Abstract
The objective of this study was to determine the role of a lactococcal branched-chain amino acid aminotransferase gene, ilvE, in the production of branched-chain fatty acids. Lactococcus lactis subsp. lactis LM0230 and an ilvE deletion mutant, JLS450, produced branched-chain fatty acids from amino and α-keto acids at levels above α-keto acid spontaneous degradation and the fatty acids' flavor thresholds. The deletion mutant produced the same amounts of branched-chain fatty acids from precursor amino acids as did the parent. This was not the case, however, for the production of branched-chain fatty acids from the corresponding precursor α-keto acids. The deletion mutant produced a set of fatty acids different from that produced by the parent. We concluded from these observations that ilvE plays a role in the specific type of fatty acids produced but has little influence on the total amount of fatty acids produced by lactococci.
Lactic acid bacteria (LAB) are important for flavor production in fermented milk products. Fatty acids (FAs) are an important class of compounds produced by these organisms, contributing to distinctive flavor profiles in cheese (7, 25). Typical flavors of FAs at high concentrations are considered detrimental, but mixtures of FAs at low concentrations are beneficial to cheese flavor (21), especially Italian varieties (17).
FA production occurs via three known mechanisms: (i) glycolysis (14), (ii) lipolysis (8), and (iii) microbial metabolism of amino acids and α-keto acids (16). Branched-chain fatty acids (BCFAs) cannot be produced via glycolysis or lipolysis because milk fat lacks BCFAs; yet their concentration increases during aging. The mechanism for their production is unexplained.
The environment of cheese as a medium for bacteria is acidic and of suboptimal water activity and temperature, and it lacks fermentable carbohydrates. Stuart et al. (22) demonstrated that lactococci metabolize amino acids during carbohydrate starvation under laboratory conditions that partially mimic cheese. Presumably, this metabolism may produce FAs via amino acid catabolism. This metabolism is relevant because amino acid metabolism generates ATP via substrate level phosphorylation and regenerates protons to maintain the cellular oxidation-reduction potential (15). Amino acid catabolism is one plausible explanation for the occurrence of BCFAs in cheese due to the conditions and the metabolism of the associated bacteria.
Aminotransferases (ATases) initiate catabolism of aromatic, branched-chain, and sulfur-containing amino acids to cheese flavor compounds (11, 28). α-Keto acids are the intermediates and amino group acceptors in an ATase reaction (28); consequently, they are of interest in determining the role of these reactions in FA production. A direct role for the ATases in FA production, except for methionine catabolism (4, 10), is unknown in LAB. Analysis of the genome sequences of Lactococcus lactis IL-1403 (2) (GenBank accession no. AE005176) and Lactococcus lactis subsp. cremoris SK11 (http://www.jgi.doe.gov/JGI_microbial/html/index.html) determined that each contains 12 ATases. However, detailed expression studies of each ATase in these organisms are lacking. Overlapping ATase substrate specificity (6, 20, 27), the large number of ATases in the lactococcal genomes, and the lack of defined conditions leading to expression increase the complexity involved in assigning a specific role for individual ATases.
Previous studies focused on relating lactococcal ATases to flavor compound generation. Deletion of the lactococcal aromatic ATase (araT) reduces catabolism of leucine (20), while addition of α-ketoglutarate leads to an increase in aroma compounds (26). These studies independently verified the activities of lactococcal ATases or FA production but did not associate these two events, making it impossible to define the causal relationship of ATases and FA production. To determine the influence of the IlvE ATase in L. lactis subsp. lactis LM0230, a deletion mutant was previously tested for growth (1).
The role of individual ATases in the metabolism of amino and keto acids is unclear. Use of an ATase deletion mutant allows a direct assessment of the impact of the loss of the ATase on this metabolism. While other studies found IlvE to be involved in anabolism and growth, ATases are bidirectional and they may also be involved in the catabolism of amino and α-keto acids. Based on previous studies, we hypothesized that ilvE modulates the production of BCFAs in lactococci. This study found that deletion of ilvE did not significantly (P > 0.05) change the total concentration of BCFAs produced from amino acids but did significantly (P < 0.05) change the type of FAs produced at pH 5.2 with 4% NaCl and no carbohydrate in the assay buffer.
Strains and media.
