Abstract
Three Pseudomonas strains were tested for the ability to sense and respond to nitrobenzoate and aminobenzoate isomers in chemotaxis assays. Pseudomonas putida PRS2000, a strain that grows on benzoate and 4-hydroxybenzoate by using the β-ketoadipate pathway, has a well-characterized β-ketoadipate-inducible chemotactic response to aromatic acids. PRS2000 was chemotactic to 3- and 4-nitrobenzoate and all three isomers of aminobenzoate when grown under conditions that induce the benzoate chemotactic response. P. putida TW3 and Pseudomonas sp. strain 4NT grow on 4-nitrotoluene and 4-nitrobenzoate by using the ortho (β-ketoadipate) and meta pathways, respectively, to complete the degradation of protocatechuate derived from 4-nitrotoluene and 4-nitrobenzoate. However, based on results of catechol 1,2-dioxygenase and catechol 2,3-dioxygenase assays, both strains were found to use the β-ketoadipate pathway for the degradation of benzoate. Both strains were chemotactic to benzoate, 3- and 4-nitrobenzoate, and all three aminobenzoate isomers after growth with benzoate but not succinate. Strain TW3 was chemotactic to the same set of aromatic compounds after growth with 4-nitrotoluene or 4-nitrobenzoate. In contrast, strain 4NT did not respond to any aromatic acids when grown with 4-nitrotoluene or 4-nitrobenzoate, apparently because these substrates are not metabolized to the inducer (β-ketoadipate) of the chemotaxis system. The results suggest that strains TW3 and 4NT have a β-ketoadipate-inducible chemotaxis system that responds to a wide range of aromatic acids and is quite similar to that present in PRS2000. The broad specificity of this chemotaxis system works as an advantage in strains TW3 and 4NT because it functions to detect diverse carbon sources, including 4-nitrobenzoate.
With few exceptions, nitroaromatic compounds are man-made chemicals that have been used extensively only for the last century. Nitrobenzene, nitrotoluenes, nitrobiphenyls, nitrophenols, and nitrobenzoates have been used as industrial solvents and in the production of dyes, polymers, fungicides, pesticides, herbicides, and explosives (19, 39). In addition, soil and groundwater contamination by trinitrotoluene and dinitrotoluenes has resulted from their extensive production and use as explosives during World Wars I and II. Various nitroaromatic compounds have been shown to be toxic, mutagenic, and possibly carcinogenic to a range of organisms, from bacteria to humans (19, 39, 48), and as a result, seven nitroaromatic compounds are included on the U.S. Environmental Protection Agency's November 2002 list of priority pollutants (http://www.epa.gov/region1/npdes/permits/generic/prioritypollutants.pdf).
Since these xenobiotic compounds have been present in the environment for a relatively short period of time, microorganisms have not had long to evolve pathways for their degradation. Although nonspecific reduction of nitroaromatic compounds appears to be quite common (4, 37), until recently most nitroaromatic compounds were thought to be nonbiodegradable. It has only been within the last 15 years that bacteria capable of growth on compounds such as nitrobenzene and nitrotoluenes have been isolated, and such strains have been isolated only from locations with a history of exposure to nitroaromatic contamination (27, 45, 46).
The role of chemotaxis in the biodegradation of pollutants and man-made chemicals has not been extensively explored. Organic chemicals such as sugars and amino acids are strong chemoattractants for the enteric bacteria (23). In contrast, Pseudomonads and related nonenteric bacteria have been shown to be attracted to aromatic acids, many of which are growth substrates for particular strains. Pseudomonas putida PRS2000 has been the model organism for studying chemotaxis to aromatic acids (12-14, 16, 17). Attractants for PRS2000 include the growth substrates benzoate, p-hydroxybenzoate, and benzoylformate as well as the nonmetabolizable compounds 2-hydroxybenzoate (salicylate), m- and p-toluate, 3- and 4-chlorobenzoate, and 3-fluorobenzoate (16, 17). Other aromatic attractants for various Pseudomonads and related Proteobacteria include the aromatic hydrocarbons naphthalene (9, 32, 43), toluene and benzene (32), and the herbicide 2,4-dichlorophenoxyacetic acid (18). Recent reports have demonstrated chemotaxis to nitrocatechol, nitrobenzoate, and nitrophenols by a Ralstonia sp. (2, 30, 42). The aim of this study was to determine whether nitrobenzoates and aminobenzoates are recognized as chemoattractants by the aromatic acid chemotaxis system in P. putida PRS2000 and whether these compounds serve as attractants for specific Pseudomonas strains capable of 4-nitrobenzoate degradation. The results reported here indicate that nitrobenzoates and aminobenzoates are detected as chemoattractants by strains that do not degrade these compounds, as well as by strains that can grow on and mineralize 4-nitrotoluene and 4-nitrobenzoate. This finding is significant because bacterial strains with the ability to detect the presence of 4-nitrobenzoate may have an increased growth and survival advantage and could contribute to the biodegradation of 4-nitrotoluene and 4-nitrobenzoate in the environment (31, 33).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. putida PRS2000 is a wild-type strain that grows on benzoate and 4-hydroxybenzoate by using the β-ketoadipate pathway (29). P. putida TW3 (38) and Pseudomonas sp. strain 4NT (11) are wild-type isolates that each grow on 4-nitrotoluene and 4-nitrobenzoate. Prior to the initiation of experiments, highly motile populations of all three strains were enriched on diluted L-agar swarm plates (1, 17). A variation of auxanography (36) was used to test for growth of PRS2000, 4NT, and TW3 on aminobenzoates, nitrobenzoates, and 4-nitrotoluene. Overnight cultures grown in MSB minimal medium (47) with succinate as the carbon source were harvested by centrifugation, washed with MSB, resuspended in MSB to a concentration of approximately 1010 cells/ml and spread (50 μl/plate) on MSB medium solidified with 1.8% Noble agar (Difco Laboratories, Detroit, Mich.). Potential growth substrates (2-, 3-, and 4-aminobenzoate; 2-, 3-, and 4-nitrobenzoate; and 4-nitrotoluene) were applied as crystals at the edge of the plate. To control plates, 4-hydroxybenzoate crystals were added as the carbon source. Plates were incubated at 30°C for up to 1 week. Growth on auxanography plates was verified in liquid MSB containing crystals of the substrate. For chemotaxis assays, P. putida PRS2000 was grown to mid-logarithmic phase in MSB with 5 mM benzoate, 5 mM 4-hydroxybenzoate, or 10 mM succinate. P. putida TW3 and Pseudomonas sp. strain 4NT were grown under similar conditions or with crystals of 4-nitrobenzoate or 4-nitrotoluene as the carbon source.
