Abstract
ATP, which is present in the extracellular matrix of multicellular organisms and in the extracellular fluid of unicellular organisms, has been shown to function as a signaling molecule in animals. The concentration of extracellular ATP (xATP) is known to be functionally modulated in part by ectoapyrases, membrane-associated proteins that cleave the γ- and β-phosphates on xATP. We present data showing a previously unreported (to our knowledge) linkage between apyrase and phosphate transport. An apyrase from pea (Pisum sativum) complements a yeast (Saccharomyces cerevisiae) phosphate-transport mutant and significantly increases the amount of phosphate taken up by transgenic plants overexpressing the gene. The transgenic plants show enhanced growth and augmented phosphate transport when the additional phosphate is supplied as inorganic phosphate or as ATP. When scavenging phosphate from xATP, apyrase mobilizes the γ-phosphate without promoting the transport of the purine or the ribose.
Apyrases are enzymes with the unifying characteristic of being able to hydrolyze both the γ- and the β-phosphate on ATP or ADP (Plesner, 1995). Most apyrases are expressed as plasma membrane-associated proteins with their hydrolytic activity facing into the ECM (Wang and Guidotti, 1996). The pea (Pisum sativum) apyrase used in this investigation, psNTP9, was originally characterized as a 47-kD nuclear NTPase because it was initially purified from nuclei (Chen et al., 1987), because it was shown to be localized in the nucleus by immunocytochemistry (Tong et al., 1993), and because it had potential nuclear-localization and DNA-binding sequences (Hsieh et al., 1996). However, the fact that it also had a signal peptide (Hsieh et al., 1996) led us to investigate whether some fraction of the pea apyrase is extracellular, and the results shown here indicate that it is.
Recent reports of ATP in the ECM of multicellular organisms (Sedaa et al., 1990) and in the extracellular fluid of unicellular organisms (Boyum and Guidotti, 1997) have prompted investigations into the fate of ATP outside of the cell. One conclusion is that xATP is hydrolyzed by apyrases. The physiological relevance of xATP degradation has been demonstrated in at least two systems, synaptic xATP hydrolysis following nerve stimulation to inactivate xATP as a neurotransmitter (Todorov et al., 1997), and xATP degradation during thrombosis (Marcus et al., 1997). The products of ATP hydrolysis do not accumulate in the extracellular fluid but are presumed to be recouped by purine transporters. Work in animal systems has shown that adenosine derived from xATP by the joint action of extracellular apyrases and ecto-5′-nucleotidases may be transported into cells by a sodium/adenosine cotransporter (Che et al., 1992). We present evidence from a plant system that shows that extracellular apyrases have a biological function in addition to xATP catabolism. We also demonstrate that apyrase from pea augments the uptake of Pi, as well as the Pi derived from xATP, without promoting the transport of the purine or ribose.
MATERIALS AND METHODS
Expression, Growth, and Transport in Yeast
The yeast (Saccharomyces cerevisiae) mutant NS219 was provided by Satoshi Harashima (Osaka University, Japan). The coding region of psNTP9, the pea (Pisum sativum) apyrase cDNA, was subcloned into pYES2 (Invitrogen, Carlsbad, CA) downstream of the GAL1 promoter and was transformed into the mutant by a PEG lithium acetate method (Elble, 1992). For time-course growth assays, single colonies of transformants were grown in a low-Pi (100 μm) synthetic-defined induction medium containing 2% Gal, 0.1% Glu, and 5% glycerol, and growth was monitored spectrophotometrically. Western analysis was performed on 30 μg of total protein isolated from saturated cultures grown in synthetic-defined induction medium using a polyclonal antiapyrase antibody raised against the purified pea protein (Tong et al., 1993). For growth assays in which the concentration of Pi was varied, 10,000 cells from a log-phase culture were added to an induction medium in which the Pi concentration had been adjusted. After 48 h of growth, the turbidity of the cultures was analyzed spectrophotometrically.
