Abstract
1,2,3,4-Diepoxybutane (DEB) is a key carcinogenic metabolite of the important industrial chemical 1,3-butadiene. DEB is a bifunctional alkylating agent capable of reacting with DNA and proteins. Initial DNA alkylation by DEB produces N7-(2′-hydroxy-3′,4′-epoxybut-1′-yl)-guanine monoadducts, which can react with another nucleophilic site to form cross-linked adducts. A recent report revealed a strong correlation between cellular expression of the DNA repair protein O6-alkylguanine DNA alkyltransferase (AGT) and the cytotoxic and mutagenic activity of DEB, suggesting that DEB induces AGT-DNA cross-links (J. G. Valadez et al., Activation of bis-electrophiles to mutagenic conjugates by human O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 17 (2004) 972–982). The purpose of our study was to analyze the formation and structures of DEB-induced AGT-DNA conjugates and to identify specific amino acid residues within the protein involved in cross-linking. DNA-protein cross-link formation was detected by SDS-PAGE when 32P-labeled double-stranded oligodeoxynucleotides were exposed to DEB in the presence of either wild-type hAGT or a C145A hAGT mutant. Capillary HPLC-electrospray ionization mass spectrometry (ESI-MS) analysis of hAGT that had been treated with N7-(2′-hydroxy-3′,4′-epoxybut-1′-yl)-deoxyguanosine (dG monoepoxide) revealed the ability of the protein to form either one or two butanediol- dG cross-links, corresponding to mass shifts of +353 and +706 Da, respectively. HPLC-ESI+-MS/MS sequencing of the tryptic peptides obtained from dG monoepoxide-treated protein indicated that the two cross linking sites were the alkyl acceptor site, Cys145 and a neighboring active site residue, Cys150. The same two amino acid residues of hAGT became covalently cross-linked to DNA following DEB treatment. Modification of Cys145 was further confirmed by HPLC-ESI+-MS/MS analysis of dG monoepoxide-treated synthetic peptide GNPVPILIPCHR which represents the active site tryptic fragment of AGT (C = Cys145 ). The replacement of the catalytic cysteine residue with alanine in the C145A hAGT mutant abolished DEB-induced cross-linking at this site, while the formation of conjugates via neighboring Cys150 was retained. The exact chemical structure of the cross-linked lesion was established as 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol by HPLC-ESI+-MS/MS analysis of the amino acids resulting from the total digestion of modified proteins analyzed in parallel with an authentic standard. AGT-DNA cross-linking is a likely mechanism of DEB-mediated cytotoxicity in cells expressing this important repair protein.
Keywords: 1, 2, 3, 4-Diepoxybutane, 1, 3-Butadiene, O6-Alkylguanine DNA Alkyltransferase, Cross-Linking, Bioactivation, Mutagenicity
Introduction
The DNA repair protein O6-alkylguanine DNA alkyltransferase (AGT) transfers the O6-alkyl group from promutagenic O6-alkylguanine lesions in DNA to an active site cysteine (Cys145 in human protein), restoring normal guanine and preventing mutagenesis (1). Crystal structures of DNA-bound AGT support a repair mechanism involving protein binding to the minor groove of double-stranded DNA and subsequent “flipping” of nucleotides out of the base stack into the AGT active site (2). This conformational change places O6-alkylguanine group <3 Å from the side chain of Cys145 within the active site. Cys145 is activated to a thiolate anion because of its interactions with a bound Zn (II) cation and the formation of a Glu-His-water-Cys H-bond network (3,4). Nucleophilic attack of Cys145 at the O6-alkyl lesion results in the formation of a thioether at Cys145 and restores intact dG within DNA. Because alkylation of the catalytic residue of AGT is irreversible, this repair process is non-enzymatic in nature, consuming a molecule of AGT in each alkyl transfer reaction (3). Cys145 alkylation destabilizes the native fold of AGT, targeting the protein for ubiquitination and subsequent degradation by the 26S proteasome (3,5).
AGT can repair a wide variety of O6-alkylguanines, including those resulting from exposure to anticancer chemotherapeutics (dacarbazine, temozolomide, bis-chloroethyl-nitrosourea). AGT overexpression in tumors can be responsible for the acquired resistance of cancer cells to DNA-alkylating agents (6,7). A reversal of AGT-mediated drug resistance can be achieved by co-administration of O6-benzylguanine, an AGT substrate that inactivates the protein via transfer of the O6-benzyl group to Cys145 (8).
In a sharp contrast with the protective effects of AGT against alkylation damage by many DNA alkylating agents, AGT expression in cell culture enhances the cytotoxic and mutagenic effects of 1,2,3,4-diepoxybutane (DEB) (9,10). DEB is the suspected carcinogenic metabolite of 1,3-butadiene (BD), a major industrial chemical used in the manufacturing of rubber and plastics and a recognized human carcinogen (11,12). The metabolic activation of BD to DEB is catalyzed by CYP2E1 and CYP2A6 monooxygenases. The first epoxidation step yields (R)- and (S)-3,4-epoxy-1-butene (EB) (13,14). EB can then be hydrolyzed to 1-butene-3,4-diol or can undergo a second oxidation to yield R,R; S,S; and meso-DEB (15). Although DEB is a relatively minor metabolite, available experimental evidence suggests that it is responsible for many of the adverse effects of BD. Biological studies indicate that DEB is 50–100-fold more genotoxic and mutagenic than its monoepoxide analogues, EB and 3,4-epoxy-1,2-butanediol (16,17). Efficient metabolism of BD to DEB in target tissues of laboratory mice is thought to be responsible for susceptibility of this species to BD carcinogenesis (18).
The enhanced biological activity of DEB in the presence of AGT protein (9) may result from chemical cross-linking of AGT to DNA, by analogy with other bis-alkylating agents (Scheme 1) (9,22). DEB is a bifunctional electrophile capable of sequentially alkylating two nucleophilic sites within DNA and/or proteins (19–21). Evidence for the formation of DNA-protein cross-links of DEB in vivo has been provided by alkaline elution assays (23), however, no structural information for these lesions was available. The goal of the present work was to investigate the molecular mechanisms of AGT-mediated cytotoxicity and mutagenicity of DEB by determining DNA-AGT cross-link structures and identifying the amino acid residues within AGT involved in cross-linking.
Scheme 1.
DNA-AGT cross-linking by dihaloethanes and DEB.
Materials and Methods
Chemicals and Reagents
Racemic DEB was purchased from Sigma-Aldrich (Milwaukee, WI), 2′-deoxyguanosine was obtained from TCI (Tokyo, Japan). L-cysteine and Boc-L-cysteine were purchased from Fluka (Buchs, Switzerland). Adenosine 5′-[γ-32P]triphosphate was purchased from Perkin-Elmer (Boston, MA), and T4 polynucleotide kinase was obtained from New England Biolabs (Beverly, MA). Trypsin, carboxypeptidase Y, and proteinase K were purchased from Worthington Biochemical Corporation (Lakewood, NJ). Synthetic DNA oligodeoxynucleotides and the peptide GNPVPILIPCHR corresponding to positions 136–147 of hAGT were prepared at the University of Minnesota’s Microchemical Facility (Minneapolis, MN). Recombinant C-terminal histidine-tagged wild-type hAGT and an N-terminal histidine-tagged C145A hAGT mutant (Tables 1 and 2) were prepared as previously described (24,25).
Table 1.
