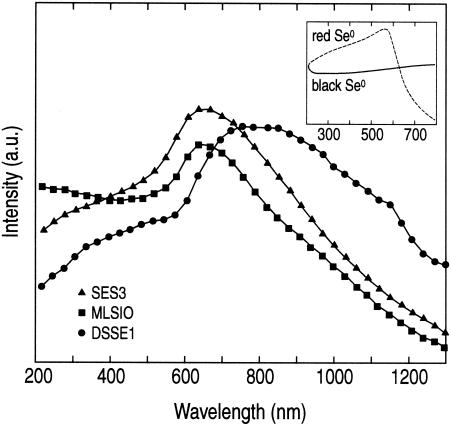
The typical structural arrangements of Se atoms in the various allotropes of elemental selenium are rings or chains of Se atoms, which can have different molecular compositions or chain lengths, ranging from Se3 to Se12 (18). Figure 6 shows the UV-visible absorption spectra for the three Se samples obtained from S. barnesii, B. selenitireducens, and S. shriftii. These spectra are compared with the spectra of chemically formed Se(0) (Fig. 6, inset), which include the red, amorphous Se(0) form and the black, vitreous Se(0) form. The spectral properties of the samples obtained from the bacteria were considerably different than the spectral properties of the chemically formed Se(0). Indeed, rather than being uniform, the spectra obtained for the three biologically derived samples varied considerably from species to species, and the Se nanosphere samples from S. shriftii exhibited the most unusual and broadest absorption spectrum at wavelengths greater than 600 nm.
FIG. 6.
UV-visible spectra of purified Se nanospheres from S. barnesii SES3, B. selenitireduncens MLS10, and S. shriftii DSSE1. (Inset) Comparative spectra of chemically produced red, amorphous Se(0) and black, vitreous Se(0). a.u., arbitrary units.
All the biological Se samples exhibited low bandgap features. We defined bandgap as the minimum energy needed to promote a valence electron into a conducting electron. The bandgap calculated for the red, amorphous Se(0) form was 2.1 eV, which was considerably higher than the bandgaps for Se nanospheres derived from S. barnesii (1.62 eV), S. shriftii (1.52 eV), and B. selenitirducens (1.67 eV). The bandgap variation seen in the biological samples was due to the different molecular configurations of the internal crystalline structure. The structural confinement of Se atoms within the 200- to 400-nm spheres caused the decrease in the bandgap and resulted from increased π-π and π-d transitions. This phenomenon is akin to that which has been observed elsewhere from clustering effects when increased Se ring-ring interactions caused a red shift (lower energy) in the absorption spectrum (22). This was apparent in the spectral differences among the three biological samples when they were compared to the chemically formed red, amorphous Se(0) (Fig. 6). By contrast, the black, vitreous Se(0) showed the typical broad, featureless absorption expected for large, amorphous, synthetically formed selenium clusters. The S. shriftii sample had another distinguishing feature. There were two optical absorption peaks at 780 and 820 nm, indicating that there was a bimodal distribution. This revealed the presence of another Se molecular species within this bacterium's Se nanospheres (see below).
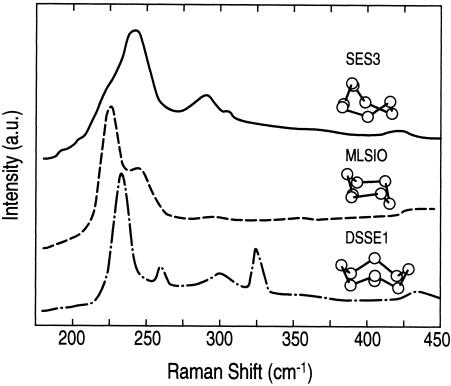
The three microbial samples also exhibited Raman spectra having distinctly different features (Fig. 7). Two of the samples (S. barnesii and B. selenitireducens) had similar dominant Se6 units within their Se particles, while the third sample (S. shriftii) had a dominant Se8 unit of the D4d space group. The data also showed that poly(Se) was formed; the peak at 260 cm−1 was indicative of single-chain Se, while the peak at 234 cm−1 was a feature of Se polymer formation in addition to ring structures. The polymer is formed by close interactions between the Se8 rings via van der Waals bonds (5). Se6 vibrates when it is in the presence of a composite structure at 288 and 301 cm−1, which are the causes of the bimodal distribution which we observed in the absorption spectra. The S. barnesii samples had a slightly different vibrational spectrum than the B. selenitireducens samples, which also had a Se6 structure, but they differed in the configuration of the Se6 chains. In B. selenitireducens, Se6 vibrational modes A1g and Eg were observed at 244 and 225 cm−1, respectively. These modes were dominated by the more stable D3d (chair) structure. While S. barnesii also forms Se6, in this case it produced the spectrum typical of Se6 with the more unstable C2v (boat) structure; these modes were observed at 228 and 241 cm−1, findings which compare favorably with previously described findings. However, these vibrations can also be strongly linked to Se polymer structures, as previously described (4, 31).