Chemotaxis assays.
Soft agar swarm plates consisted of MSB medium containing 0.1% rather than 1% Hutner's mineral base and 0.3% Noble agar (7). Benzoate was provided as a potential attractant in swarm plates at a final concentration of 1 mM, and 4-nitrobenzoate was provided at a final concentration of 0.5 mM. Cells were inoculated at the center of the plates; plates were incubated at 30°C overnight. Modified capillary assays were carried out as previously described (9, 32). Capillaries (1 μl) contained attractant in either 2% low-melting-temperature agarose (NuSieve GTG Agarose; FMC Bioproducts, Rockland, Maine) dissolved in a chemotaxis buffer (50 mM potassium phosphate buffer [pH 7.0], 10 μM disodium EDTA, 0.05% glycerol) (32) or as crystals in chemotaxis buffer. Freshly grown cells were harvested in log phase (when the optical density at 660 nm [OD660] was between 0.3 and 0.5), suspended in chemotaxis buffer to an OD660 of approximately 0.05, and placed in a chamber formed by a microscope slide, a glass U-tube, and a coverslip. A capillary containing the attractant was inserted into the pool of cells. Control capillaries contained 2% low-melting-temperature agarose in the chemotaxis buffer. Cell behavior was observed at a magnification of ×40. Chemotaxis was measured quantitatively by using a temporal assay that monitors adaptation to the chemoattractant (44). Briefly, cells were harvested by centrifugation and resuspended in the chemotaxis buffer to an OD660 of approximately 0.05. After addition of attractant, cells were either viewed directly or videotaped, and the time required for approximately 50% of the population to return to prestimulus behavior was determined.
Preparation of cell extracts.
Pseudomonas cultures (150 ml) grown in MSB containing the appropriate carbon source were harvested in the exponential phase of growth (OD660 of 0.4 to 0.6). Cells were washed once with MSB, and pellets were stored at −20°C until just prior to disruption. Thawed cells were resuspended in a solution of 50 mM Tris-HCl (pH 7.5)-1 mM dithiothreitol. Chilled cell suspensions were disrupted by sonication, and cell debris was removed by centrifugation.
Enzyme assays.
Catechol 1,2- and catechol 2,3-dioxygenase activities were assayed spectrophotometrically at 260 and 375 nm as previously described (24, 28). Assays were carried out in 1-ml quartz cuvettes containing 50 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, and 1 mM catechol. In each case, 1 unit is defined as the amount of enzyme that oxidizes 1.0 μmol of catechol per min at 25°C. Protein concentrations were determined by using the method of Bradford (3) with bovine serum albumin as standard.
RESULTS
Growth of Pseudomonas strains on nitrobenzoates and aminobenzoates.
P. putida PRS2000 is known to be chemotactic to aromatic acids that serve as growth substrates as well as several others that do not (16, 17). We tested this strain for the ability to grow with nitro- and aminobenzoates and 4-nitrotoluene and found that none of these compounds were utilized as carbon sources (Table 1). The two 4-nitrotoluene-degrading strains P. putida TW3 and Pseudomonas sp. strain 4NT grew with 4-nitrobenzoate and 4-nitrotoluene as previously reported (11, 38) but not with 2- or 3-nitrobenzoate or 2-, 3-, or 4-aminobenzoate (Table 1).
TABLE 1.
Growth of Pseudomonas strains on various aromatic compoundsa
| Substrate | Mean doubling time (min) ± SD of strain:
|
||
|---|---|---|---|
| PRS2000 | 4NT | TW3 | |
| Benzoate | 77 ± 11 | 54 ± 2 | 113 ± 8 |
| 4-Hydroxybenzoate | 123 ± 24 | 118 ± 14 | 92 ± 9 |
| 4-Nitrobenzoate | − | 180 ± 36 | 124 ± 13 |
| 3-Nitrobenzoate | − | − | − |
| 2-Nitrobenzoate | − | − | − |
| 4-Aminobenzoate | − | −b | −c |
| 3-Aminobenzoate | − | − | − |
| 2-Aminobenzoate | − | − | − |
| 4-Nitrotoluene | − | 179 ± 23 | 296 ± 16 |
Growth was assessed by auxanography as described in Materials and Methods. Positive growth was verified in liquid culture (MSB containing 5 mM benzoate or 4- hydroxybenzoate or saturating 4-nitrobenzoate or 4-nitrotoluene at 30°C with shaking at 250 rpm); −, no growth.