HPLC-ESI+-MS analysis of hAGT tryptic peptides.
| Position | Peptide | [M + H]+calculated | [M + 2H]2+calculated | Observed Ions |
|---|---|---|---|---|
| 1–9 | MDKDCEMKR | 1156.4 | 578.7 | 1156.7, 578.9 |
| 10–18 | TTLDSPLGK | 931.5 | 466.3 | 931.7, 466.3 |
| 19–32 | LELSGCEQGLHEIK | 1555.8 | 778.4 | 778.8 |
| 33–36 | LLGK | 430.3 | 215.7 | 430.4, 215.6 |
| 37–96 | GTSAADAVEVPAPAAVLGG PEPLMQCTAWLNAYFHQP EAIEEFPVPALHHPVFQQES FTR |
6469.2 | 3235.1 | ND |
| 97–101 | QVLWK | 673.4 | 337.2 | 673.5 |
| 102–104 | LLK | 373.3 | 187.1 | 373.3, 187.1 |
| 105–107 | VVK | 345.3 | 173.1 | ND |
| 108–125 | FGEVISYQQLAALAGNPK | 1906.0 | 953.5 | 953.5 |
| 126–128 | AAR | 317.2 | 159.0 | 317.2 |
| 129–135 | AVGGMAR | 661.4 | 331.2 | 661.4, 331.2 |
| 136–147 | GNPVPIKIPCHR | 1315.7 | 658.4 | 1315.7, 658.6 |
| 148–165 | VVCSSGAVGNYSGGLAVK | 1667.8 | 834.4 | 834.6 |
| 166–175 | EWLLAHEGHR | 1247.6 | 624.3 | 1247.7, 624.5 |
| 176–193 | LGKPGLGGSSGLAGAWLK | 1668.9 | 835.0 | 1669.0, 835.2 |
| 194–207 | GAGATSGSHHHHHH | 1429.6 | 715.3 | 1429.6, 715.4 |
Table 2.
HPLC-ESI+-MS analysis of C145A hAGT tryptic peptides.
| Position | Peptide | [M + H]+calculated | [M + 2H]2+calculated | Observed Ions |
|---|---|---|---|---|
| 1–9 | MRGSHHHHHHGSMDKDCEMKR | 2553.1 | 1277.1 | ND |
| 10–18 | TTLDSPLGK | 931.5 | 466.3 | 931.6, 466.3 |
| 19–32 | LELSGCEQGLHEIK | 1555.8 | 778.4 | 778.4 |
| 33–36 | LLGK | 430.3 | 215.7 | 430.4 |
| 37–96 | GTSAADAVEVPAPAAVLGG PEPLMQCTAWLNAYFHQP EAIEEFPVPALHHPVFQQES FTR |
6469.2 | 3235.1 | ND |
| 97–101 | QVLWK | 673.4 | 337.2 | 673.5 |
| 102–104 | LLK | 373.3 | 187.1 | 373.4 |
| 105–107 | VVK | 345.3 | 173.1 | ND |
| 108–125 | FGEVISYQQLAALAGNPK | 1906.0 | 953.5 | 953.6 |
| 126–128 | AAR | 317.2 | 159.0 | ND |
| 129–135 | AVGGAMR | 661.4 | 331.2 | 661.4 |
| 136–147 | GNPVPILIPAHR | 1283.8 | 642.4 | 1283.8, 642.5 |
| 148–165 | VVCSSGAVGNYSGGLAVK | 1667.8 | 834.4 | 834.4 |
| 166–175 | EWLLAHEGHR | 1247.6 | 624.3 | 1247.6, 624.5 |
| 176–193 | LGKPGLGGSSGLAGAWLK | 1668.9 | 835.0 | 1669.1, 835.4 |
| 194–207 | GAGATSGSPPAGRN | 1199.6 | 600.4 | 1199.7, 600.6 |
Note: Numbering begins at M.
Detection of DEB-Induced AGT- DNA Cross-Links by SDS-PAGE
DNA 18-mer 5′-GGA GCT GGT GGC GTA GGC-3′ (+ strand) and the complimentary (−) strand were purified using an Agilent Technologies HPLC system (model 1100) incorporating a UV diode array detector and a semi-micro UV cell. A Supelcosil LC-18-DB semipreparative column (25 cm × 10 mm, 5 μm) was eluted with 100 mM TEAA, pH 7.0 (A) and 50% acetonitrile in 100 mM TEAA, pH 7.0 (B) at a flow rate of 3 mL/min. The gradient program began at 7.5% B, followed by a linear increase to 26% B in 42 min, and further to 60% B in 28 min. Under these conditions, DNA 18-mers eluted as sharp peaks around 40 min. HPLC pure (+) strand (200 pmol) was 5′-end labeled with 32P in the presence of [γ-32P]ATP/T4 polynucleotide kinase (26). The resulting [γ-32P] 5′-end labeled oligodeoxynucleotides were purified by 8% denaturing PAGE and desalted by SPE using Waters SepPak C18 cartridges. Once spiked with the corresponding unlabeled DNA (12 nmol), the radiolabeled (+) strand was annealed to its complimentary strand (12.2 nmol) in AGT buffer (50 mM TRIS (pH 7.6) and 0.1 mM EDTA). The 32P-labeled duplex (0.93 nmol) was incubated with hAGT (2.0 μg, 0.093 nmol) in the presence of 100, 500, and 3000 molar equivalents of racemic DEB (9.3 nmol, 46.5 nmol, and 279 nmol respectively). In a separate experiment, C145A hAGT (2.0 μg, 0.087 nmol ) was incubated with 3000 molar equivalents of racemic DEB. Following 3 h incubation at 37 °C, the reaction mixtures were separated by 12% SDS-PAGE, and the radiolabeled products were visualized using a Bio-Rad Molecular Imager FX. DEB-induced DNA-protein cross-linking was quantified by volume analysis employing Bio-Rad Quantity One Software, with cross-linking efficiency determined by the intensity ratio of the low-mobility cross-link band versus the band corresponding to single-stranded DNA.
Preparation of dG Monoepoxide
2′-Deoxyguanosine (59.0 mg, 0.22 mmol) was suspended in 3 mL of 10 mM TRIS-HCl buffer (pH 7.2), and 34 μL (0.44 mmol) of racemic DEB was added. The resulting mixture was incubated overnight at 37 °C. N7-(2′-hydroxy-3′,4′-epoxybut-1′-yl)-deoxyguanosine (dG monoepoxide) was purified by semipreparative HPLC on an Agilent 1100 HPLC system. A Supelcosil LC-18-DB column (25 cm × 10 mm, 5 μm) was eluted with a gradient of water (A) and acetonitrile (B) at a flow rate of 3 mL/min. Solvent composition was changed linearly from 3 to 7 % B in 8 min, further to 8% B in 3 min, and held at 8% B for 9 min. Under these conditions, dG monoepoxide eluted as a broad peak at ~16 min. dG monoepoxide was characterized by tandem mass spectrometry (ESI+-MS/MS: m/z 354.0 [M + H]+ → 238.0 [M + H – dR]+, 152.0 [Gua + H]+). UV absorption spectrum of dG monoepoxide (λmax = 274 nm at pH 7.0) was reminiscent of other N7-alkylguanosines, including N7-methyl dG. Thus, the extinction coefficient for N7-methyl dG (ε 274 = 9,000 at pH 7.0) was used to estimate standard solution concentrations of HPLC-purified dG monoepoxide. dG monoepoxide was stable in an aqueous solution at −20 °C for up to a month as determined by ESI+-MS analysis.
Preparation of Cys-Gua-BD
2′-Deoxyguanosine (29.5 mg, 0.11 mmol) was combined with racemic DEB (17 μL, 0.22 mmol) in 1 mL of 10 mM TRIS-HCl buffer (pH 7.2), and the resulting mixture was incubated overnight at 37 °C. Boc-L-cysteine (24.4 mg, 0.11 mmol) was added, and the mixture was left for an additional 48 h at 37 °C. The reaction mixture was then centrifuged at 1,000 × g for 2 min, and the supernatant was dried in a speed-vac concentrator. The resulting white powder was dissolved in 200 μL of TFA and allowed to stand at room temperature for 15 min to remove the Boc protecting group. The structure of 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol (Cys-Gua-BD) was confirmed by HPLC-ESI+-MS/MS (m/z 359.7 [M + H]+ → 208.0 [M+ H – Gua]+, 190.0 [M+ H – Gua – H2O]+).