The acid-phosphatase assay was performed on cells grown in a high-Pi (10 mm) synthetic-defined induction medium. Cells were grown to an A660 of 1.0, and 1 mL was pelleted. The pellet was resuspended in an acetate buffer (pH 4.0) containing the chromogenic substrate complex nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate (KPL, Gaithersburg, MD). Acid-phosphatase activity was monitored by the development of a blue color. Time-course transport assays using 32Pi (Amersham) were performed on cells grown to an A660 of 1.0 in synthetic-defined induction medium containing 100 μm Pi. For each sample, 1 mL of cells grown in synthetic-defined induction medium was pelleted and resuspended in 250 μL of fresh synthetic-defined induction medium containing 1 μCi 32Pi and 100 μm Pi. The transport reaction was stopped at various times by the addition of 1 mL of ice-cold water, followed by four washes in ice-cold water. Kinetic assays were performed in the same manner as the time course of transport, but the concentration of Pi in the synthetic-defined induction medium was varied, and all reactions were stopped 8 min after the addition of radioactivity. In separate experiments transport reactions were performed in the presence of 2% Glu for the duration of the reaction. Cells used for these experiments were grown in the low-Glu induction medium until the time of the experiment, because Glu was found to repress expression of the psNTP9 gene under the control of the Gal promoter in pYES2. The kinetic data were fitted using regression analysis.
Transgenic Plant Construction
psNTP9 was subcloned as a SalI to XbaI fragment into pKYLX71 (Schardl et al., 1987) cut with XhoI and XbaI. This plasmid was transformed into Agrobacterium tumefaciens GV3101 [pMP90] (Knocz and Schell, 1986), which was used to infect root calli from the Wassilewskija ecotype of Arabidopsis under kanamycin selection (Valvekens et al., 1992). Four individual lines obtained from separate calli were propagated to the T3 generation.
Subcellular Apyrase Distribution in Pea
Etiolated pea plumules served as the tissue source for nuclei and cytoplasm isolation, as described previously (Chen and Roux, 1986). Plasma membrane was prepared from 30 g of root tissue. Western analysis was performed on 15 to 30 μg of protein from the cytoplasm, plasma membrane, and nuclei using a polyclonal antiapyrase antibody raised against the purified pea protein (Tong et al., 1993). To determine the orientation of the pea apyrase in the pea plasma membrane, outside-out vesicles were prepared, and the accessibility of the enzyme was determined by selective trypsin proteolysis or membrane shaving, followed by activity assays and western blotting.
Apyrase Activity Measurement and Immunochemistry in Transgenic Arabidopsis
Approximately 0.5 g of the total tissue from 3-week-old plants was frozen and powdered. ECM material was then extracted by the method of Barcelo et al. (1987). Apyrase activity was determined using the phosphomolybdate assay (Chen and Roux, 1986). Western analysis was performed on 20 μg of the total ECM protein using the pea apyrase antibody. Immunoblots were developed with an alkaline phosphatase substrate system.
Pi-Uptake Experiments and Growth Assays
In all experiments the growth medium contained no sugar, and plants were grown in sterile culture at 22°C under 150 to 200 μE of continuous light. Unless otherwise noted, a standard 0.8% agar medium containing 100 μm Pi was used for uptake assays (Somerville and Ogren, 1982). Plants used for the Pi-uptake experiments were grown singly in 1 mL of the standard agar medium for 15 d prior to the experiment. On the day of the experiment, 10 μCi of 32Pi was applied to the side of the culture dish and allowed to diffuse through the agar. In kinetic studies additional Pi was added with the 32P to the final concentration specified. The lids of the tissue-culture dishes were removed to facilitate transpiration. After 18 h the plants were removed from the medium. The aerial portions of the plant not in contact with the agar were weighed and counted by liquid scintillation. For each plant the entire root system was carefully pulled from the agar and washed in ice-cold water prior to scintillation counting. For kinetic analysis the data were fitted using linear regression. In experiments involving uptake from radioactive adenyl phosphates, 0.8% agar Murashige and Skoog basal salt medium (Sigma) was used. The procedures were the same as those used for Pi uptake; however, only the aerial portions of the plants were counted.
In growth assays involving the response of plants to Pi, the standard 0.8% agar medium was used (Somerville and Ogren, 1982), with appropriate modifications made to the potassium phosphate concentration. Wild-type Arabidopsis (ecotype Wassilewskija) and transgenics were plated on 10 mL of this medium. In growth experiments involving nucleotides, Murashige and Skoog agar medium was used. adenosine nucleotide phosphates were spread onto the medium to a final concentration of 300 μm; for Pi treatments, the final concentration was 1.3 mm (1 mm in the Murashige and Skoog basal salt mixture plus 300 μm in the supplement). Leaf-area assays were performed on an automated imager (Alpha Imager 2000, Alpha Innotech, San Leandro, CA) and were calculated as the sum of pixel areas for 400 to 600 plants per treatment.