Reported in reference 11.
Reported in reference 38.
Benzoate and 4-nitrobenzoate chemotaxis by strains TW3 and 4NT.
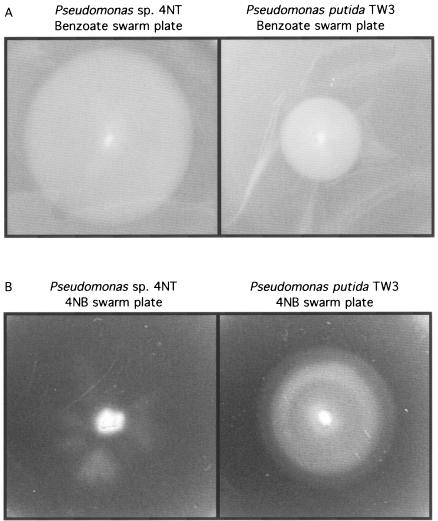
Chemotaxis to benzoate and 4-nitrobenzoate was initially tested by swarm plate assays. Both strains TW3 and 4NT were chemotactic to benzoate, as seen by the swarm patterns (Fig. 1A). The larger swarm formed by strain 4NT is consistent with its faster growth rate on benzoate (Table 1). Even though both strains are able to grow with 4-nitrobenzoate, only strain TW3 showed a chemotactic response in the swarm plate assay (Fig. 1B).
FIG. 1.
Chemotactic responses of Pseudomonas sp. strain 4NT and P. putida TW3 to 1 mM benzoate (A) and 0.5 mM 4-nitrobenzoate (B) in swarm plates.
Nitrobenzoates and aminobenzoates are attractants for PRS2000, TW3, and 4NT.
Previous studies have shown that PRS2000 expresses an inducible chemotactic response to benzoate and other aromatic acids (12, 16, 17). After growth of PRS2000 in minimal medium with benzoate (induced) or succinate (uninduced), cells were harvested by centrifugation and resuspended in buffer and chemotaxis was assayed in two ways. The smooth swimming response assay (44) provides a quantitative temporal assay that measures the strength of the attractant response. In this assay, cells were visualized microscopically immediately after the addition of a potential attractant. Upon addition of an attractant, the cells respond by swimming smoothly, i.e., cells that are responding change direction much less frequently as they bias the rotation of the flagellar motor. The time required for approximately 50% of the cells to adapt (return to unstimulated behavior) is the smooth swimming response time. The length of time taken for the cells to adapt is a general measure of the strength of the attractant.
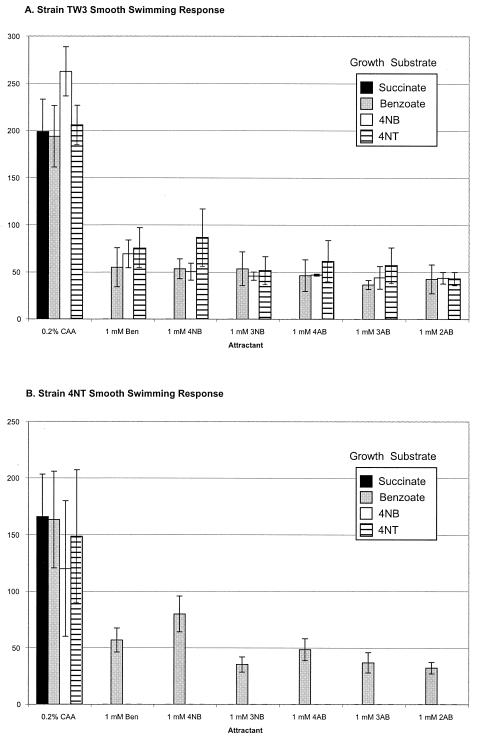
When grown with benzoate, PRS2000 responded to benzoate as expected, and one-half of the population adapted within approximately 1 min (Table 2). This result is consistent with the results of computer-assisted motion analysis studies, which demonstrated that PRS2000 adapted within 1 to 2 min after stimulation with benzoate (12). Benzoate-grown PRS2000 was also attracted to 3- and 4-nitrobenzoate (Table 2). Similarly, strain TW3 also responded to benzoate and to 3- and 4-nitrobenzoate when grown with benzoate but not when grown with succinate. TW3 was also attracted to the same subset of chemicals after growth with 4-nitrobenzoate or 4-nitrotoluene (Fig. 2A). In contrast, strain 4NT was chemotactic to benzoate and 3- and 4-nitrobenzoate only after growth with benzoate (Fig. 2B). After growth with succinate, all three strains responded only to the addition of Casamino Acids (Fig. 2; Table 2), indicating that the chemotactic response to aromatic compounds was inducible in all of the Pseudomonas strains. We tested the ability of the three strains to detect the structurally related aminobenzoate isomers and found that 2-, 3-, and 4-aminobenzoate were attractants for all three strains when grown under the conditions that elicited benzoate and nitrobenzoate chemotaxis (Fig. 2; Table 2).
TABLE 2.