Modification of synthetic peptide GNPVPILIPCHR with dG monoepoxide
Synthetic peptide GNPVPILIPCHR representing positions 136–147 of hAGT (40.0 μg, 30.42 nmol) was reacted with 5 molar equivalents of dG monoepoxide in 400 μL of 10 mM TRIS-HCl buffer, pH 7.2. The mixture was incubated under argon at 37 °C for 3 h. The peptide-guanine DEB conjugate was isolated via HPLC using an Agilent 1100 HPLC system. A Supelcosil LC-18-DB column (25 cm × 4.6 mm, 5 μm) was eluted with 0.1% TFA in water (A) and 0.1% TFA in acetonitrile (B) at a flow rate of 0.2 mL/min. The solvent composition began at 5% B and was changed linearly to 80% B in 45 min. Under these conditions, the peptide-guanine cross-link eluted as a broad peak at ~13 min.
dG Monoepoxide-Induced Cross-Linking of AGT for MS Analysis
Recombinant hAGT (77.6 μg, 3.55 nmol) or C145A hAGT (77.5 μg, 3.37 nmol) was incubated with 50 molar equivalents of dG monoepoxide in 200 μL of 10 mM TRIS-HCl buffer (pH 7.2) under argon for 3 h at 37 °C. Following incubation, the proteins were isolated by HPLC using an Agilent 1100 HPLC system. A Zorbax 300 SB-C3 column (2.1 mm × 150 mm, 5 μm) was eluted with 0.05% TFA in water (A) and 0.05% TFA in acetonitrile (B) at a flow rate of 0.2 mL/min. The elution program began at 30% B and was increased to 80% B in 30 min. Under these conditions, both native and modified AGT proteins eluted as a single peak at 27 min. The collected protein fractions were dried, dissolved in 0.05% TFA, and aliquots were taken for HPLC-ESI+-MS analysis. To identify the residues of hAGT and C145A hAGT involved in cross-linking to guanine, dG monoepoxide-treated protein samples (~70 μg each) were dried, dissolved in 200 μL of 100 mM ammonium bicarbonate buffer (pH 7.9), and trypsin (7.0 μg, 0.30 nmol) was added to initiate proteolytic digestion. The resulting solutions were incubated at 37 °C for 24 h. The proteolytic mixtures were then dried, reconstituted in 50 μL of 0.5% formic acid/0.01% TFA in water, and subjected to HPLC-ESI+-MS/MS analysis as described below. To confirm the structures of dG monoepoxide-induced cross-links, hAGT and C145A hAGT tryptic peptides were subjected to total digestion to amino acids. hAGT and C145A hAGT tryptic peptides (from ~50 μg protein) were dissolved in 100 μL of water and filtered through Microcon YM-10 membrane filters to remove trypsin, followed by a 24 h incubation of the filtrate with carboxypeptidase Y (1.0 μg, 16.4 pmol) and proteinase K (1 μg, 34.6 pmol) at room temperature. Once dried, the samples were reconstituted in 40 μL water and analyzed by HPLC-ESI+-MS/MS as detailed in a later section.
dG Monoepoxide-Induced Cross-Linking of AGT in the Presence of L-Cysteine (Competition Experiments)
hAGT (20.0 μg, 0.914 nmol) was treated with 50 equivalents of racemic dG monoepoxide (45.7 nmol, 0.23 mM) in the presence of 0, 500, 1250, and 2500 equivalents of L-cysteine (2.3, 5.7, and 11.4 mM respectively) in 200 μL of 10 mM TRIS-HCl buffer, pH 7.2. Following a 3 h incubation under argon at 37 °C, the samples were acidified via addition of 50 μL 1.0% formic acid and directly analyzed by HPLC-ESI+-MS as detailed below.
DEB-Induced Cross-Linking of AGT to Double-Stranded Oligodeoxynucleotides for MS Analysis
The DNA 18-mer (5′-GGA GCT GGT GGC GTA GGC-3′) (+ strand) and the complementary (−) strand (5′-GCC TAC GCC ACC AGC TCC-3′) were prepared by standard phosphoramidite chemistry on a DNA synthesizer. To obtain double-stranded DNA, (+) and (−) strand (20 nmol each) were combined, dried under vacuum, and dissolved in 10 mM TRIS-HCl buffer, pH 7.2. The solution was heated to 90 °C and allowed to slowly cool to room temperature to anneal the complementary strands. hAGT (44.0 μg, 2.01 nmol) or C145A hAGT (46.0 μg, 2.00 nmol) was incubated under argon for 3 h at 37 °C with 10 nmol of double-stranded DNA in 10 mM TRIS-HCl buffer (pH 7.2) containing racemic DEB (1 μmol, 2.5 mM). Following incubation, the samples were heated to 70 °C for 1 h to release N7-alkylated guanines. Once dry, the samples were dissolved in 100 mM ammonium bicarbonate buffer (pH 7.9) and trypsin (5 μg, 0.21 nmol) was added. The samples were digested overnight at 37 °C. The resulting tryptic peptides were dried, reconstituted in 50 μL 0.5% formic acid/0.01% TFA in water, and analyzed by HPLC-ESI+-MS/MS.
Capillary HPLC-ESI+-MS/MS Analysis of dG Monoepoxide and DEB-Induced AGT-Guanine Cross-Links
HPLC-ESI+-MS analysis of hAGT and C145A hAGT proteins was performed using an Agilent 1100 capillary HPLC-ion trap MS system operated in the ESI+ mode. The spectra were obtained by performing full scan MS within the m/z range of 100–1500. Chromatographic separation was achieved with an Agilent Zorbax 300 SB-C3 column (150 mm × 0.5 mm, 5 um) eluted at a flow rate of 15 uL/min. The mobile phase consisted of 0.05% TFA in water (A) and 0.05% TFA in acetonitrile (B). The elution program started at 30% B for 5 min, followed by a linear increase to 80% B in 25 min. Under these conditions, hAGT and C145A hAGT (both native and alkylated) eluted as sharp peaks at ~16 min. Deconvolution of the resulting charge envelopes was performed using the Agilent ion trap deconvolution software.
HPLC-ESI+-MS/MS analysis of hAGT and C145A hAGT tryptic peptides was performed using the same Agilent 1100 capillary HPLC-ion trap MS system. Operating in ESI+ mode, Auto MS2 was used to select and fragment the following doubly-charged ions: m/z 658.4 (unmodified peptide G136NPVPILIPCHR147); m/z 776.9 (G136NPVPILIPCHR147 peptide cross-linked to guanine); m/z 710.4 (G136NPVPILIPCHR147 peptide containing a DEB-induced trihydroxybutyl adduct); m/z 834.4 (unmodified peptide V148VCSSGAVGNYSGGLAVK165); m/z = 952.9 (V148VCSSGAVGNYSGGLAVK165 peptide cross-linked to guanine); m/z 886.4 (V148VCSSGAVGNYSGGLAVK165 peptide containing a DEB-induced trihydroxybutyl adduct); and m/z 642.4 (unmodified peptide G136NPVPILIPAHR147 from C145A hAGT).
Chromatographic separation of tryptic peptides was achieved using an Agilent Zorbax SB-C18 column (150 mm × 0.5 mm, 5 μm) eluted at a flow rate of 15 μL/min. The mobile phase consisted of 0.5% formic acid/0.01% TFA in water (A) and 0.5% formic acid/0.01% TFA in acetonitrile (B). The solvent composition began at 3% B and was held constant for the first 3 min. The gradient was then increased to 5% B in 7 min, held at 5% B for 10 min, increased to 35% B in 95 min, and further to 75% B in 10 min. Full scan data obtained from these and control experiments was used in the identification of other tryptic peptides. This same method was employed for the analysis of synthetic peptide GNPVPILIPCHR containing a dG monoepoxide-induced cross-link to guanine and for DEB-induced AGT-DNA cross-links.