FIG. 7.
Raman spectra at 514.5 nm of purified Se nanospheres from S. barnesii SES3, B. selenitireducens MLS10, and S. shriftii DSSE1. a.u., arbitrary units.
DISCUSSION
In this study we documented the occurrence of both intracellular and extracellular Se granules in three phylogenetically and physiologically distinct bacteria that are able to respire Se oxyanions. Thus, this phenomenon appears to be widespread among such bacteria. Indeed, it is not just confined to the bacteria that are capable of dissimilatory reduction of Se oxyanions, as it has also been reported in Se-resistant bacteria. For example, electron micrographs of Wollinella succinogenes (43), Enterobacter cloacae (23), and Stenotrophomonas maltophilia (8) have all shown that spherical granules of Se(0) that were similar sizes were formed after cells were exposed to Se(IV) or Se(VI). Intracellular Se(0) granules have also been reported in cells of the photosynthetic bacterium Chromatium vinosum, although in this case the granules were generated by the light-induced oxidation of H2Se rather than by the reduction of Se oxyanions (25).
It is noteworthy that the formation of Se nanospheres is not confined to biological systems since Se particles that are similar sizes can be generated by chemical synthesis. Thus, monoclinic Se(0) nanospheres were formed by chemical reduction of selenite with cytochrome c3 (1). Nanospheres composed of amorphous Se(0) appeared as temporary intermediates in the synthesis of trigonal Se(0) used to form Se nanowires (10, 11). These Se allotropes were produced under harsher conditions in which hydrazine was used for reduction of Se(IV) at ∼100°C. We did not observe Se(0) nanosphere formation when we used milder ascorbate reduction of Se(IV) at room temperature, a reaction that resulted instead in the formation of the black, vitreous Se(0) allotrope. However, neither of these previously described artificial chemical synthesis studies presented data on the spectral properties of the Se nanospheres themselves that would allow us to infer the internal arrangement(s) of their Se atoms. This can also be said to be the case for all of the studies that documented formation of Se(0) granules by the Se-resistant bacteria mentioned above; the authors made no further attempts to purify these materials or to examine their inherent spectral properties.
Elemental selenium is known to form three specific types of structural subsets, a six-member ring, an eight-member ring, and infinite α-helical chains with a Se-Se distance of approximately 2.37 Å and a bond angle of 103° (44). Thus, we propose that the nanospheres are composed of interconnected three-dimensional nets of selenium in which both chain and ring structural aspects are maintained, factors that should result in the spherical shape. Similar types of two-dimensional and three-dimensional Se nets have been observed for alkali metal-boron-selenium compounds (13) and have been proposed for partially disordered phases of Se (20). Finally, it has even been proposed that Se chains with terminal Cl atoms are able to interact with other Se chains to form such Se-based three-dimensional networks (19), which might account in part for the presence of Cl in Fig. 2B.
In our investigations we noted that extracellular Se(0) accumulation was far more common than intracellular Se(0) accumulation (Fig. 1 and 3) and that this was even more pronounced when nitrate-grown cells were washed and then fed Se(IV) (Fig. 2). It does not seem possible that the large external accumulations of Se(0) granules on the exterior cell surfaces of all of the microorganisms studied could have been derived from primary cytoplasmic synthesis and subsequent export. Such a system for respiratory reduction of Se(VI) to Se(0) would be, at the least, extremely discomforting for individual cells, and because of the size of the internal Se particles, the particles should be released only upon cell lysis. Relatively little is known about dissimilatory selenate reductases of prokaryotes, but like other respiratory enzymes, they are usually associated with the peripheral cell envelope. Only the respiratory selenate reductase of Thauera selenatis has been characterized, and it is located in the periplasm (35), which supports the idea that the bulk of the Se(VI) and Se(IV) reduction to Se(0) occurs on or outside the envelope. To date, no work has been done on dissimilatory selenite reductases.