Temporal response of PRS2000 to nitro- and aminobenzoatesa
| Attractant | Mean smooth swimming response time (s) ± SD (no. of independent assays) after growth with:
|
|
|---|---|---|
| Succinate | Benzoate | |
| None (buffer control) | − (3) | − (3) |
| Casamino Acids (0.2%) | 185 ± 30 (11) | 142 ± 19 (11) |
| Benzoate (1 mM) | − (2) | 66 ± 21 (9) |
| 4-Nitrobenzoate (1 mM) | − (3) | 100 ± 11 (8) |
| 3-Nitrobenzoate (1 mM) | − (3) | 57 ± 12 (9) |
| 2-Nitrobenzoate (1 mM) | − (5) | − (2) |
| 4-Aminobenzoate (1 mM) | − (5) | 64 ± 12 (6) |
| 3-Aminobenzoate (1 mM) | − (5) | 40 ± 7 (6) |
| 2-Aminobenzoate (1 mM) | − (4) | 41 ± 6 (6) |
Amount of time for approximately 50% of the cells to adapt to the added attractant; −, no response detected.
FIG. 2.
Chemotactic responses of P. putida TW3 (A) and Pseudomonas sp. strain 4NT (B) in smooth swimming response assays after growth with 10 mM succinate, 5 mM benzoate, or saturating amounts of 4-nitrobenzoate (4NB) or 4-nitrotoluene (4NT). The response in this assay is the amount of time in seconds needed for approximately 50% of the cells to adapt to the added attractant. The abbreviations for the attractants in the chemotaxis buffer are as follows: 0.2% CAA, 0.2% Casamino Acids; Ben, benzoate; 4NB, 4-nitrobenzoate; 3NB, 3-nitrobenzoate; 4AB, 4-aminobenzoate; 3AB, 3-aminobenzoate; 2AB, 2-aminobenzoate. No response was seen with chemotaxis buffer alone or with 1 mM 2-nitrobenzoate (data not shown).
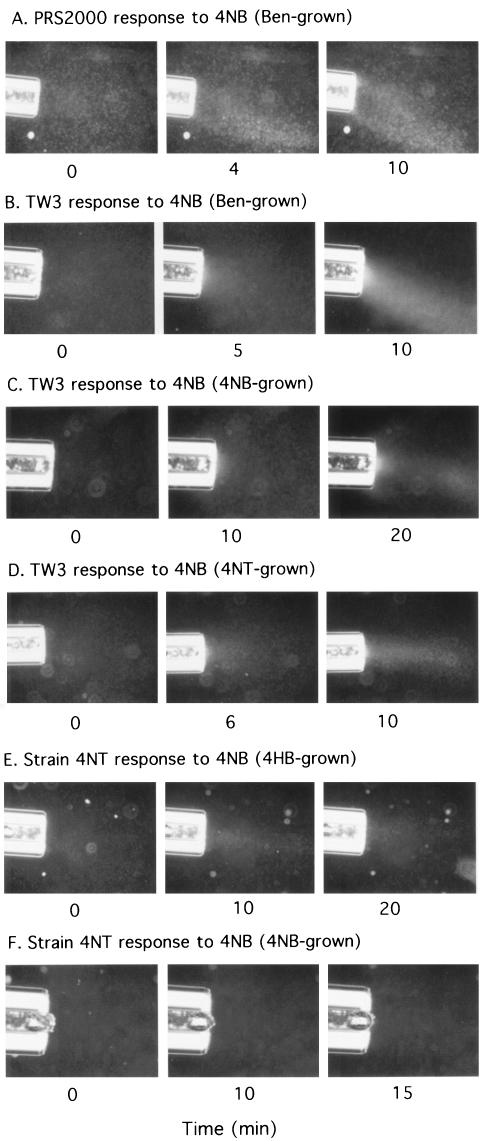
Chemotactic responses were verified with the modified capillary assay, which allows a direct visualization of the accumulation of cells in response to an attractant present in a 1-μl capillary. Benzoate- and 4-hydroxybenzoate-grown PRS2000 cells accumulated at the mouths of capillaries containing either 5 mM benzoate or 4-nitrobenzoate (Fig. 3A; Table 3). No response to either benzoate or 4-nitrobenzoate was detected with succinate-grown cells (Table 3). Similar results were seen with strain TW3 (Fig. 3B; Table 3). 4-Nitrobenzoate- and 4-nitrotoluene-grown TW3 cells were also clearly attracted to benzoate and 4-nitrobenzoate (Fig. 3C and D; Table 3). In contrast, strain 4NT responded to benzoate and 4-nitrobenzoate after growth with benzoate or 4-hydroxybenzoate but not 4-nitrobenzoate or 4-nitrotoluene (Fig. 3E and F; Table 3).
FIG. 3.
Time courses of the chemotactic responses of the three Pseudomonas strains to 4-nitrobenzoate in modified capillary assays (magnification, ×40). Capillaries (1 μl) contained crystals of 4-nitrobenzoate in chemotaxis buffer. (A) Benzoate-grown PRS2000; (B) benzoate-grown TW3; (C) 4-nitrobenzoate-grown TW3; (D) 4-nitrotoluene-grown TW3; (E) 4-hydroxybenzoate-grown strain 4NT; (F) 4-nitrobenzoate-grown strain 4NT. No responses were seen when capillaries contained only agarose and chemotaxis buffer or silica in chemotaxis buffer (data not shown).
TABLE 3.