HPLC-ESI+-MS/MS analysis of the amino acids present in total digests of alkylated AGT was performed with an Agilent 1100 capillary HPLC-ion trap MS system. Operating in positive ion mode, Auto MS2 was used to isolate and fragment the following singly-charged ions: m/z 359.1 (1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol); m/z 420.1 (1-(O-tyrosyl)-4-(guan-7-yl)-2,3-butanediol); m/z 385.4 (1-(N-lysyl)-4-(guan-7-yl)-2,3-butanediol); m/z 413.4 (1-(N-arginyl)-4-(guan-7-yl)-2,3-butanediol); and m/z 393.4 (1-(N-histidyl)-4-(guan-7-yl)-2,3-butanediol). Chromatographic separation was achieved using a Phenomenex Synergi C18 column (250 mm × 0.5 mm, 4 um) eluted at a flow rate of 10 uL/min. The mobile phase consisted of 15 mM ammonium acetate, pH 5.0 in water (A) and 3:1 methanol/acetonitrile (B). The solvent composition was changed from 2–8% B in 10 min, increased to 12% B in 22 min, and further to 30% B in 10 min. Under these conditions, Cys-Gua-BD eluted as a sharp peak at ~15 min.
Results
SDS-PAGE Analysis of DEB-Induced AGT-DNA Cross-Links
The ability of DEB to induce AGT-DNA cross-links was first examined by SDS-PAGE (Figure 1). DNA duplexes composed of the 5′-32P-end labeled 18-mer 5′-GGA GCT GGT GGC GTA GGC-3′ (+ strand) and its complementary (−) strand (5′-GCC TAC GCC ACC AGC TCC-3′) were incubated with recombinant human AGT protein in the presence of increasing amounts of racemic DEB (100, 500, or 3000 molar equivalents). SDS-PAGE analysis of the resulting reaction mixtures provided evidence for the formation of hAGT-DNA conjugates as indicated by the appearance of a new DNA band with reduced mobility as compared with free 18-mer (Figure 1). This slowly moving band was absent in control experiments lacking AGT, DEB, or the DNA oligomer (lanes 1–3 in Figure 1). The degree of DEB-induced hAGT-DNA cross-linking varied between 1.5 and 3%, depending on DEB concentration (lanes 4–6). Interestingly, AGT-DNA complex formation was also observed for the C145A hAGT mutant lacking the catalytic cysteine residue (lane 7), suggesting that AGT-DNA cross-linking by DEB can take place at an alternative site within the protein.
Figure 1.
12% SDS-PAGE analysis of 32P-end labeled DNA duplexes (5′-GGA GCT GGT GGC GTA GGC-3′, (+) strand) following incubation with DEB and hAGT (lanes 4–6) or C145A hAGT (lane 7). AGT-DNA cross-links are observed as slowly moving bands on the gel. Lanes 1–3 serve as negative controls.
Capillary HPLC-ESI+-MS Analysis of dG Monoepoxide-Treated AGT: Whole Protein MS
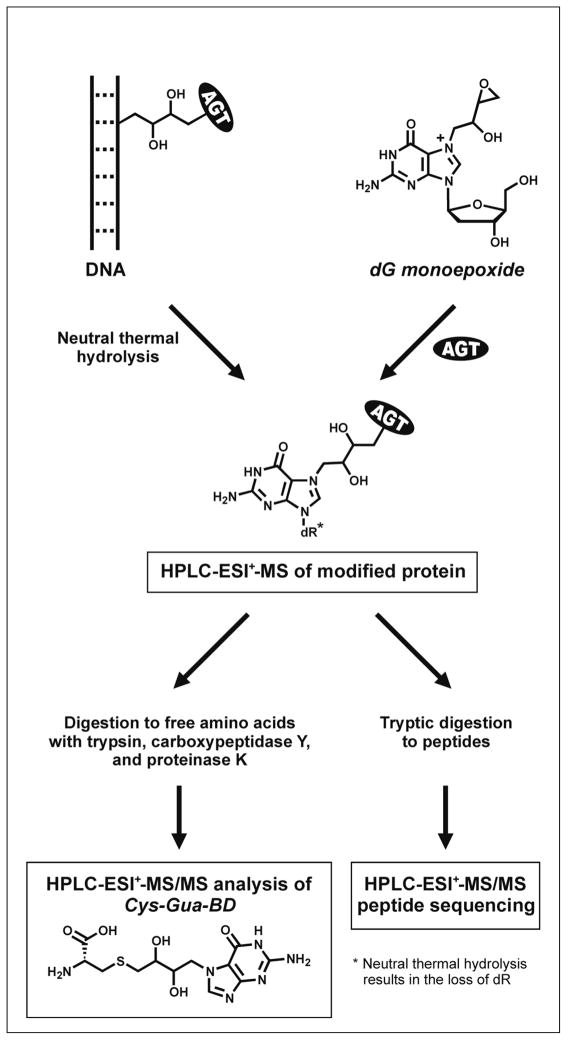
A mass spectrometry based strategy was employed to identify the cross-linking sites within the protein and to determine the covalent structures of DEB-induced DNA-AGT conjugates (Scheme 2). Initial DNA alkylation by DEB is known to produce N7-(2′-hydroxy-3′,4′-epoxybut-1′-yl)-guanine (dG monoepoxide) adducts, which result from the nucleophilic attack of N7-guanine at the terminal carbon of DEB (> 90% of total adducts) (27). Therefore, our initial studies of DNA-protein cross-linking by DEB employed synthetic N7-(2′-hydroxy-3′,4′-epoxybut-1′-yl)-deoxyguanosine as a model for monoalkylated DNA (Scheme 2, right). We then investigated AGT cross-linking to double-stranded DNA in the presence of DEB (Scheme 2, left). In DNA experiments, N7-alkylguanine residues were released from the DNA backbone by neutral thermal hydrolysis to facilitate the mass spectral analysis of protein conjugates (Scheme 2).
Scheme 2.
Mass spectrometric analysis of DEB-induced AGT-DNA conjugates.
HPLC-ESI+-MS analysis of native hAGT on an ion trap mass spectrometer reveals a single protein peak eluting at 16.6 min (Figure 2A). The ESI+ spectrum across this peak contains m/z signals corresponding to multiple charge states (+18 to +30) of a protein with a mass of 21 880 Da, consistent with the calculated molecular weight of 21 876 Da (see Supporting Information, S-1). HPLC-ESI+-MS analysis of hAGT following incubation with excess dG monoepoxide reveals the presence of native hAGT (M = 21 877 Da), as well as monoalkylated protein containing a single butanediol-dG cross-link (calculated M = 22 229 Da, observed M = 22 230 Da) and a protein containing two butanediol cross-links to dG (calculated M = 22 582 Da, observed M = 22 586 Da) (Figure 2B, insert). For each additional adduct, a mass increase of ~353 Da is observed, corresponding to the addition of a molecule of dG monoepoxide. dG monoepoxide-induced modification of hAGT was not blocked by the presence of free L-cysteine (Table 3). Over 1000-fold molar excess of L-cysteine was required to achieve a small decrease of AGT-dG conjugate yields, accompanied by the formation of disulfide adducts between the free L-cysteine and cysteine residues within the protein (Table 3). These experiments suggest that free cysteine cannot prevent the formation of DNA-protein cross-links by DEB, consistent with our previous data which revealed a lack of reactivity between glutathione and DEB under physiological conditions (Loeber and Tretyakova, unpublished results).
Figure 2.
HPLC-ESI+-MS analysis of dG monoepoxide-induced butanediol cross-links to hAGT. (A) HPLC-ESI+-MS of untreated hAGT. Top: Total ion chromatogram; Bottom: ESI+ mass spectrum of the 16.6 min protein peak; Inset: Deconvoluted mass spectrum of the 16.6 min peak (observed M = 21 880 Da, calculated M = 21 876 Da). (B) HPLC-ESI+-MS of hAGT following incubation with 50 molar equivalents dG monoepoxide. Top: Total ion chromatogram; Bottom: ESI+ mass spectrum of the 16.3 min protein peak; Inset: Deconvoluted mass spectrum of the 16.3 min peak: A = hAGT containing a single cross-link to dG (observed M = 22 230 Da, calculated M = 22 229 Da); B = double cross-link to dG (observed M = 22 586 Da, calculated M = 22 582 Da); C = unmodified hAGT (observed M = 21 877 Da, calculated M = 21 876 Da).
Table 3.