L. lactis subsp. lactis LM0230 was obtained from the Utah State University culture collection. The ilvE deletion mutant, JLS450 (1), was kindly provided by James L. Steele, University of Wisconsin at Madison. Stock cultures were prepared by growing the organisms twice in 10 ml of Elliker broth (Difco laboratories, Detroit, Mich.) at 30°C for 24 h. Cultures were frozen at −70°C in 10% nonfat dry milk containing 30% glycerol. Before each experiment, frozen stock cultures were thawed and subcultured twice at 30°C for 24 h in 10 ml of Elliker broth.
Cell preparation for FA assays.
Amino acids, α-keto acids, FA standards, and buffers were purchased from Sigma (Sigma-Aldrich, St. Louis, Mo.). Cells were harvested from 10 ml of broth by centrifugation (3,500 × g for 15 min at 4°C), washed twice with 10 ml of sterile 0.05 M potassium phosphate buffer (pH 7.2), and resuspended in 5 ml of 0.15 M 2-(N-morpholino)ethane-sulfonic acid-sodium salt buffer (pH 5.2) containing 4% NaCl to a final cell density of 0.2 at A600. The suspended cells were incubated in sterile buffer (negative control), individual amino acids, individual α-keto acids, and mixtures of the amino acids or α-keto acids as described below. The amino acids used were glutamic acid, isoleucine, leucine, and valine. The α-keto acids used were α-ketoisocaproate (KIC), α-ketoglutarate (KGL), α-ketoisovalerate (KIV), and α-keto-β-methylvalerate (KMV). These components were used individually as substrates at a final concentration of 1 mM in the assay buffer containing the cells.
Based on previous studies with LAB (18, 19) and previous work (9), the cell-substrate mixtures were incubated at 30°C for 3 h and filter sterilized with 0.2-μm-pore-size syringe filters (Nalge Company, Rochester, N.Y.). The collected filtrates were used for FA analysis.
Gas chromatography.
FAs were extracted and determined in the sterile buffer filtrates as described by de Jong and Badings (3), and instrument operating conditions were as described previously (9). Gas chromatography was done on the Shimadzu (Columbia, Md.) gas chromatograph model GC17-A, equipped with a flame ionization detector. Samples were separated with a fused silica DB-FFAP column (J & W Scientific, Folsom, Calif.). Peak areas were determined from chromatograms by using the Shimadzu VP 4.2 software. FAs were identified by using coinjection of known standards and quantified based on internal standards as described previously (9). Analyses were replicated twice, and the results were averaged before being presented as millimolar concentrations of FAs.
Statistical analysis.
Assays were replicated twice, and the data were averaged before presentation. Student's t test was used to determine significant differences in FA production between the parent and mutant and the substrates. Results with P values of ≤0.05 were considered significantly different. The standard deviation for FA production was less than 10% between replicates for both strains and all products.
Results and discussion.
Previous experiments (9) demonstrated that lactococci produced more FAs than lactobacilli from α-keto acids, while the two genera produced similar amounts of FAs from amino acids. These observations and suggestions from other reports (11, 12) led us to suppose that the initiation of the catabolic route by an ATase may be the limiting step in FA production. Despite the large number of ATases in lactococcal genomes (2) (http://www.jgi.doe.gov/JGI_microbial/html/index.html) and the overlapping substrate specificity of ATases (6, 20, 27), we hypothesized that deletion of a single enzyme would decrease the total concentration of FAs. Therefore, a direct comparison with a branched-chain ATase (ilvE) deletion mutant (JLS450) of L. lactis subsp. lactis LM0230 in an assay system was used to investigate FA production from branched-chain amino acids (BCAAs) and α-keto acids.
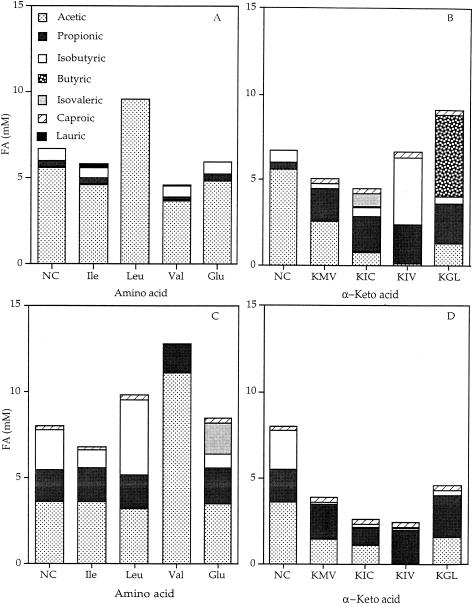
As noted previously (9), the cells incubated in a buffer without a substrate produced FAs (Fig. 1). This is probably due to the utilization of intracellular proteins and amino acids in the absence of external substrates. We also observed that the absolute concentrations of some products were higher than the substrate concentrations both in this study and in previous studies (9). This is plausible, given the degradation of amino acids and α-keto acids to shorter-chain FAs and BCFAs. If these data are analyzed as mole fractions, the total amount of carbon is smaller after the assay, despite the FA products having a higher concentration value than amino or α-keto acid substrates (9) (data not shown). Other intracellular intermediates, such as pyruvate, phosphoglycerate, or acyl coenzyme A derivatives, may be incorporated from other pathways into FA production, leading to a higher concentration for products of smaller chain length (24).