Measurement of ATP in a Defined Soil
For measurements of soil ATP, we used uncharged Sunshine Mix 2 potting soil (Hummert, Earth City, MO). The soil was hydrated to 4 times its dry weight and then autoclaved for 0.5 h. When the soil had cooled to room temperature, it was divided into pots packed with 30 g of sterile soil. One-half of the pots were inoculated with 10,000 colony-forming units of a stock soil flora derived from a single field sample at the University of Texas (Austin), and the other half were not inoculated. The pots were then wrapped in plastic, sealed, and placed in darkness at 37°C for 7 d. After 7 d a 15-g sample was removed from each pot and pressed in a syringe until 1 mL of soil fluid was collected. A dilution of this fluid was plated on Luria-Bertani medium and grown for 12 h at 37°C, and the colonies were then counted. The remaining fluid sample was centrifuged for 60 s to pellet soil debris and was then filtered through a 0.2-μm filter. This filtrate was used as the source for the firefly luciferase assay.
Luminometry was performed in triplicate on 30 μL of each sample reconstituted in 70 μL of firefly luciferase buffer (Firelight, Analytical Luminescence Laboratory, Cockeysville, MD) After the buffer was added, all samples were kept on ice. ATP standards (Sigma) were reconstituted in firefly luciferase buffer with 30% soil water from an uninoculated sample to account for the inhibitory effects of humic acid on the luciferase enzyme. Standards and sample were loaded into a 96-well plate and read on an automated luminometer (model MLX, Dynex Technologies, Chantilly, VA). Samples were processed with the addition of 50 μL of firefly luciferase, followed by a reading delay of 1.0 s and an integration time of 5 s. Output was taken as an average of the integration time and was then averaged for the triplicate. The sample handling time was less than 2 h.
RESULTS
Pea Apyrase Complements a Yeast Pi-Transport Mutant
Initial observations of heightened Pi transport by transgenic plants expressing apyrase suggested that apyrase might be involved in Pi uptake. To test the ability of an apyrase to function in Pi nutrition, we expressed a previously characterized pea NTPase gene psNTP9 (Hsieh et al., 1996), a member of the apyrase family (Handa and Guidotti, 1996), in a yeast Pho84 mutant (NS219) deficient in a Pi transporter (Bun-ya et al., 1991). The mutant showed reduced Pi acquisition and a decreased growth rate, and it constitutively expressed the Pi-repressible acid phosphatase because it could not accumulate Pi. Complementation with a functionally homologous gene resulted in increased growth, increased Pi transport, and repression of the acid phosphatase (Harrison and van Buuren, 1995). Based on a survey of the yeast genomic sequences, yeast lacks membrane-associated apyrases resembling psNTP9. Since the yeast system is naive to this apyrase, complementation of the NS219 mutant could be used to determine whether psNTP9 could function in Pi transport.
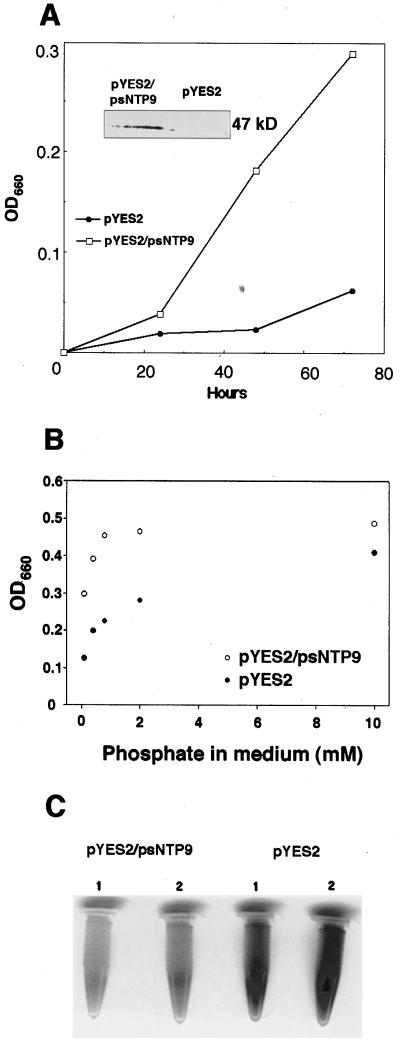
Three independent tests indicated that the apyrase gene successfully complemented the PHO84 mutant phenotype: growth assays (Fig. 1, A and B), repression of acid phosphatase activity (Fig. 1C), and labeled Pi transport (Fig. 2). The psNTP9 gene significantly improved the growth of NS219 cells (Fig. 1A) that overexpressed the apyrase (Fig. 1A, inset). Furthermore, the growth benefits conferred to the mutant by apyrase extended to Pi concentrations as high as 2 mm (Fig. 1B). When cells were incubated under acid conditions in nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate, mutant cells with high acid phosphatase activity stained dark, and wild-type cells and complemented cells stained light. The NS219 cells expressing the pea apyrase gene had less acid phosphatase activity than a mock-transformed control (Fig. 1C).