Inhibition of dG monoepoxide-induced modification of hAGT in the presence of L-cysteine.
| Equivalents L-Cysteine | % Unmodified AGT M = 21877.7 ± 0.2 Da | % AGT Butanediol Cross-Link to dG M = 22229.9 ± 0.9 Da | % AGT-Cysteine Disulfide M = 21996.5 ± 0.1 Da |
|---|---|---|---|
| 0 | 78.9 | 21.1 | 0 |
| 500 | 81.3 | 18.7 | 0 |
| 1250 | 77.6 | 11.5 | 10.9 |
| 2500 | 80.5 | 5.4 | 14.1 |
Similar analyses were conducted with the C145A hAGT active site mutant. In this protein, the Cys145 alkyl acceptor site is replaced with alanine, eliminating the possibility of cross-linking through this position. The calculated molecular weight of the C145A hAGT variant is 23 012 Da because of the addition of the N-terminal histidine tag (MRGSHHHHHHGS) (see Supporting Information, S-1). HPLC-ESI+-MS analysis of C145A AGT prior to treatment reveals a single protein species with a deconvoluted mass of 23 015 Da, which is consistent with the theoretical value (Figure 3A). HPLC-ESI+-MS analysis of dG monoepoxide-treated C145A hAGT provides evidence of cross-linking at a single site (observed M = 23 367, mass increase of 353 Da), suggesting that one of the cross-linking sites observed within the wild-type protein, namely Cys145, has been removed (Figure 3B). The residual ability of C145A hAGT mutant to form butanediol conjugates to dG is consistent with our SDS-PAGE results, which support C145A hAGT cross-linking to DNA in the presence of DEB (Figure 1, lane 7).
Figure 3.
HPLC-ESI+-MS analysis of C145A hAGT protein following treatment with dG monoepoxide. (A) HPLC-ESI+-MS of untreated C145A hAGT. Top: Total ion chromatogram; Bottom: ESI+ mass spectrum of the 16.2 min protein peak; Inset: Deconvoluted mass spectrum of the 16.2 min peak (observed M = 23 015 Da, calculated M = 23 012 Da). (B) HPLC-ESI+-MS of C145A hAGT mutant following incubation dG monoepoxide. Top: Total ion chromatogram; Bottom: ESI+ mass spectrum of the 16.1 min protein peak; Inset: Deconvoluted mass spectrum of the 16.1 min peak: A = C145A hAGT containing a single cross-link to dG (observed M = 23 367 Da, calculated M = 23 365 Da); B = unmodified protein (observed M = 23 014 Da, calculated M = 23 012 Da).
Capillary HPLC-ESI+-MS Analysis of dG Monoepoxide-Treated AGT: Peptide Mapping
Our whole protein MS results for dG monoepoxide-treated AGT variants (Figures 2 and 3) are consistent with the presence of two distinct cross-linking sites in the wild-type protein, one of which involves the alkyl acceptor site, Cys145. Further insight into the identities of AGT amino acids responsible for reaction with dG monoepoxide was provided by HPLC-ESI+-MS/MS analysis of tryptic digests of alkylated protein.
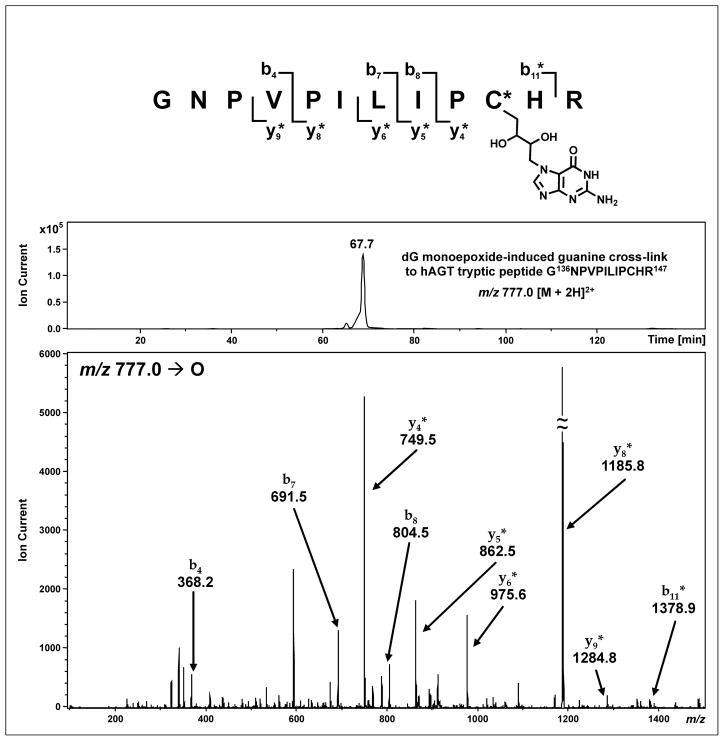
Proteolytic digestion with trypsin provides good sequence coverage for both protein variants (Tables 1 and 2). For wild-type hAGT, 14 of the 16 possible tryptic peptides were observed, including the doubly charged active site peptide G136NPVPILIPC*HR147 (C* = Cys145) at 658.4 m/z (calculated M = 1314.7 Da). HPLC-ESI+-MS/MS analysis of the tryptic digest of dG monoepoxide-treated hAGT detected a prominent doubly charged peptide (m/z 777.9 [M + 2H]2+) corresponding to a butanediol conjugate between G136NPVPILIPCHR147 and a free guanine base (calculated M = 1551.7 Da, ΔM = 237 Da) (Figure 4). As expected, the deoxyribose originating from dG monoepoxide was released by spontaneous hydrolysis of the N7-alkylated dG during tryptic digestion of the cross-linked protein. When this peptide was subjected to collision induced dissociation (CID) in the ion trap mass spectrometer, it produced MS/MS spectrum consistent with the presence of a butanediol-guanine adduct at C* (Cys 145) (Figure 4). The masses of b4-b8 ions were in agreement with theoretical values for unmodified peptide, while the mass of the b11 fragment was increased by 237 Da (observed M = 1377.9 Da vs calculated M = 1141.6 Da for the unmodified peptide), suggesting that the adduct resides between the 9th and 11th residue of the peptide (PC*H). The y4-y9 ions carry the butanediol-Gua adduct as indicated by a +237 Da mass shift, consistent with modification at the C-terminus (PC*HR). Taken together, these results are consistent with butanediol-guanine cross-linking to Cys145 (C*). Further proof of modification at this site was obtained by HPLC-ESI+-MS/MS analysis of synthetic peptide GNPVPILIPCHR containing a dG monoepoxide-induced butanediol cross-link to guanine (m/z 777.1 [M + 2H]2+) which displayed similar HPLC retention time and MS/MS fragmentation as the hAGT derived cross-link (see Supporting Information, S-2). Finally, the C145A mutant has lost the ability to form conjugates within this protein region, supporting a critical role of Cys145 in the cross-linking reaction (see below).
Figure 4.
HPLC-ESI+-MS/MS analysis of hAGT tryptic peptide G136NPVPILIPCHR147 containing a dG monoepoxide-induced butanediol cross-link between Cys145 and guanine. Top: Extracted ion chromatogram of m/z 777.0 [M + 2H]2+; Bottom: MS/MS spectrum. Modified fragment ions are indicated by “*”.
The second cross-linking site was located upon further analysis of the tryptic fragments originating from dG monoepoxide-treated wild-type AGT. HPLC-ESI+-MS/MS signal corresponding to the peptide V148VC*SSGGAVGNYS GGLAVK165 containing a single butanediol-guanine adduct was detected (m/z 953.6 [M + 2H]2+; calculated M = 1903.8) (Figure 5). The cross-linking site was mapped to Cys150 (C*) based on the MS/MS fragmentation patterns, especially the diagnostic b3 and y15 ions at 593.3 and 1366.8 m/z, respectively (Figure 5). The y15 ion mass matched the theoretical value, while the b3 ion experienced a +237 Da mass shift, suggesting that cross-linking took place within the C-terminal region of the peptide (VVC), probably at Cys150.
Figure 5.