FIG. 1.
Mean FA production by L. lactis subsp. lactis LM0230 from amino acids (A) and from α-keto acids (B), respectively, and by L. lactis subsp. lactis JLS450 from amino acids (C) and from α-keto acids (D), respectively, beyond α-keto acid autodegradation. Values represent the averages of twice-replicated assays of FA production. NC, negative control (cells in plain buffer); Ile, isoleucine; Leu, leucine; Val, valine; Glu, glutamic acid; KMV, α-keto-β-methylvalerate; KIC, α-ketoisocaproate; KIV, α-ketoisovalerate; KGL, α-ketoglutarate. The standard deviation of the total FA production and individual FAs was ≤10% within both strains.
The impact of ilvE was assessed by using two approaches that allowed analysis of the impact of ATases. First, the initiating ATase reaction was bypassed by adding α-keto acid substrates to a culture of the parent (LM0230) in the assay buffer. Second, the ilvE deletion mutant (JLS450) was incubated with BCAAs under the same conditions as those of the parent. Additions of an amino acid substrate to LM0230 and of the α-keto acid substrate to JLS450 were treated as the controls for these experiments (Fig. 1B and C).
The treatment condition using α-keto acids as the substrate for FA production by LM0230 produced approximately the same total FA concentrations as the control of amino acids, which ranged from 5 to 8 mM. LM0230 produced isobutyric acid from all the α-keto acid substrates and isovaleric acid from KIC. Bypassing the initial ATase reaction with the addition of α-keto acids to the parent significantly (P < 0.05) changed the type of BCFAs produced compared with those found in the control amino acid substrates (Fig. 1A and B). A significantly smaller (P < 0.05) amount of acetic acid was produced from all α-keto acids than from the corresponding BCAAs. Isobutyric acid was produced in significantly (P < 0.05) higher amounts from KIC and KIV than leucine and valine, respectively. Interestingly, the only condition that produced n-butyric acid was LM0230 with KGL. In the amino acid control, the total FA production ranged from 5 to 10 mM FAs (Fig. 1A) and isobutyric acid was the only BCFA produced (Fig. 1A). These observations allowed us to suppose that the ATase reactions were not the limiting step in the production of FAs from BCAAs. However, this approach provided evidence that ATases play a role in directing intermediates into specific pathways that lead to specific FA products. Subsequent to these observations, we also hypothesized that ATases play a role in the types of FAs produced.
To verify the role of ilvE, the deletion mutant (JLS450) was assayed for FA production with amino acid substrates. Deletion of ilvE did not significantly (P > 0.05) change the total FA quantities (Fig. 1C). However, deletion of ilvE changed the specific FAs produced from amino acids compared with the parent with the same substrates (Fig. 1A). With either approach, the total FA production did not decrease significantly (P > 0.05). These data agree with the previous observations that ATases were not the limiting step for producing FAs and lend support to this study's hypothesis.
The addition of α-keto acids to JLS450 allowed comparison of catabolic products independent of ATase reactions. JLS450 produced similar FA products and quantities from all α-keto acids, except for KIV, in which case acetic acid was not produced. When the ATase reactions were bypassed, the mutant produced no BCFAs from α-keto acids. The mutant also produced significantly less (P < 0.05) acetic acid than it did from the precursor BCAAs (Fig. 1 C and D). These differences suggest that α-keto acids are catabolized via multiple routes and that the specific routes are ATase dependent. This is plausible, considering the numerous possible pathways subsequent to the ATase reaction (5, 13, 15, 23, 29).