Summary of modified capillary assay results
| Attractantc | Chemotactic responsea of Pseudomonas strain:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PRS2000 grown onb:
|
TW3 grown onb:
|
4NT grown onb:
|
|||||||||||
| Suc | Ben | 4HB | Suc | Ben | 4HB | 4NB | 4NT | Suc | Ben | 4HB | 4NB | 4NT | |
| None | − | − | − | − | − | − | − | − | − | − | − | − | − |
| CAA | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Ben | − | + | + | − | + | + | + | + | − | + | + | − | − |
| 4NB | − | + | + | − | + | + | + | + | − | + | + | − | − |
Measured in modified capillary assays as described in Materials and Methods and shown in Fig. 3. +, positive response observed; −, no response observed.
Growth substrates: Suc, 10 mM succinate; Ben, 5 mM benzoate; 4HB, 5 mM 4-hydroxybenzoate; 4NB, 4-nitrobenzoate crystals; 4NT, 4-nitrotoluene crystals.
Attractants present in capillary: None, chemotaxis buffer in 2% agarose; CAA, 2% Casamino Acids in chemotaxis buffer and 2% agarose; Ben, 5 mM benzoate in chemotaxis buffer and 2% agarose; 4NB, 4-nitrobenzoate crystals in chemotaxis buffer and 2% agarose.
No response by any of the strains was detected to 2-nitrobenzoate in temporal assays or modified capillary assays (Table 2; data not shown), and no response to any of the aromatic compounds was detected with succinate-grown cells with either assay method (Fig. 2; Table 3).
Benzoate degradation pathway in strains TW3 and 4NT.
Although strains TW3 and 4NT were previously shown to grow with benzoate (11, 38), the benzoate degradation pathways present in these strains have not been described in detail. Strain TW3 was reported to have elevated catechol 1,2-dioxygenase activity after growth with benzoate compared to succinate-grown cells (22). Catechol 1,2- and catechol 2,3-dioxygenase assays were carried out with both strains after growth with either succinate or benzoate. In both strains, catechol 1,2-dioxygenase activity was induced during growth with benzoate (Table 4). Extracts of succinate-grown strain TW3 had a low but detectable level of the enzyme, while the activity was undetectable in succinate-grown strain 4NT. Catechol 2,3-dioxygenase activity was not detectable in strain 4NT grown under either condition. Very low levels of catechol 2,3-dioxygenase were detected in strain TW3 grown under both conditions (Table 4). These results indicate that strains TW3 and 4NT utilize the β-ketoadipate pathway for the degradation of benzoate.
TABLE 4.
Catechol dioxygenase activities in extracts of strains 4NT and TW3
| Growth substrate (concn) | Sp act (μmol min−1 mg−1 of protein) of indicated strain for:
|
|||
|---|---|---|---|---|
| Catechol 1,2-dioxygenase
|
Catechol 2,3-dioxygenase
|
|||
| 4NT | TW3 | 4NT | TW3 | |
| Succinate (10 mM) | −a | 0.039 | − | 0.005 |
| Benzoate (5 mM) | 1.34 | 0.330 | − | 0.015 |
−, no activity detected.
DISCUSSION
PRS2000 degrades benzoate and 4-hydroxybenzoate by using the β-ketoadipate pathway (15), and neither nitrobenzoates nor aminobenzoates are growth substrates for PRS2000 (Table 1). However, metabolism of nitrobenzoates and aminobenzoates is apparently not necessary for the behavioral response to these compounds (Table 2). Previous studies have also shown that PRS2000 is attracted to several other compounds that it is unable to metabolize, including toluates, chlorobenzoates, and salicylate (14, 16, 17), and mutants of PRS2000 that are unable to grow with benzoate and 4-hydroxybenzoate were chemotactic to benzoate, provided that β-ketoadipate was available to induce the chemotactic response (17). The chemotactic response to benzoate, 4-hydroxybenzoate, salicylate, toluates, and chlorobenzoates is induced when PRS2000 is grown with benzoate or 4-hydroxybenzoate (14, 16, 17). It is quite possible that the same chemotaxis system can detect the presence of the structurally related nitrobenzoates and aminobenzoates. Consistent with this possibility, only benzoate- or 4-hydroxybenzoate-grown cells responded to nitro- and aminobenzoates (Tables 2 and 3). The chemotactic response to 4-hydroxybenzoate is mediated by the PcaK protein in PRS2000 (13). PcaK is a member of the major facilitator superfamily (41) of transport proteins, and it also functions to transport 4-hydroxybenzoate and protocatechuate (25). In PRS2000, the pcaK gene is located nearby and coordinately regulated by β-ketoadipate with several benzoate and/or 4-hydroxybenzoate degradation genes. In this strain, β-ketoadipate, an intermediate in benzoate and 4-hydroxybenzoate degradation, induces the chemotactic response to substituted benzoates as well as several of the structural genes for the degradation of these two compounds in the presence of the PcaR activator protein (10, 16, 17, 26, 34, 40).