HPLC-ESI+-MS/MS analysis of hAGT tryptic peptide V148VCSSGGAVGNYSG GLAVK165 containing dG monoepoxide-induced butanediol cross-link between Cys150 and guanine. Top: Extracted ion chromatogram of m/z 953.6 [M + 2H]2+; Bottom: MS/MS spectrum. Modified fragment ions are indicated by “*”.
HPLC-ESI+-MS/MS analysis of the tryptic digests of the C145A hAGT mutant detected 12 of the 16 expected tryptic fragments, including the active site peptides G136NPVPILIPAHR147 (m/z 624.5 [M + 2H]2+) and V148VC*SSGGAVGNYSG GLAVK165 (m/z 834.8 [M + 2H]2+) (Table 2). Mass spectral analysis of dG monoepoxide treated C145A mutant detected an adducted peptide V148VC*SSGGAVGNY SGGLAVK165 containing a butanediol cross-link to guanine (m/z 953.4 [M + 2H]2+). This peptide adduct displayed the same HPLC retention time and CID fragmentation pattern as the product observed in wild-type hAGT exposed to dG monoepoxide (Figure 5), suggesting that these two peptide cross-links were identical. In contrast, no cross-linking to peptide G136NPVPILIPAHR147 was observed for C145A mutant protein in which the key cysteine residue was replaced with alanine (A) (results not shown), confirming that C145 participates in cross-linking to DNA.
HPLC-ESI+-MS/MS Analysis of DEB-Induced AGT Cross-Links to Double-Stranded DNA
Although our peptide mapping results using a model system composed of AGT protein and pre-formed dG monoepoxide (Scheme 2, right) are consistent with the formation of butanediol conjugates involving active site cysteines 145 and 150 (Figures 3–5), the cross-linking specificity of DEB can potentially be altered upon AGT binding to double-stranded DNA. Furthermore, protein modification by DEB may take place prior to DNA binding as proposed for dihaloethanes (22). Therefore, an additional series of experiments was conducted to map the locations of DEB-induced cross-links of AGT to double-stranded DNA. In these studies, hAGT or C145A hAGT proteins were incubated under physiological conditions with oligodeoxynucleotide duplexes (5′-GGA GCT GGT GGC GTA GGC-3′, (+) strand) in the presence of excess racemic DEB (500 molar equivalents). Samples were subjected to thermal hydrolysis to release N7-alkylated guanines, followed by enzymatic digestion with trypsin and mass spectral analysis (Scheme 2, left).
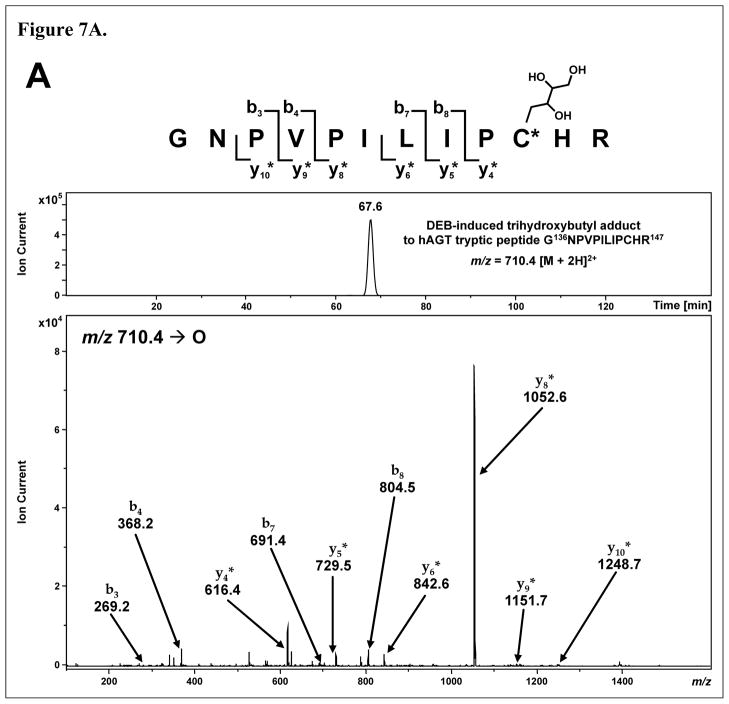
HPLC-ESI+-MS/MS analysis of the tryptic digest of wild-type hAGT which had been incubated with DEB in the presence of double stranded DNA revealed the presence of butanediol cross-links to G136NPVPILIPC*HR147 and V148VC*SSGGAVGNYSGGLA VK165, with CID fragmentation patterns supporting modifications at Cys145 and Cys150 (Figures 6A and 6B, respectively). In addition, DEB-induced trihydroxybutyl adducts were detected at Cys145 (m/z 710.4 [M + 2H]2+) and Cys150 (m/z 886.4 [M + 2H]2+) (Figure 7). The latter products likely originate from the reactions of cysteine residues of AGT with DEB to form (2′-hydroxy-3′,4′-epoxybut-1′-yl)-cysteine intermediates, followed by spontaneous hydrolysis to the corresponding 2′,3′,4′-trihydroxybutyl species.
Figure 6.
HPLC-ESI+-MS/MS analysis of hAGT tryptic peptides containing DEB-induced butanediol cross-links to guanine. (A) Top: Extracted ion chromatogram of hAGT tryptic peptide G136NPVPILIPCHR147 cross-linked to guanine (m/z 776.9 [M + 2H]2+); Bottom: MS/MS spectrum of tryptic peptide G136NPVPILIPCHR147 mapping the cross-link to Cys145. (B) Top: Extracted ion chromatogram of hAGT tryptic peptide V148VCSSGGAVGNYSGGLAVK165 cross-linked to guanine (m/z 952.9 [M + 2H]2+); Bottom: MS/MS spectrum of tryptic peptide V148VCSSGGAVGNYSGGLAVK165 mapping the cross-link to Cys150. Modified fragment ions are indicated by “*”.
Figure 7.
HPLC-ESI+-MS/MS analysis of hAGT tryptic peptides G136NPVPILIPCHR147 and V148VCSSGGAVGNYSGGLAVK165containing DEB-induced 2′,3′,4′-trihydroxybut-1′-yl adducts to active site cysteine residues Cys145 (A) and Cys150 (B). Top: Extracted ion chromatograms; Bottom: MS/MS spectra. Modified fragment ions are indicated by “*”.
Tryptic digests of the C145A mutant subjected to DEB treatment in the presence of DNA contained V148VCSSGGAVGNYSGGLAVK165 butanediol cross-links to guanine (m/z 952.9 [M + 2H]2+), which displayed the same HPLC retention time and CID fragmentation patterns as the corresponding product originating from the wild-type protein (Figure 6B). In contrast, no cross-links to the active site peptide G136NPVPILIPAHR147 were formed for C145A hAGT, supporting the direct participation of C145 in DNA-protein cross-linking.
In general, our results for AGT cross-linking to double-stranded DNA in the presence of DEB were analogous to those obtained upon AGT treatment with pre-formed dG monoepoxide. One noticeable difference involved the relative reactivity of Cys145 and Cys150 residues. While both cysteines were modified with similar efficiency in dG monoepoxide treated AGT, Cys145 was 3-fold more reactive in DNA experiments, suggesting that cross-linking specificity of DEB can be altered by DNA-protein interactions between AGT and double stranded oligodeoxynucleotides.