Figure 1.
psNTP9 complements a Pi-uptake mutant. A, pYES2 vector alone and pYES2/psNTP9 were transformed into NS219. Single colonies were picked and grown in low-Pi medium. Values represent the averages of two independent transformants. Inset shows a western blot of the psNTP9 transformant and a control (vector alone). B, Growth of the mutant transformed with the pea apyrase and a mock transformant at five Pi concentrations. Values are the averages of two independent transformants for two separate experiments. Cultures were inoculated with 104 yeast cells from a log-phase culture. C, Acid-phosphatase activity in NS219 yeast: two independent NS219 transformants harboring the vector alone show the dark staining of the mutant, whereas two independent NS219 transformants carrying the pea apyrase gene psNTP9 show the light staining of the wild type.
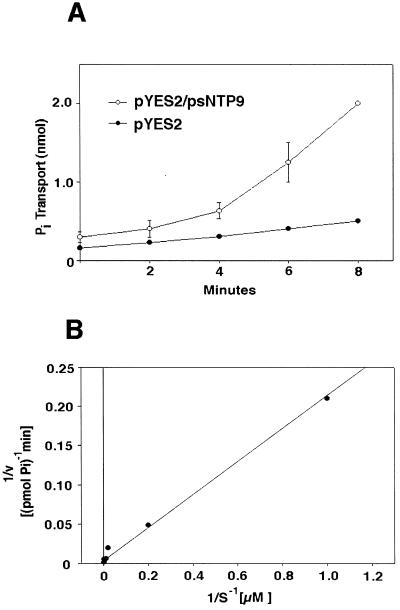
Figure 2.
Pi uptake by NS219 in the presence and absence of the psNTP9 gene. A, Two independent pYES2/psNTP9 transformants were tested. Values reported are in nanomoles per milliliter and are the averages of duplicate samples. B, Lineweaver-Burk plot of Pi uptake versus the Pi concentration of the culture medium. Linear-regression analysis was used to fit the data and a first-order polynomial describing the line of best fit was used to approximate the Km.
Although the pea gene partially restored many wild-type properties to the PHO84 mutant, pleiotropic effects of apyrases, such as increased turnover of cellular phosphate pools or changes in Pi metabolism resulting from feedback effects, could also explain the reversion. Complementation was also confirmed directly by testing Pi uptake in NS219 cells expressing psNTP9 (Fig. 2A). Cells expressing the pea apyrase gene followed Michaelis-Menten kinetics and had an estimated Km of 24 μm in the induction medium containing Gal (Fig. 2B). When 2% Glu was substituted for Gal for the duration of the transport assay, the Km was estimated at 14 μm.
Detection of the Pea Apyrase in Nuclei and in Purified Plasma Membrane
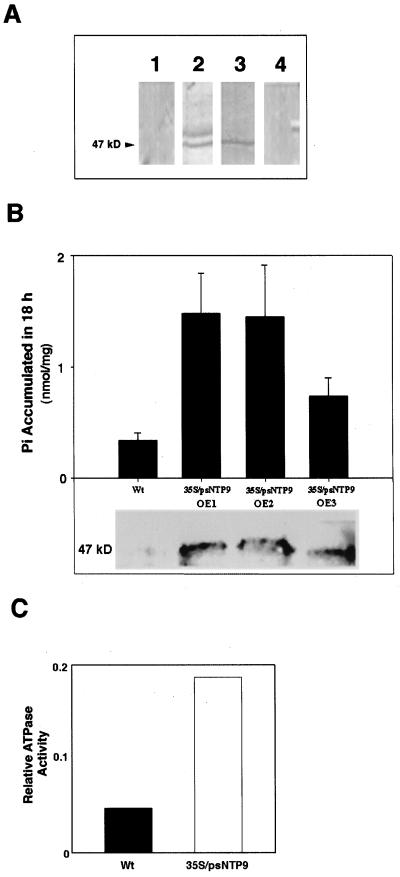
An immunoblot assay showed that pea apyrase was associated with nuclei and with purified plasma membranes but not with the cytoplasm (Fig. 3A). Protease treatment destroyed both apyrase activity and antigenicity in outside-out plasma membrane vesicles. After trypsin treatment the exterior face of the vesicle showed 30% of the ecto-ATPase activity of the untreated sample. Endo-ATPase activities were retained after trypsin treatment, indicating that the digestion occurred exclusively on the exterior face of the membrane.