To further assess the role of IlvE, a comparison of the catabolic products from α-keto acids by the parent and the mutant was done (Fig. 1B and D). We expected that the deletion of ilvE and the bypassing of the ATase reactions would yield similar FAs from α-keto acids. Interestingly, JLS450 produced significantly (P < 0.05) smaller amounts of BCFAs from α-keto acids than did LM0230. Deletion of ilvE also abolished production of n-butyric acid from KGL. While it is unclear why n-butyric acid was produced from KGL by LM0230, it is possible that subsequent metabolism of these components followed different pathways via interconversion of α-keto acids and acyl coenzyme A, leading to different products. These data indicate that ilvE is involved in the production of n-butyric and isobutyric acids. These unexpected differences point toward the interactions of metabolic pathways that occur subsequent to the ATase reaction.
Atiles et al. (1) deleted ilvE and found that addition of α-keto acids did not support growth of JLS450; they hypothesized that the deletion blocked BCAA biosynthesis. While this study did not test the role of ilvE in growth, we observed that amino acids and α-keto acids were catabolized to FAs in JLS450, suggesting that ilvE plays a role in production of specific FAs via a complex web of intermediates in addition to a role in growth on α-keto acids.
Yvon et al. (27) deleted bcaT of L. lactis subsp. cremoris, thought to be involved in BCAA catabolism, and expectedly observed a reduction in BCFA production. Conversely, this study found the ilvE deletion of L. lactis subsp. lactis led to an increase in isobutyric and isovaleric acids from isoleucine and leucine compared with that produced by the parent. Observations from this study may be explained by the activities of at least 11 other ATases (2) that may catabolize leucine and isoleucine to BCFAs (6).
Considering the observations from this study, the complementary activity of multiple ATases with BCAAs, and the bidirectional activity of ATases, it is plausible that IlvE and other ATases direct intermediates into different pathways that lead to specific FAs. Deletion or inactivation of any single component of the metabolic web changes the specific products without completely abolishing the catabolic capabilities or reducing the total amount of products.
The observations from this study indicate that the activity of ATases is not the limiting step in producing FAs from amino acids or α-keto acids. However, ATases appear to play a role in directing intermediate substrates toward specific FAs. IlvE was directly involved in the production of isobutyric, n-butyric, and isovaleric acids from amino and α-keto acids in L. lactis subsp. lactis. Additional studies are needed to delineate the roles of other ATases in FA production.
Acknowledgments
Mention of companies and products does not constitute endorsement by Utah State University or Utah Agricultural Experiment Station over similar products not mentioned.
Footnotes
Contribution number 7537 of the Utah Agricultural Experiment Station.
REFERENCES
- 1.Atiles, M. W., E. G. Dudley, and J. L. Steele. 2000. Gene cloning, sequencing, and inactivation of the branched-chain aminotransferase of Lactococcus lactis LM0230. Appl. Environ. Microbiol. 66:2325-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis subsp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Jong, C., and H. T. Badings. 1990. Determination of free fatty acids in milk and cheese: procedures for extraction, clean up, and capillary gas chromatographic analysis. J. High Resolut. Chromatogr. 13:94-98. [Google Scholar]
- 4.Dias, B., and B. Weimer. 1998. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl. Environ. Microbiol. 64:3320-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickinson, J. R., M. M. Lanterman, D. J. Danner, B. M. Pearson, P. Sanz, S. J. Harrison, and M. J. E. Hewlins. 1997. A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 272:26871-26878. [DOI] [PubMed] [Google Scholar]
- 6.Engels, W. J. M., A. C. Alting, M. M. T. G. Arntz, H. Gruppen, A. G. J. Voragen, G. Smit, and S. Visser. 2000. Partial purification and characterization of two aminotransferases from Lactococcus lactis subsp. cremoris B78 involved in the catabolism of methionine and branched chain amino acids. Int. Dairy J. 10:443-452. [Google Scholar]
- 7.Ferchichi, M., D. Hemme, M. Nardi, and N. Pamboukdjian. 1985. Production of methanethiol from methionine by Brevibacterium linens CNRZ 918. J. Gen. Microbiol. 131:715-723. [DOI] [PubMed] [Google Scholar]
- 8.Fox, P. F., J. Law, P. L. H. McSweeney, and J. Wallace. 1993. Biochemistry of cheese ripening, vol. 1. Chapman and Hall, London, United Kingdom.
- 9.Ganesan, B., B. C. Weimer, B. Dias, R. Koka, and K. Seefeldt. Monocarboxylic acid production by lactococci and lactobacilli. Int. Dairy J., in press.