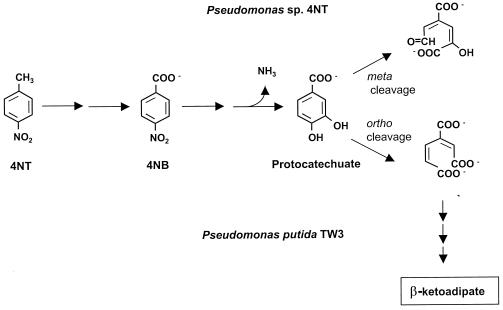
P. putida TW3 and Pseudomonas sp. strain 4NT degrade 4-nitrotoluene through 4-nitrobenzoate to protocatechuate, but after this step the pathways in the two strains differ (Fig. 4). In strain 4NT, protocatechuate is degraded via a meta ring fission pathway (11), while in TW3, protocatechuate is degraded by using the β-ketoadipate pathway (38). Results of catechol dioxygenase assays indicate that strains 4NT and TW3 both degrade benzoate by using the β-ketoadipate pathway (Table 4). The results of chemotaxis assays suggest that, similar to PRS2000, strains TW3 and 4NT may have a β-ketoadipate-inducible chemotaxis system responding to benzoate and structurally related chemicals. Thus, strain TW3 responds to benzoate, nitrobenzoates, and aminobenzoates when grown with benzoate, 4-nitrobenzoate, and 4-nitrotoluene, all of which are degraded with the formation of β-ketoadipate. In contrast, β-ketoadipate is not an intermediate in the degradation of 4-nitrotoluene and 4-nitrobenzoate in strain 4NT, and this strain responds to benzoate, nitrobenzoates, and aminobenzoates only after growth with benzoate. A similar phenomenon was described for Pseudomonas strains carrying the TOL plasmid (14). These strains, although chemotactic to benzoate when lacking the TOL plasmid, are not chemotactic to benzoate when harboring the plasmid. This is because benzoate is preferentially degraded through a meta cleavage pathway in the TOL+ strain, and consequently, β-ketoadipate is not produced as an intermediate. Therefore, under these conditions the chemotactic response is not induced. These strains exhibited normal chemotaxis to aromatic acids when grown with 4-hydroxybenzoate, which is degraded via the β-ketoadipate pathway in both TOL+ and TOL− strains.
FIG. 4.
Pathways for 4-nitrotoluene and 4-nitrobenzoate degradation in P. putida TW3 and Pseudomonas sp. strain 4NT. Note that P. putida TW3 utilizes ortho cleavage and Pseudomonas sp. strain 4NT carries out meta cleavage of protocatechuate.
Although the majority of our studies utilized benzoate-, 4-nitrobenzoate- and 4-nitrotoluene-grown cells, strains TW3 and 4NT were also found to be chemotactic to the same set of substituted benzoates after growth with 4-hydroxybenzoate (Table 3). Most Pseudomonads carry both the catechol and protocatechuate branches of the β-ketoadipate pathway (15), and the chemotactic responses of 4-hydroxybenzoate-grown TW3 and 4NT are most likely due to the formation of β-ketoadipate from 4-hydroxybenzoate.
The genes from strain TW3 that encode the conversion of 4-nitrotoluene to 4-nitrobenzoate and from 4-nitrobenzoate to protocatechuate have been cloned and sequenced (20-22), and there are no obvious chemotaxis genes located within either cluster. Apparently, the corresponding genes from strain 4NT have been identified, but results have not yet been published (21). In strains TW3 and 4NT, the chemotactic response to substituted benzoates may be mediated by a PcaK-like protein as in PRS2000 (13), but such a protein has not yet been identified in these strains.
Ralstonia sp. strain SJ98 has been shown to be chemotactic to the nitroaromatic growth substrates 4-nitrocatechol, 2-nitrobenzoate, 4-nitrobenzoate, 4-nitrophenol, and 3-methyl-4-nitrophenol (2, 42). Although the pathways for the degradation of these compounds have not been characterized, protocatechuate was identified as an intermediate in 4-nitrobenzoate degradation and 2-nitrobenzoate was converted to 2-aminobenzoate (42). The mode of protocatechuate cleavage has not been reported, and the inducibility of the chemotactic response was not investigated. It is therefore difficult to conclude whether the chemotaxis system for the detection of nitroaromatic compounds in Ralstonia sp. strain SJ98 is similar to that in the three Pseudomonads described here.
The evolution of catabolic pathways is thought to proceed in a variety of ways. For example, genes can be recruited from existing pathways and selection of modified enzymes with extended substrate ranges can increase the number of substrates degraded. Also, modular genetic elements can combine to form complete pathways (49). Later, regulatory elements can be recruited for optimization of gene expression, and this may reduce the chance of a particular strain being outcompeted at times when no selective compounds are present (5, 6, 8, 49). In the case of the 4-nitrotoluene degradation pathway in strain TW3, the enzymes used for the conversion of 4-nitrotoluene to 4-nitrobenzoate appear to have been acquired from a TOL-plasmid upper pathway (22) with the exception of the NAD(P)+-independent alcohol dehydrogenase, which has no homologue in the TOL pathway (21). The genes required for the conversion of 4-nitrobenzoate to protocatechuate are apparently unlinked to those for the formation of 4-nitrobenzoate from 4-nitrotoluene in strain TW3, and they have no close homologues in the GenBank database other than those encoding isofunctional nitrobenzoate degradation proteins (20).