We can make a simple calculation to determine what fraction of the total dissolved Se oxyanion pool is converted to exterior deposits of Se(0) associated with the cell surface, as shown by a cell of S. barnesii in Fig. 3A, on which we counted 30 externally attached Se(0) granules. If it is assumed that each granule is sphere with a 300-nm diameter, then each nanosphere has a volume of ∼1.4 × 10−20 m3. The specific gravity of Se(0) is 4.5 g ml−1 (or 4.5 g/10−6 m3), and therefore 1.0 × 10−20 m3 contains 4.5 × 10−5 ng of Se, or there is 6.3 × 10−5 ng of Se per nanosphere or 8.7 × 10−7 nmol of Se per nanosphere. If it is assumed that the cell density at the end of exponential growth was ∼1011 cells/liter (26) and that each bacterium had 30 Se(0) spheres on its surface (as shown in Fig. 3A), this Se would represent ∼2.4 mmol of Se(0) or about one-quarter of the available 10 mM Se(VI) initially present in the medium. The common occurrence of even larger Se(0) aggregates in the TEM fields (Fig. 2A) suggests that the amount of reduced Se was vastly larger than this estimate. Because the Se nanospheres eventually slough off the individual cells, the Se(0) particles represent the ultimate repository for all of the Se(VI) added at the start of incubation. This also suggests that the external and internal Se(0) nanospheres arise independent of each other and by different mechanisms. The external nanosphere accumulation is probably directly tied to the respiratory Se reductases.
What then could be the function of the internal, well-structured accumulations of selenium? It is possible that they are receptacles for storage of internally reduced Se(VI) or Se(IV) ions that have bypassed the membrane-associated respiratory reductases of the cell envelope and have entered the cell via sulfate or perhaps nitrite transporters. Considering the fact the organisms were grown in the presence of high concentrations (∼10 mM) of Se oxyanions, it is quite likely that some of these toxicants made it past the outer respiratory membrane barrier. Thus, the cells would need to perform a detoxification reaction with excess internal Se oxyanions by reducing them to Se(0) and storing them in segregated areas within the cells. It is not clear from the TEM images, however, that the internal Se nanospheres were surrounded by a distinct internal membrane. In the case of B. selenitireducens, storage in this manner could have a selective advantage by forming a reservoir of Se(0) when external electron acceptors are scarce because this organism can perform dissimilatory reduction of Se(0) to H2Se (15).
In summary, we studied elemental selenium aggregations produced by the metabolic activities of three different species of anaerobic bacteria that respire or breathe oxyanions of selenium. The complexity of the nanosphere UV-visible (Fig. 6) and Raman spectra (Fig. 7) showed that more than a single Se molecular chain structure was present, and indeed at times several structures were observed. In turn, the biologically formed Se nanospheres had very different spectral properties than chemically formed Se(0) (Fig. 6).
Production of coordinated nanoclusters with a specific molecular orientation has been well-studied by synthetic chemists. In this case, however, nature has provided an intriguing situation, in which different bacterial species produce selenium nanoclusters that have different structural orientations and optical properties. What can account for this variation? We believe that the differences are likely caused by a corresponding diversity of enzymatic reactions (with different electrochemical potentials) that concurrently take place on the outer membranes of the various bacteria. Thus, various constitutive enzymes (e.g., selenate, selenite, nitrate, nitrite, and trimethylamine oxide reductases) may be present to different extents in the cell envelopes of the different organisms, and they can likely shift their electrons to Se(IV), thereby reducing this form to Se(0) (6, 29, 33). It remains to be determined if the diverse properties of the biologically based Se nanospheres can be reliably reproduced biosynthetically and, if they can be, whether they have a practical application in the field of nanotechnology.