Capillary HPLC-ESI-MS Analysis of Total Digests of dG Monoepoxide-Treated AGT and DEB-Induced AGT-DNA Cross-Links
In order to confirm that AGT-DNA cross-linking by DEB took place exclusively at the cysteine residues within the protein, HPLC-MS/MS analysis of the total enzymatic digests of alkylated proteins was performed. Following the cross-linking reaction, AGT proteins were subjected to complete digestion in the presence of carboxypeptidase Y and proteinase K, followed by HPLC-ESI+-MS/MS analysis of the resulting amino acids. The mass spectrometer was set up to monitor the formation of 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol (m/z 359.1), 1-(O-tyrosyl)-4-(guan-7-yl)-2,3-butanediol (m/z 420.1), 1-(N-lysyl)-4-(guan-7-yl)-2,3-butanediol (m/z 385.4), 1-(N-arginyl)-4-(guan-7-yl)-2,3-butanediol (m/z 413.4), and 1-(N-histidyl)-4-(guan-7-yl)-2,3-butanediol (m/z 393.4). HPLC-ESI+-MS/MS of a digest of dG monoepoxide treated hAGT detected a prominent peak co-eluting with the authentic standard of 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol (Cys-Gua-BD, m/z 359.7 [M + H]+) (Figure 8A). MS/MS fragmentation of m/z 359.7 yielded the product ions corresponding to the loss of guanine (m/z 208.0 [M + H – Gua]+) and the removal of guanine and a molecule of water (m/z 190.0 [M + H – Gua – H20]+). This compound had the same MS/MS fragmentation pattern and HPLC retention time as synthetically prepared standard of Cys-Gua-BD. In contrast, no amino acid-guanine cross-links involving other nucleophilic residues (Lys, Arg, His, Tyr) were detected (results not shown). Similar results were obtained for the C145A hAGT mutant (Figure 8B) and for AGT-DNA cross-linking in the presence of DNA (not shown), confirming that the cross-linking reaction is specific for cysteine residues within the protein.
Figure 8.
HPLC-ESI+-MS/MS analysis of 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol (Cys-Gua-BD) in total digests of AGT protein treated with dG monoepoxide. (A) Extracted ion chromatogram of Cys-Gua-BD (m/z 359.7 [M + H]+) resulting from the total digestion of dG monoepoxide-treated hAGT; Inset: MS/MS fragmentation of Cys-Gua-BD. (B) Extracted ion chromatogram of Cys-Gua-BD (m/z 359.4 [M + H]+) resulting from the total digestion of dG monoepoxide-treated C145A hAGT; Inset: MS/MS fragmentation of Cys-Gua-BD.
Discussion
Our results presented above provide direct evidence for AGT-DNA cross-linking by DEB. The resulting AGT-DNA conjugates involve the side chain sulfhydryls of Cys145 or Cys150 within the active site of hAGT and the N7 position of guanine in duplex DNA and have the structure of 1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol.
As indicated by the crystal structure of human AGT, this protein contains two distinct domains – an N-terminal domain (residues 1–85) and a C-terminal domain (residues 86–207) (4). The N-terminal domain is comprised of a four-stranded anti-parallel β-sheet packed against a single α-helix (Helix 1). The C-terminal domain, which contains both the O6-alkylguanine-binding channel and helix-turn-helix DNA-binding motif, is made up of a short two-stranded parallel β-sheet and five α-helicies. Human AGT contains a total of five cysteine residues: Cys5, Cys24, Cys62, Cys145, and Cys150. Cysteines 145 and 150 are located in the protein’s C-terminal domain within the active site, with Cys145 directly participating in the alkyl transfer reaction. In contrast, Cys5, Cys24, and Cys62 are all located in N-terminal domain, far removed from the site of DNA binding.
Our results indicate that there are two distinct sites within AGT protein that can form cross-links to DNA in the presence of DEB (Figures 2 and 3), and that AGT-DNA cross-linking by DEB is specific for active site cysteines 145 and 150 (Figures 4–6). In contrast, no cross-linking was detected at Cys5, Cys24, or Cys62. The apparent inability of these three residues to cross-link DNA can be explained by the fact that Cys5 and Cys24 (along with His29 and His85) are involved in coordination with Zn (II) and that Cys62 is located in Helix 1 of the N-terminal domain which does not appear to be solvent-accessible (2,4).
The preferential alkylation of Cys145 is not unexpected because of its unusually low pKa (4 – 5) and high reactivity towards electrophiles (28). Previous studies examining DNA-AGT cross-linking by dibromoethane and other dihaloethanes suggest that these compounds first alkylate the side chain thiol of Cys145 to afford a half-mustard, which then reacts with nucleophilic sites in DNA to form a covalent DNA-AGT complex (Scheme 1A) (10). The reactivity of dihaloethanes towards DNA without AGT-induced activation to an episulfonium ion is minimal.
Unlike dihaloalkanes, DEB also produces a large number of cross-links at Cys150. The AGT mutant in which Cys145 was replaced with alanine (C145A) was capable of forming conjugates with DNA in the presence of DEB by utilizing the side chain of Cys150 (Figures 1 and 3). This is in contrast with dibromoethane, which participated in cross-linking to Cys150 only under extensive treatment conditions (22,29).
A likely explanation for a greater contribution of Cys150 to cross-linking to DNA by DEB involves a different sequence of events leading up to cross-link formation (Scheme 1). In contrast with dihaloalkanes, DEB has a high reactivity towards the N-7 position of guanine in DNA (19,27), giving rise to N7-(2′-hydroxy-3′,4′-epoxybut-1′-yl)-guanine monoadducts (Scheme 1B). AGT binding to a double stranded DNA substrate brings Cys150 in close proximity to DNA (2). In the crystal structure of C145S hAGT bound to a double-stranded oligonucleotide containing O6-methylguanine, Cys150 is located on the outer perimeter of the active site pocket adjacent to residues Ser151 and Tyr114, both of which directly contact the DNA backbone (2). Upon binding to duplex DNA, Cys150 may nucleophilically attack DEB-induced N7-(2′-hydroxy-3′,4′-epoxybut-1′-yl)-guanine monoepoxide lesions to form DNA-protein adducts (Scheme 1B). A similar specificity for cross-linking to DNA via Cys150 and Cys150 is also observed for antitumor nitrogen mustards, another group of bifunctional electrophiles with high reactivity towards guanine in DNA (Loeber and Tretyakova, manuscript in preparation). However, initial alkylation of AGT protein by DEB cannot be presently ruled out. Both Cys145 and Cys150 can be directly alkylated by DEB as demonstrated by the formation of (2′,3′,4′-trihydroxybut-1′-yl) adducts at these sites (Figure 7). These lesions may originate as 2′-hydroxy-3′,4′-epoxybut-1′-yl monoadducts, followed by spontaneous hydrolysis of the epoxide in the presence of water.
Although our study employed large excesses of dG monoepoxide and DEB to afford the levels of cross-linking detectible by HPLC-ESI+-MS, these results may still hold biological significance because the bulky DNA-protein cross-links are likely to cause significant disruption of cellular processes, including DNA replication and transcription. Studies are currently underway to further elucidate the mechanisms of AGT-DNA cross-link formation and to develop sensitive and specific methodology to enable their detection in vivo. These experiments will help explain the enhanced cytotoxicity and mutagenicity of DEB observed in cells expressing the AGT protein (9).
Supplementary Material
Acknowledgments
We thank Dr. Karin Musier-Forsyth and Dr. Songon An (University of Minnesota) for their help with the SDS-PAGE analysis of DEB-induced AGT-DNA cross-links and Dr. F. Peter Guengerich (Vanderbilt University) for fruitful discussions. This research is supported by a grant from the National Cancer Institute (CA095039). Rachel Loeber is a trainee of the NIH Chemistry-Biology Interface Training Grant (T32-GM08700).
Abbreviations
- AGT
O6-alkylguanine DNA alkyltransferase
- Cys-Gua-BD
1-(S-cysteinyl)-4-(guan-7-yl)-2,3-butanediol
- DBE
1,2-dibromomethane
- DEB
1,2,3,4-diepoxybutane
- dG monoepoxide
N7-(2′-hydroxy-3′,4′-epoxybut-1′-yl)-deoxyguanosine
- EB
3,4-epoxy-1-butene
- EBD
3,4-epoxy-1,2-butanediol
- ESI-MS
electrospray ionization mass spectrometry
- hAGT
human recombinant AGT
- HPLC-ESI+-MS/MS
high performance liquid chromatography-electrospray tandem mass spectrometry
- LC-MS
liquid chromatography-mass spectrometry
Footnotes
Supporting Information Available: Amino acid sequences of hAGT and C145A AGT and HPLC-ESI+-MS/MS analysis of synthetic peptide GNPVPILIPCHR containing a butanediol cross-link to guanine are available free of charge via the Internet at http://pubs.acs.org.