Chemotaxis, the ability of motile bacteria to detect and respond to specific chemicals in the environment, can increase an organism's chances of locating useful sources of carbon and energy. Thus, chemotaxis can provide an additional growth and survival advantage to bacteria and may even contribute to the dissemination of catabolic pathways, many of which are encoded by transmissible plasmids (14, 33). During the evolution of a new biodegradation pathway, it is not clear how and when specific genes for auxiliary aspects of catabolism, such as chemotaxis, are introduced. Since most catabolic genes are recruited from preexisting pathways, it seems likely that an organism would also acquire chemoreceptor functions from existing systems. It appears that an aromatic acid chemotaxis system like that in P. putida PRS2000 is also present in strains TW3 and 4NT. As pointed out earlier (14), the β-ketoadipate-inducible aromatic acid chemotaxis system in Pseudomonads is likely to be induced most of the time during the life cycle of the bacterial cells due to the abundance of plant-derived phenylpropanoids and hydroaromatic compounds in soils that are degraded through β-ketoadipate (35). The broad specificity of the system works as an advantage in the last two strains, since an existing chemotaxis system, apparently used for benzoate and 4-hydroxybenzoate chemotaxis in PRS2000, functions to detect the useful carbon source 4-nitrobenzoate.
Acknowledgments
I thank Peter Williams and Jim Spain for providing Pseudomonas putida TW3 and Pseudomonas sp. strain 4NT, respectively; Carrie Harwood and Andrew Hawkins for helpful discussions; and Carrie Harwood and two anonymous reviewers for valuable suggestions to improve the manuscript.
REFERENCES
- 1.Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. [DOI] [PubMed] [Google Scholar]
- 2.Bhushan, B., S. K. Samanta, A. Chauhan, A. K. Chakraborti, and R. K. Jain. 2000. Chemotaxis and biodegradation of 3-methyl-4-nitrophenol by Ralstonia sp. SJ98. Biochem. Biophys. Res. Commun. 275:129-133. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Cerniglia, C. E., and C. C. Somerville. 1995. Reductive metabolism of nitroaromatic and nitropolycyclic aromatic hydrocarbons, p. 99-115. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds, vol. 49. Plenum Press, New York, N.Y.
- 5.Chakrabarty, A. M. 1996. Microbial degradation of toxic chemicals: evolutionary insights and practical considerations. ASM News 62:130-137. [Google Scholar]
- 6.de Lorenzo, V., and J. Perez-Martin. 1996. Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol. Microbiol. 19:1177-1184. [DOI] [PubMed] [Google Scholar]
- 7.Ditty, J. L., and C. S. Harwood. 1999. Conserved cytoplasmic loops are important for both the transport and chemotaxis functions of PcaK, a protein from Pseudomonas putida with 12-membrane-spanning regions. J. Bacteriol. 181:5068-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez, S., V. Shingler, and V. de Lorenzo. 1994. Cross-regulation by XylR and DmpR activators of Pseudomonas putida suggests that transcriptional control of biodegradative operons evolves independently of catabolic genes. J. Bacteriol. 176:5052-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimm, A. C., and C. S. Harwood. 1997. Chemotaxis of Pseudomonas putida to the polyaromatic hydrocarbon naphthalene. Appl. Environ. Microbiol. 63:4111-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, Z., and J. E. Houghton. 1999. PcaR-mediated activation and repression of pca genes from Pseudomonas putida are propagated by its binding to both the -35 and the -10 promoter elements. Mol. Microbiol. 32:253-263. [DOI] [PubMed] [Google Scholar]
- 11.Haigler, B. E., and J. C. Spain. 1993. Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl. Environ. Microbiol. 59:2239-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harwood, C. S., K. Fosnaugh, and M. Dispensa. 1989. Flagellation of Pseudomonas putida and analysis of its motile behavior. J. Bacteriol. 171:4063-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harwood, C. S., N. N. Nichols, M.-K. Kim, J. L. Ditty, and R. E. Parales. 1994. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J. Bacteriol. 176:6479-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood, C. S., and L. N. Ornston. 1984. TOL plasmid can prevent induction of chemotactic responses to aromatic acids. J. Bacteriol. 160:797-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:533-590. [DOI] [PubMed] [Google Scholar]
- 16.Harwood, C. S., R. E. Parales, and M. Dispensa. 1990. Chemotaxis of Pseudomonas putida toward chlorinated benzoates. Appl. Environ. Microbiol. 56:1501-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harwood, C. S., M. Rivelli, and L. N. Ornston. 1984. Aromatic acids are chemoattractants for Pseudomonas putida. J. Bacteriol. 160:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins, A. C., and C. S. Harwood. 2002. Chemotaxis of Ralstonia eutropha JMP134(pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Appl. Environ. Microbiol. 68:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higson, F. K. 1992. Microbial degradation of nitroaromatic compounds. Adv. Appl. Microbiol. 37:1-19. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, M. A., and P. A. Williams. 2001. Cloning and characterization of the pnb genes, encoding enzymes for 4-nitrobenzoate catabolism in Pseudomonas putida TW3. J. Bacteriol. 183:1225-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James, K. D., M. A. Hughes, and P. A. Williams. 2000. Cloning and expression of ntnD, encoding a novel NAD(P)+-independent 4-nitrobenzyl alcohol dehydrogenase from Pseudomonas sp. strain TW3. J. Bacteriol. 182:3136-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James, K. D., and P. A. Williams. 1998. ntn genes determine the early steps in the divergent catabolism of 4-nitrotoluene and toluene in Pseudomonas sp. strain TW3. J. Bacteriol. 180:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manson, M. D., J. P. Armitage, J. A. Hoch, and R. M. Macnab. 1998. Bacterial locomotion and signal transduction. J. Bacteriol. 180:1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngai, K.-L., E. L. Neidle, and L. N. Ornston. 1990. Catechol and chlorocatechol 1,2-dioxygenases. Methods Enzymol. 188:122-126. [DOI] [PubMed] [Google Scholar]
- 25.Nichols, N. N., and C. S. Harwood. 1997. PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179:5056-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols, N. N., and C. S. Harwood. 1995. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida β-ketoadipate pathway. J. Bacteriol. 177:7033-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishino, S. F., J. C. Spain, and Z. He. 2000. Strategies for aerobic degradation of nitroaromatic compounds by bacteria: process discovery to field application, p. 7-61. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. CRC Press, Boca Raton, Fla.