Figure 3.
Expression of apyrase in pea and in transgenic lines. A, Immunoblot analysis of subcellular fractions from etiolated pea plants. Lane 1, Cytoplasm; lane 2, purified plasma membrane; lane 3, purified nuclei; and lane 4, preimmune control of nuclei. B, Top, The total Pi accumulated in the shoots of the wild type (Wt) and three independent transformants in an 18-h 32P-uptake assay tested at 2 mm Pi. Bottom, A corresponding immunoblot performed on equal amounts of protein isolated from the ECM of 3-week-old wild-type Arabidopsis and the psNTP9 transgenics. C, Assay of ATPase activity in the ECM fraction of OE1.
Enhanced Growth of Plants Overexpressing Apyrase Correlates with Increased Pi Uptake
Our findings in yeast corroborated the observations made with transgenic Arabidopsis plants. Three of the four transgenic plant lines constitutively expressed psNTP9 under the control of the cauliflower mosaic virus 35S promoter, and over an 18-h period showed 2 to 5 times as much Pi accumulation in shoots as the wild type (Fig. 3B). Apyrase-expressing plants also showed 4 times as much ATPase activity in the ECM as the wild type (Fig. 3C). Some percentage of the apyrase can be found in crude fractions of proteins ionically extracted from wall fragments in pea epicotyls (data not shown) and in Arabidopsis seedlings. Because this fraction could be more conveniently prepared than a plasma membrane fraction from Arabidopsis, it was used to assay the level of ECM apyrase expression in the transgenic Arabidopsis plants. Using an antibody raised against the pea apyrase (Tong et al., 1993), we detected a high level of the 47-kD gene product in the extracellular fraction of three transgenic lines compared with that detectable in immunoblots in the lane loaded with the same protein quantity from wild-type plants (Fig. 3B, bottom).
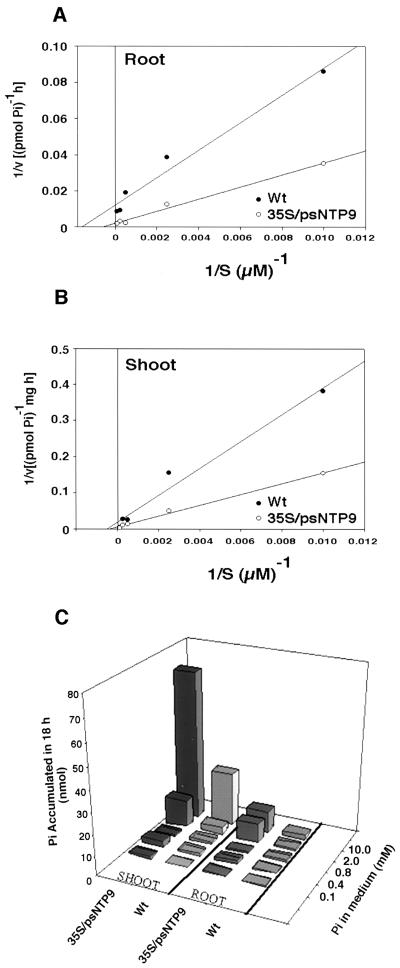
Kinetic studies of the transgenic and wild-type plants showed large differences in the relative rate of Pi accumulation in the low-affinity range. On a per-root basis the average Vmax was 77.51 pmol Pi h−1 for wild-type plants and 932 pmol Pi h−1 for the transgenic plants. Whereas the roots of transgenic plants accumulated Pi at 12 times the rate of the wild type (Fig. 4A), these differences applied to only plants grown in high-Pi medium, because the affinity for Pi in the transgenic plants was substantially reduced. The Km of wild-type and transgenic plants was 625 and 3770 μm, respectively. In contrast, the shoots of both transgenic and wild-type plants retained the same relative affinity for Pi at high concentrations, but the transgenics accumulated Pi at about 3 times the rate of the wild type, with a Vmax of 181 and 50 pmol Pi mg−1 h−1 (Fig. 4B).