- 10.Gao, S., E. S. Mooberry, and J. L. Steele. 1998. Use of 13C nuclear magnetic resonance and gas chromatography to examine methionine catabolism by lactococci. Appl. Environ. Microbiol. 64:4670-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao, S., D. H. Oh, J. R. Broadbent, M. E. Johnson, B. C. Weimer, and J. L. Steele. 1997. Aromatic amino acid catabolism by lactococci. Lait 77:371-381. [Google Scholar]
- 12.Gao, S., and J. L. Steele. 1998. Purification and characterization of oligomeric species of an aromatic amino acid aminotransferase from Lactococcus lactis subsp. lactis S3. J. Food Biochem. 22:197-211. [Google Scholar]
- 13.Guillouet, S., A. A. Rodal, G.-H. An, P. A. Lessard, and A. J. Sinskey. 1999. Expression of the Escherichia coli catabolic threonine dehydratase in Corynebacterium glutamicum and its effect on isoleucine production. Appl. Environ. Microbiol. 65:3100-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper, W. J., T. Kristoffersen, and J. Y. Wang. 1978. Formation of free fatty acids during the ripening of fat modified cheese slurries. Milchwissenschaft 33:604-608. [Google Scholar]
- 15.Harwood, C. S., and E. Canale-Parola. 1981. Adenosine 5′-triphosphate-yielding pathways of branched-chain amino acid fermentation by a marine spirochete. J. Bacteriol. 148:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemme, D., C. Bouillanne, F. Metro, and M.-J. Desmazeaud. 1982. Microbial catabolism of amino acids during cheese ripening. Sci. Aliments 2:113-123. [Google Scholar]
- 17.Langsrud, T., and G. W. Reinbold. 1973. Flavor development and microbiology of Swiss cheese-a review. J. Milk Food Technol. 36:593-609. [Google Scholar]
- 18.Nakae, T., and J. A. Elliott. 1965. Production of volatile fatty acids by some lactic acid bacteria. II. Selective formation of volatile fatty acids by degradation of amino acids. J. Dairy Sci. 48:293-299. [DOI] [PubMed] [Google Scholar]
- 19.Nakae, T., and J. A. Elliott. 1965. Volatile fatty acids produced by some lactic acid bacteria. I. Factors influencing production of volatile fatty acids from casein hydrolysate. J. Dairy Sci. 48:287-292. [DOI] [PubMed] [Google Scholar]
- 20.Rijnen, L., S. Bonneau, and M. Yvon. 1999. Genetic characterization of the major lactococcal aromatic aminotransferase and its involvement in conversion of amino acids to aroma compounds. Appl. Environ. Microbiol. 65:4873-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandine, W. E., and P. R. Elliker. 1970. Microbially induced flavours and fermented food flavour in fermented dairy products. J. Agric. Food Chem. 18:557-562. [Google Scholar]
- 22.Stuart, M. R., L. S. Chou, and B. C. Weimer. 1999. Influence of carbohydrate starvation and arginine on culturability and amino acid utilization of Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 65:665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tauch, A., T. Hermann, A. Burkovski, R. Krämer, A. Pühler, and J. Kalinowski. 1998. Isoleucine uptake in Corynebacterium glutamicum ATCC 13032 is directed by the brnQ gene product. Arch. Microbiol. 169:303-312. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, J., and T. D. Thomas. 1977. Phosphoenolpyruvate and 2-phosphoglycerate: endogenous energy sources for sugar accumulation by starved cells of Streptococcus lactis. J. Bacteriol. 130:583-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weimer, B., K. Seefeldt, and B. Dias. 1999. Sulfur metabolism in bacteria associated with cheese. Antonie van Leeuwenhoek 76:247-261. [PubMed] [Google Scholar]
- 26.Yvon, M., S. Berthelot, and J. C. Gripon. 1999. Adding α-keto glutarate to semi-hard cheese curd highly enhances the conversion of amino acids to aroma compounds. Int. Dairy J. 8:889-898. [Google Scholar]
- 27.Yvon, M., E. Chambellon, A. Bolotin, and F. Roudot-Algaron. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yvon, M., S. Thirouin, L. Rijnen, D. Fromentier, and J. C. Gripon. 1997. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl. Environ. Microbiol. 63:414-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Y.-X., C. D. Denoya, D. D. Skinner, R. W. Fedechko, H. A. I. McArthur, M. R. Morgenstern, R. A. Davies, S. Lobo, K. A. Reynolds, and R. C. Hutchinson. 1999. Genes encoding acyl-CoA dehydrogenase (AcdH) homologues from Streptomyces coelicolor and Streptomyces avermitilis provide insights into the metabolism of small branched-chain fatty acids and macrolide antibiotic production. Microbiology 145:2323-2334. [DOI] [PubMed] [Google Scholar]