- 28.Nozaki, M., H. Kagamiyama, and O. Hayaishi. 1963. Metapyrocatechase I. Purification, crystallization and some properties. Biochem. Z. 338:582-590. [PubMed] [Google Scholar]
- 29.Ornston, L. N., and D. Parke. 1976. Properties of an inducible uptake system for β-ketoadipate in Pseudomonas putida. J. Bacteriol. 125:475-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey, G., A. Chauhan, S. K. Samanta, and R. K. Jain. 2002. Chemotaxis of a Ralstonia sp. SJ98 toward co-metabolizable nitroaromatic compounds. Biochem. Biophys. Res. Commun. 299:404-409. [DOI] [PubMed] [Google Scholar]
- 31.Pandey, G., and R. K. Jain. 2002. Bacterial chemotaxis toward environmental pollutants: role in bioremediation. Appl. Environ. Microbiol. 68:5789-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parales, R. E., J. L. Ditty, and C. S. Harwood. 2000. Toluene-degrading bacteria are chemotactic to the environmental pollutants benzene, toluene, and trichloroethylene. Appl. Environ. Microbiol. 66:4098-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parales, R. E., and C. S. Harwood. 2002. Bacterial chemotaxis to pollutants and plant-derived aromatic molecules. Curr. Opin. Microbiol. 5:266-273. [DOI] [PubMed] [Google Scholar]
- 34.Parales, R. E., and C. S. Harwood. 1993. Regulation of the pcaIJ genes for aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 175:5829-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parke, D., D. A. D'Argenio, and L. N. Ornston. 2000. Bacteria are not what they eat: that is why they are so diverse. J. Bacteriol. 182:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parke, D., and L. N. Ornston. 1984. Nutritional diversity of Rhizobiaceae revealed by auxanography. J. Gen. Microbiol. 130:1743-1750. [Google Scholar]
- 37.Preuβ, A., and P.-G. Rieger. 1995. Anaerobic transformation of 2,4,6-trinitrotoluene and other nitroaromatic compounds, p. 69-85. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds, vol. 49. Plenum Press, New York, N.Y.
- 38.Rhys-Williams, W., S. C. Taylor, and P. A. Williams. 1993. A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas. J. Gen. Microbiol. 139:1967-1972. [DOI] [PubMed] [Google Scholar]
- 39.Rieger, P.-G., and H.-J. Knackmuss. 1995. Basic knowledge and perspectives on biodegradation of 2,4,6-trinitrotoluene and related nitroaromatic compounds in contaminated soil, p. 1-18. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds, vol. 49. Plenum Press, New York, N.Y.
- 40.Romero-Steiner, S., R. E. Parales, C. S. Harwood, and J. E. Houghton. 1994. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for complete degradation of p-hydroxybenzoate. J. Bacteriol. 176:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saier, M. H. J., J. T. Beatty, A. Goffeau, K. T. Harley, W. H. Heijne, S. C. Huang, D. L. Jack, P. S. Jahn, K. Lew, J. Liu, S. S. Pao, I. T. Paulsen, T. T. Tseng, and P. S. Virk. 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1:257-279. [PubMed] [Google Scholar]
- 42.Samanta, S. K., B. Bhushan, A. Chauhan, and R. K. Jain. 2000. Chemotaxis of a Ralstonia sp. SJ98 toward different nitroaromatic compounds and their degradation. Biochem. Biophys. Res. Commun. 269:117-123. [DOI] [PubMed] [Google Scholar]
- 43.Samanta, S. K., and R. K. Jain. 2000. Evidence for plasmid-mediated chemotaxis of Pseudomonas putida towards naphthalene and salicylate. Can. J. Microbiol. 46:1-6. [DOI] [PubMed] [Google Scholar]
- 44.Shioi, J., C. V. Dang, and B. L. Taylor. 1987. Oxygen as attractant and repellent in bacterial chemotaxis. J. Bacteriol. 169:3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spain, J. C. 1995. Bacterial biodegradation of nitroaromatic compounds under aerobic conditions, p. 19-35. In J. C. Spain (ed.), Biodegradation of nitroaromatic compounds, vol. 49. Plenum Press, New York, N.Y.
- 46.Spain, J. C. 1995. Biodegradation of nitroaromatic compounds. Annu. Rev. Microbiol. 49:523-555. [DOI] [PubMed] [Google Scholar]
- 47.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 48.Tokiwa, H., and Y. Ohnishi. 1986. Mutagenicity and carcinogenicity of nitroarenes and their sources in the environment. CRC Crit. Rev. Toxicol. 17:23-60. [DOI] [PubMed] [Google Scholar]
- 49.van der Meer, J. R., W. M. de Vos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]