Figure 4.
Kinetic analysis of Pi uptake into wild-type (Wt) and transgenic Arabidopsis plants. A, Lineweaver-Burk plot of Pi uptake into roots. Values on the x axis are the averages of Pi accumulated per minute for three transgenic and three wild-type roots at each of the five Pi concentrations tested. Linear-regression analysis was used to fit the data, and a first-order polynomial describing the line of best fit was used to approximate the Km. B, Lineweaver-Burk plot of Pi uptake into shoots. The data were acquired and analyzed in the same manner as for the root kinetics but were normalized for weight because of the differences in shoot size. C, To permit comparisons between the uptake of shoots and roots, the total Pi accumulation during the 18-h experiment is presented for each tissue. For each concentration the average uptake of the three plants is presented.
A comparison of the total Pi transported into the entire root or shoot over a range of high Pi concentrations revealed that in both roots and shoots the transgenic plant accumulated an average of 3 times as much Pi as the wild type (Fig. 4C). The most dramatic difference in accumulation between transgenic and wild-type plants was at 2 mm Pi. At this concentration the apyrase-expressing plants accumulated 9 times as much Pi in the root and 3.6 times as much in the shoot compared with the wild type. When partitioning of Pi between root and shoot in the transgenic and wild type was compared, both wild-type and transgenic plants tended to accumulate more Pi in the shoot at higher concentrations of Pi. At 100 μm external Pi, 63% of the total Pi transported into the transgenic plants was in the shoot. In wild-type plants given the same concentration of exogenous Pi, 53% of the total Pi transported into the plant was in the shoot. At 10 mm Pi, 87% of the total Pi was found in the shoots of transgenic plants and 92% in the shoots of wild-type plants.
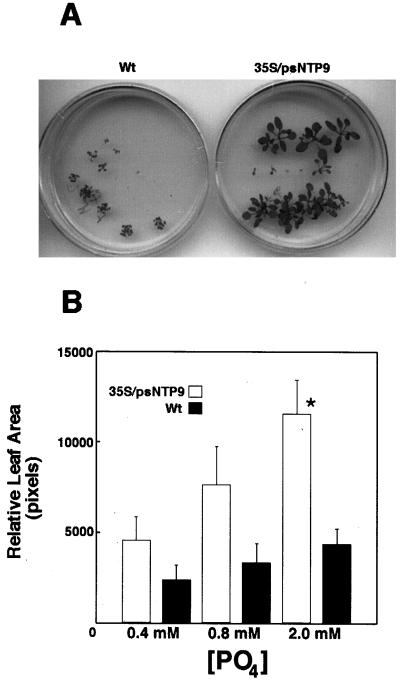
We examined the transgenic plants for phenotypes that might relate to the presence of the apyrase gene and found that those grown in high Pi were significantly larger than wild-type plants (Fig. 5A). Differences in growth became statistically significant when the Pi concentration was approximately 2 mm (Fig. 5B), at which point the average leaf area of the transgenic plants was 3 times that of the wild type. The growth differences between transgenic and wild-type plants at 2 mm Pi was correlated with the increased Pi uptake of transgenics at this same concentration (Fig. 4C). The growth and transport phenotypes were visible only in media depleted of sugar.
Figure 5.
A, Representative 20-d-old wild-type (Wt) and transgenic Arabidopsis plants grown in 2 mm Pi. B, Leaf area of transgenics compared with wild type. Although there was a trend for increased growth with increased Pi, the difference was only statistically significant at 2 mm. Data were analyzed for significance in a Student's t test (n = 18; *P < 0.05; error bars = se).