Our findings illustrate the relevance of screening the natural world for novel biological nanostructures that have unique physical properties that cannot currently be produced by purely chemical means. Although the formation of Se(0) by microorganisms has been known for some time, the physical properties of the aggregates have not been examined previously. In this work we found unique signatures associated with the spectra from anaerobes isolated from freshwater, hypersaline, and alkaline saline habitats. Since the microbial diversity of Se-respiring prokaryotes is much broader than these three representative species and includes hyperthermophiles capable of respiring Se oxyanions at ∼90°C (16, 30, 39), a survey of unexamined cultures for Se nanosphere formation and the associated optical properties of the nanospheres is warranted. Furthermore, if Se(0) nanospheres with characteristic biological optical spectra are found in suboxic Se-contaminated sediments, then the data could be taken as strong evidence that the nanospheres had a microbiological origin rather than a chemical origin (24). Since the concentration of selenium oxyanions even in contaminated water is usually low (e.g., <1 μM), it would be best to first attempt this in evaporation ponds where the brines have high levels of selenium oxyanions (∼40 μM) and the underlying sediments have marked selenate reductase activity (26, 27). On a much more speculative level, such a characteristic biosignature imposed on a trace element like selenium may conceivably be useful in the search for evidence of extinct microbial communities in sedimentary deposits of extraterrestrial origin (i.e., Mars).
Acknowledgments
We thank M. J. Oremland, H. L. Ehrlich, L. G. Miller, L. Young, S. Fendorf, and J. F. Stolz for their constructive comments and D. Moyles for expert assistance with the TEM.
R.S.O. was funded by the USGS National Research Program and a NASA Exobiology grant, and T.J.B. was supported by Natural Science and Engineering Research Council of Canada (NSERC) and US-DOE-NABIR grants. The TEM was performed at the NSERC Guelph Regional STEM Facility at the University of Guelph, whose maintenance is partially funded by an NSERC Major Facilities Access grant to T.J.B. P.A. and S.C. were supported by the NSF-funded Nanoscale Science and Engineering Center at Rensselaer Polytechnic Institute.
REFERENCES
- 1.Abdelouas, A., W. L. Gong, W. Lutze, J. A. Shelnutt, R. Franco, and I. Moura. 2000. Using cytochrome c3 to make selenium nanowires. Chem. Mater. 12:1510-1512. [Google Scholar]
- 2.Boyington, J. C., V. N. Gladyshev, S. V. Khangulov, T. C. Stadtman, and P. D. Sun. 1997. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 275:1305-1308. [DOI] [PubMed] [Google Scholar]
- 3.Brandt, W., and M. Oremland. 1976. Positron measurements of selenium recrystallization rates. Phys. Lett. 57A:387-389. [Google Scholar]
- 4.Brassington, N. J., H. G. M. Edwards, and V. Fawcett. 1987. 1987. The vibrational Raman spectra of selenium trioxide ag. Spectrochem. Acta 43A:451-454. [Google Scholar]
- 5.Carini, G., M. Cutroni, G. Galli, P. Migliardo, and F. Wanderlingh. 1980. Raman scattering in a-Se bulk near Tg. Solid State Commun. 33:1139-1141. [Google Scholar]
- 6.DeMoll-Decker, H., and J. M. Macy. 1993. The periplasmic nitrite reductase of Thauera selenatis may catalyze the reduction of selenite to elemental selenium. Arch. Microbiol. 160:241-247. [Google Scholar]
- 7.Dowdle, P. R., and R. S. Oremland. 1998. Microbial oxidation of elemental selenium in soil slurries and bacterial cultures. Environ. Sci. Technol. 32:3749-3755. [Google Scholar]
- 8.Dungan, R. S., S. R. Yates, and W. T. Frankenberger, Jr. 2003. Transformations of selenate and selenite by Stenotrophomonas maltophilia isolated from a seleniferous agricultural drainage pond sediment. Environ. Microbiol. 5:287-295. [DOI] [PubMed] [Google Scholar]
- 9.Ganther, H. 1968. Selenotrisulfides. Formation by the reaction of thiols with selenous acid. Biochemistry 7:2898-2905. [DOI] [PubMed] [Google Scholar]
- 10.Gates, B., B. Mayers, B. Cattle, and Y. Xia. 2002. Synthesis and characterization of uniform nanowires of trigonal selenium. Adv. Funct. Mater. 12:219-227. [Google Scholar]
- 11.Gates, B., B. Mayers, B. Grossman, and Y. Xia. 2002. A sonochemical approach to the synthesis of crystalline selenium nanowires in solutions and on solid supports. Adv. Mater. 14:1749-1752. [Google Scholar]
- 12.Glasauer, S., P. G. Weidler, S. Langley, and T. J. Beveridge. 2003. Controls on Fe reduction and mineral formation by a subsurface bacterium. Geochim. Cosmochim. Acta 67:1277-1288. [Google Scholar]
- 13.Hammerschmidt, A., A. Lindemann, M. Doch, C. Koster, C., and B. Krebs. 2002. 2D-polymeric anion networks: the two novel perselenoborates BaB2Se6 and Ba2B4Se13. Z. Anorg. Alleg. Chem. 628:1561-1567. [Google Scholar]
- 14.Heider, J., and A. Bock. 1993. Selenium metabolism in micro-organisms. Adv. Microb. Physiol. 35:71-109. [DOI] [PubMed] [Google Scholar]
- 15.Herbel, M. J., J. Switzer Blum, S. E. Borglin, and R. S. Oremland. 2003. Reduction of elemental selenium to selenide: experiments with anoxic sediments and bacteria that respire Se-oxyanions. Geomicrobiol. J. 20:587-602.