Reference List
- 1.Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 2.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 3.Rasimas JJ, Dalessio PA, Ropson IJ, Pegg AE, Fried MG. Active-site alkylation destabilizes human O6-alkylguanine DNA alkyltransferase. Protein Sci. 2004;13:301–305. doi: 10.1110/ps.03319404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels DS, Mol CD, Arvai AS, Kanugula S, Pegg AE, Tainer JA. Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding. EMBO J. 2000;19:1719–1730. doi: 10.1093/emboj/19.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Federwisch M, Hassiepen U, Bender K, Dewor M, Rajewsky MF, Wollmer A. Recombinant human O6-alkylguanine-DNA alkyltransferase (AGT), Cys145-alkylated AGT and Cys145 --> Met145 mutant AGT: comparison by isoelectric focusing, CD and time-resolved fluorescence spectroscopy. Biochem J. 1997;324 (Pt 1):321–328. doi: 10.1042/bj3240321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolan ME, Mitchell RB, Mummert C, Moschel RC, Pegg AE. Effect of O6-benzylguanine analogues on sensitivity of human tumor cells to the cytotoxic effects of alkylating agents. Cancer Res. 1991;51:3367–3372. [PubMed] [Google Scholar]
- 7.Dolan ME, Pegg AE, Biser ND, Moschel RC, English HF. Effect of O6-benzylguanine on the response to 1,3-bis(2-chloroethyl)-1-nitrosourea in the Dunning R3327G model of prostatic cancer. Cancer Chemother Pharmacol. 1993;32:221–225. doi: 10.1007/BF00685839. [DOI] [PubMed] [Google Scholar]
- 8.Wanner MJ, Koch M, Koomen GJ. Synthesis and antitumor activity of methyltriazene prodrugs simultaneously releasing DNA-methylating agents and the antiresistance drug O6-benzylguanine. J Med Chem. 2004;47:6875–6883. doi: 10.1021/jm049556d. [DOI] [PubMed] [Google Scholar]
- 9.Valadez JG, Liu L, Loktionova NA, Pegg AE, Guengerich FP. Activation of bis-electrophiles to mutagenic conjugates by human O6-alkylguanine-DNA alkyltransferase. Chem Res Toxicol. 2004;17:972–982. doi: 10.1021/tx049897u. [DOI] [PubMed] [Google Scholar]
- 10.Guengerich FP. Principles of covalent binding of reactive metabolites and examples of activation of bis-electrophiles by conjugation. Arch Biochem Biophys. 2005;433:369–378. doi: 10.1016/j.abb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 11.Melnick RL, Kohn MC. Mechanistic data indicate that 1,3-butadiene is a human carcinogen. Carcinogenesis. 1995;16:157–163. doi: 10.1093/carcin/16.2.157. [DOI] [PubMed] [Google Scholar]
- 12.Rice JM, Boffetta P. 1,3-Butadiene, isoprene and chloroprene: reviews by the IARC monographs programme, outstanding issues, and research priorities in epidemiology. Chem Biol Interact. 2001;135–136:11–26. doi: 10.1016/s0009-2797(01)00175-2. [DOI] [PubMed] [Google Scholar]
- 13.Malvoisin E, Roberfroid M. Hepatic microsomal metabolism of 1,3-butadiene. Xenobiotica. 1982;12:137–144. doi: 10.3109/00498258209046787. [DOI] [PubMed] [Google Scholar]
- 14.Himmelstein MW, Turner MJ, Asgharian B, Bond JA. Metabolism of 1,3-butadiene: inhalation pharmacokinetics and tissue dosimetry of butadiene epoxides in rats and mice. Toxicology. 1996;113:306–309. doi: 10.1016/0300-483x(96)03462-2. [DOI] [PubMed] [Google Scholar]
- 15.Krause RJ, Elfarra AA. Oxidation of butadiene monoxide to meso- and (+/−)-diepoxybutane by cDNA-expressed human cytochrome P450s and by mouse, rat, and human liver microsomes: evidence for preferential hydration of meso-diepoxybutane in rat and human liver microsomes. Arch Biochem Biophys. 1997;337:176–184. doi: 10.1006/abbi.1996.9781. [DOI] [PubMed] [Google Scholar]
- 16.Sasiadek M, Norppa H, Sorsa M. 1,3-Butadiene and its epoxides induce sister-chromatid exchanges in human lymphocytes in vitro. Mutat Res. 1991;261:117–121. doi: 10.1016/0165-1218(91)90057-s. [DOI] [PubMed] [Google Scholar]
- 17.Cochrane JE, Skopek TR. Mutagenicity of butadiene and its epoxide metabolites: I. Mutagenic potential of 1,2-epoxybutene, 1,2,3,4-diepoxybutane and 3,4-epoxy-1,2- butanediol in cultured human lymphoblasts. Carcinogenesis. 1994;15:713–717. doi: 10.1093/carcin/15.4.713. [DOI] [PubMed] [Google Scholar]
- 18.Henderson RF, Thornton-Manning JR, Bechtold WE, Dahl AR. Metabolism of 1,3-butadiene: species differences. Toxicology. 1996;113:17–22. doi: 10.1016/0300-483x(96)03422-1. [DOI] [PubMed] [Google Scholar]
- 19.Park S, Tretyakova N. Structural characterization of the major DNA-DNA cross-link of 1,2,3,4-diepoxybutane. Chem Res Toxicol. 2004;17:129–136. doi: 10.1021/tx0342058. [DOI] [PubMed] [Google Scholar]
- 20.Park S, Hodge J, Anderson C, Tretyakova NY. Guanine-adenine cross-linking by 1,2,3,4-diepoxybutane: potential basis for biological activity. Chem Res Toxicol. 2004;17:1638–1651. doi: 10.1021/tx0498206. [DOI] [PubMed] [Google Scholar]
- 21.Park S, Anderson C, Loeber R, Seetharaman M, Jones R, Tretyakova N. Interstrand and intrastrand DNA-DNA cross-linking by 1,2,3,4-diepoxybutane: role of stereochemistry. J Am Chem Soc. 2005;127:14355–14365. doi: 10.1021/ja051979x. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Hachey DL, Valadez G, Williams KM, Guengerich FP, Loktionova NA, Kanugula S, Pegg AE. Characterization of a mutagenic DNA adduct formed from 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J Biol Chem. 2004;279:4250–4259. doi: 10.1074/jbc.M311105200. [DOI] [PubMed] [Google Scholar]
- 23.Jelitto B, Vangala RR, Laib RJ. Species differences in DNA damage by butadiene: role of diepoxybutane. Arch Toxicol Suppl. 1989;13:246–249. doi: 10.1007/978-3-642-74117-3_42. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Xu-Welliver M, Kanugula S, Pegg AE. Inactivation and degradation of O6-alkylguanine-DNA alkyltransferase after reaction with nitric oxide. Cancer Res. 2002;62:3037–3043. [PubMed] [Google Scholar]
- 25.Edara S, Kanugula S, Goodtzova K, Pegg AE. Resistance of the human O6-alkylguanine-DNA alkyltransferase containing arginine at codon 160 to inactivation by O6-benzylguanine. Cancer Res. 1996;56:5571–5575. [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; 1989. [Google Scholar]
- 27.Tretyakova NY, Sangaiah R, Yen TY, Swenberg JA. Synthesis, characterization, and in vitro quantitation of N-7-guanine adducts of diepoxybutane. Chem Res Toxicol. 1997;10:779–785. doi: 10.1021/tx970004q. [DOI] [PubMed] [Google Scholar]
- 28.Guengerich FP, Fang Q, Liu L, Hachey DL, Pegg AE. O6-alkylguanine-DNA alkyltransferase: low pKa and high reactivity of cysteine 145. Biochemistry. 2003;42:10965–10970. doi: 10.1021/bi034937z. [DOI] [PubMed] [Google Scholar]
- 29.Liu L, Pegg AE, Williams KM, Guengerich FP. Paradoxical enhancement of the toxicity of 1,2-dibromoethane by O6-alkylguanine-DNA alkyltransferase. J Biol Chem. 2002;277:37920–37928. doi: 10.1074/jbc.M205548200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.