Transgenic Plants Can Utilize Pi Supplied as ATP
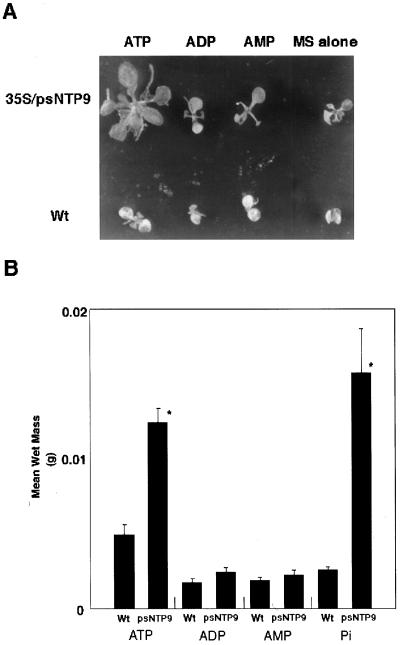
Since apyrases seem to function in the breakdown of xATP, we decided to test the ability of root-fed adenosine nucleotide phosphates to substitute for additional Pi. The basal Pi concentration, a growth-sustaining concentration at which there was no statistically significant difference between the growth of wild-type and transgenic plants, was 1 mm. Transgenic and wild-type plants were fed an additional 0.3 mm ATP, ADP, or AMP, and growth was measured using total mass as an indicator. Transgenic plants on ATP showed enhanced growth (Fig. 6A). The xATP-fed transgenic plants had an average wet weight double that of the wild type, a growth stimulation nearly equivalent to that observed from the addition of free Pi (Fig. 6B). Whereas xATP had dramatic effects on growth, xADP and xAMP did not. In contrast to the transgenic plants, none of the treatments, including additional Pi, significantly affected wild-type growth, as determined by Student's t test.
Figure 6.
Growth of wild-type (Wt) versus transgenic plants expressing psNTP9 in the presence of exogenous nucleoside phosphates. A, Representative 3-week-old Arabidopsis plants grown with three different treatments compared with Murashige and Skoog medium (MS) alone. B, Plants from the treatments were grown for 17 d and then weighed. All treatments were cross-analyzed for significance in a Student's t test. Bars marked by an asterisk differ significantly from those without an asterisk (n > 20 for all groups; P < 0.05; error bars = se).
Transgenic Plants Preferentially Transport the γ-Phosphate of ATP
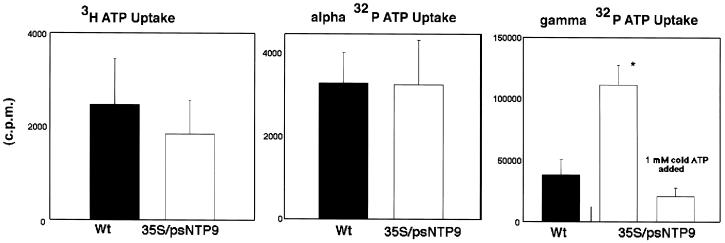
The inability of apyrase to translocate xAMP was demonstrated by the low level of radiolabel accumulated in transgenic plants fed [2,8-3H]ATP and [α-32P]ATP (Fig. 7). However, in feeding experiments in which the γ-phosphate was labeled, transgenic plants accumulated 3 times as much as the wild type. In separate experiments nonradioactive xATP was able to competitively inhibit the uptake of γ-32P from xATP.
Figure 7.
Transport of the products of ATP hydrolysis by transgenic and wild-type (Wt) plants. [2,8-3H]ATP, [α-32P]ATP, and [γ-32P]ATP were fed to 15-d-old plants in separate treatments. All treatments were analyzed for significance in a Student's t test (n > 4–6 for all groups; *P < 0.05; error bars = se).
Soil ATP
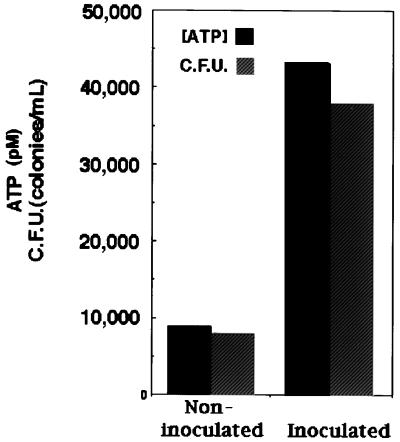
Soil treated with microbes showed increased microbial growth and an accumulation of ATP after 7 d, whereas those that were not inoculated accumulated far less ATP. In both instances the presence of ATP was correlated positively with the number of colony-forming units (Fig. 8).
Figure 8.
Soil ATP measurement with respect to microbial count (C.F.U. = colony-forming units). Bars are average ATP concentrations in soil water taken from three to five pots.
DISCUSSION
The growing evidence for xATP in animal systems has cast ectoapyrases as likely regulators of extracellular energy charge. Although this role has functional significance for purinergic receptor signaling, it may not be the exclusive function of apyrases. Experiments using pea apyrase in both yeast and transgenic Arabidopsis have revealed a new function for these enzymes. Our evidence from the use of this gene shows that plant apyrases, like their animal counterparts, are located in the ECM and hydrolyze xATP. More importantly, these experiments suggest that apyrases play a role in the mobilization of Pi and phosphate derived from xATP.