- 16.Huber, R., M. Sacher, A. Vollman, H. Huber, and D. Rose. 2000. Respiration of arsenate and selenate by hyperthermophilic archaea. Syst. Appl. Microbiol. 23:305-314. [DOI] [PubMed] [Google Scholar]
- 17.Kessi, J., M. Ramuz, E. Wejrli, M. Spycher, and R. Bachofen. 1999. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl. Environ. Microbiol. 65:4734-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohara, S., S. Goldbach, N. Koura, M.-L. Sabsoungi, and L. A. Curtiss. 1998. Vibrational frequencies of small selenium molecules. Chem. Phys. Lett. 287:282-288. [Google Scholar]
- 19.Kondakova, O. A., A. S. Zyubin, S. A. Dembovsky, and N. S. Kurnakov. 2001. Quantum-chemical modeling of chlorine-doped and hypervalent defects participation in reconstruction of the a-Se structure. J. Optoelectr. Adv. Mater. 3:847-853. [Google Scholar]
- 20.Koslowski, T., M. Koblischke, and A. Blumen. 2002. Modified small-world networks as models of liquid and amorphous selenium. Phys. Rev. B 66:064205-1-064205-7. [Google Scholar]
- 21.Laddaga, R. A., and R. A. MacLeod. 1982. Effects of wash treatments on the ultrastructure and lysozyme penetrability of the outer membrane of various marine and two terrestrial gram-negative bacteria. Can. J. Microbiol. 28:318-324. [Google Scholar]
- 22.Lin, Z., Z. Wang, W. Chen, L. Lin, G. Li, Z. Liu, H. Han, and Z. Wang. 1996. Absorption and Raman spectra of Se8-ring clusters in zeolite 5A. Solid State Commun. 100:841-843. [Google Scholar]
- 23.Losi, M. E., and W. T. Frankenberger, Jr. 1997. Reduction of selenium oxyanions by Enterobacter cloacae SLD1a-1: isolation and growth of the bacterium and its expulsion of selenium particles. Appl. Environ. Microbiol. 63:3079-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myneni, S. C. B., T. K. Tokunaga, and G. E. Brown, Jr. 1997. Abiotic selenium redox transformations in the presence of Fe(II, III) oxides. Science 278:1106-1109. [Google Scholar]
- 25.Nelson, D. C., W. H. Casey, J. D. Sison, E. R. Mack, A. Ahmad, and J. S. Pollack. 1996. Selenium uptake by sulfur-accumulating bacteria. Geochim. Cosmochim. Acta 60:3531-3539. [Google Scholar]
- 26.Oremland, R. S., J. T. Hollibaugh, A. S. Maest, T. S. Presser, L. Miller, and C. Culbertson. 1989. Selenate reduction to elemental selenium by anaerobic bacteria in sediments and culture: biogeochemical significance of a novel, sulfate-independent respiration. Appl. Environ. Microbiol. 55:2333-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oremland, R. S., N. A. Steinberg, N. A., A. S. Maest, L. G. Miller, and J. T. Hollibaugh. 1990. Measurement of in situ rates of selenate removal by dissimilatory bacterial reduction in sediments. Environ. Sci. Technol. 24:1157-1164. [Google Scholar]
- 28.Oremland, R. S., J. Switzer Blum, C. W. Culbertson, P. T. Visscher, L. G. Miller, P. Dowdle, and F. E. Strohmaier. 1994. Isolation, growth, and metabolism of an obligately anaerobic, selenate-respiring bacterium, strain SES-3. Appl. Environ. Microbiol. 60:3011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oremland, R. S., J. S. Blum, A. Burns Bindi, P. R. Dowdle, M. Herbel, and J. F. Stolz. 1999. Simultaneous reduction of nitrate and selenate by cell suspensions of selenium-respiring bacteria. Appl. Environ. Microbiol. 65:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oremland, R. S., and J. F. Stolz. 2000. Dissimilatory reduction of selenate and arsenate in nature, p. 199-224. In D. R. Lovley (ed.), Environmental metal-microbe interaction. ASM Press, Washington, D.C.
- 31.Poborchin, V. V., A. V. Kolobov, H. Oyanagi, S. G. Romanov, and K. Tomaka. 1997. Structure of selenium incorporated into nanochannels of mordenite: dependence on ion exchange and method of incorporation. Chem. Phys. Lett. 280:10-16. [Google Scholar]
- 32.Presser, T. S. 1998. The Kesterson effect. Environ. Manag. 18:437-454. [Google Scholar]
- 33.Rech, S. A., and J. M. Macy. 1992. The terminal reductases for selenate and nitrate respiration in Thauera selenatis are two distinct enzymes. J. Bacteriol. 174:7316-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shisler, J. L., T. G. Senkevich, M. J. Berry, and B. Moss. 1998. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotrophic poxvirus. Science 279:102-105. [DOI] [PubMed] [Google Scholar]
- 35.Shröder, I., S. Rech, T. Krafft, and J. M. Macy. 1997. Purification and characterization of the selenate reductase from Thauera selenatis. J. Biol. Chem. 272:23765-23768. [DOI] [PubMed] [Google Scholar]
- 36.Smith, R. L., F. S. Strohmaier, F. S., and R. S. Oremland. 1985. Isolation of anaerobic oxalate degrading bacteria from freshwater lake sediments. Arch. Microbiol. 14:8-13. [Google Scholar]
- 37.Stadtman, T. C. 1996. Selenocysteine. Annu. Rev. Biochem. 65:83-100. [DOI] [PubMed] [Google Scholar]
- 38.Steinberg, N. A., and R. S. Oremland. 1990. Dissimilatory selenate reduction potentials in a diversity of sediment types. Appl. Environ. Microbiol. 56:3550-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stolz, J. F., and R. S. Oremland. 1999. Bacterial respiration of selenium and arsenic. FEMS Microbiol. Rev. 23:615-627. [DOI] [PubMed] [Google Scholar]
- 40.Stolz, J. F., D. J. Ellis, J. Switzer Blum, D. Ahmann, R. S. Oremland, and D. R. Lovley. 1999. Sulfurospirillum barnesii sp. nov., Sulfurospirillum arsenophilus sp. nov., and the Sulfurospirillum clade in the epsilon proteobacteria. Int. J. Syst. Bacteriol. 49:1177-1180. [DOI] [PubMed] [Google Scholar]
- 41.Switzer Blum, J., A. Burns Bindi, J. Buzzelli, J. F. Stolz, and R. S. Oremland. 1998. Bacillus arsenicoselenatis sp. nov. and Bacillus selenitireducens sp. nov.: two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171:19-30. [DOI] [PubMed] [Google Scholar]
- 42.Switzer Blum, J., J. F. Stolz, A. Oren, and R. S. Oremland. 2001. Selenihalanaerobacter shriftii gen. nov., sp. nov., a halophilic anaerobe from Dead Sea sediments that respires selenate. Arch. Microbiol. 175:208-219. [DOI] [PubMed] [Google Scholar]
- 43.Tomei, F. A., L. L. Barton, C. L. Lemanski, and T. G. Zocco. 1992. Reduction of selenate and selenite to elemental selenium by Wolinella succinogenes. Can. J. Microbiol. 38:1328-1333. [Google Scholar]
- 44.Wells, A. F. 1984. Structural inorganic chemistry, 5th ed., p. 699-747. Claredon Press, Oxford, United Kingdom.