Many apyrases are ectoapyrases, which means that they are anchored in the plasma membrane with their active site facing into the ECM (Komoszynski and Wojtczak, 1996; Wang and Guidotti, 1996). Our results indicate that a significant fraction of the protein encoded by psNTP9 can also be classified as ectoapyrase. As judged by immunoblots, highly purified plasma membranes from pea roots contain the apyrase oriented with its activity accessible to proteolytic removal. These results are consistent with the interpretation that, like most of the animal apyrases characterized thus far, the hydrophobic signal peptide at the N terminus of the pea apyrase specifies a transmembrane domain and orients the enzyme in the plasma membrane so that its activity is in the ECM. Although animal ectoapyrases are considered to be localized primarily in the plasma membrane, in some systems a fraction of the total apyrase population is found free in the ECM (Todorov et al., 1997). This observation led us to determine whether a similar situation exists in plants. The results shown in Figure 3B indicate that transgenic Arabidopsis plants had the 47-kD apyrase in the ECM, whereas the wild type did not. Although it is possible that the extracellular apyrase may be secreted, an alternative explanation could be that some fraction of the plasma membrane-anchored protein may be proteolytically removed from the plasma membrane and released into the wall.
Our data indicate that the psNTP9 gene product can function in Pi nutrition in two heterologous systems. In yeast the apyrase promotes growth of the NS219 mutant at Pi concentrations up to 2 mm, above which point the low-affinity system probably masks the apyrase. This finding is consistent with PHO84 gene-transport activities and growth assays originally reported. In the initial characterization of the yeast PHO mutants (Toh-E and Oshima, 1974; Bun-ya et al., 1991), experiments were performed in two standard Pi-containing media: one at 10 mm (low-affinity) and one at 0.1 mm (high-affinity). Although pea apyrase does not have the multiple membrane-spanning regions of plant Pi transporters recently cloned (Muchal et al., 1996; Leggecie et al., 1997), the pea gene for psNTP9 complements NS219, giving the mutant a Km in the high-affinity range.
It is possible that apyrase itself may oligomerize to form a pore in the membrane. Recent work with the mammalian homolog of psNTP9 indicated that this ectoapyrase tetramerizes in the membrane to form a structure reminiscent of several channels (Wang et al., 1998). Alternatively, apyrase could interact with a transport system that functions as a complex. A similar scenario has been suggested by others to explain the partial complementation of high-affinity mutants with low-affinity transporters (Leggecie et al., 1997). Indeed, extracellular proteins are postulated to bind Pi as part of a Pi-transport system (Mass et al., 1979).
Although the complementation of NS219 is for a high-affinity transporter, growth differences in the complemented mutant persist at external Pi concentrations as high as 2 mm. Similarly, significant differential growth is seen at Pi levels near or at 2 mm in plants, the concentration at which apyrase-expressing plants accumulated 9 times as much Pi in the root and 3.6 times as much in the shoot compared with wild type. However, the accumulation of Pi in the transgenic Arabidopsis plants is unlike that seen in pho2, a shoot Pi-hyperaccumulator mutant (Delhaize and Randall, 1995), because Pi intoxication is not seen. The phenotype of the transgenics is also different from what might be expected of a plant overexpressing the gene at the pho1 locus. The pho1 mutant is defective in the ability to transfer Pi from the roots to the xylem (Poirier et al., 1991).
Plants ectopically expressing the pea apyrase show increased Pi accumulation in roots and shoots. Apyrase functions in a transport system that is common to the root and xylem. It also seems likely that elements of this shared system are present in yeast, given the ability of apyrase to complement NS219. The differences in the kinetics of transport may be a function of the transport complex stoichiometry. In such a scenario, increased apyrase levels may modulate the affinity of a pre-existing low-affinity system by channeling Pi or phosphate derived from ATP. In the transgenics the dependency of growth and transport phenotypes on Suc-depleted medium suggests that the Pi complex in which apyrase participates is tightly linked to photosynthate availability.
Physiologically, the expression of an extracellular enzyme that can hydrolyze only γ- and β-phosphates on nucleotides is difficult to explain, given the abundance of other substrate-versatile phosphatases. Indeed, both plants and yeast have evolved highly effective means of scavenging organophosphate using acid and alkaline phosphatases (Reid and Bieleski, 1970; Yoshida et al., 1989). Why are apyrases needed at all? The immediate answer may be that apyrases, with an ATPase kcatKm of 108 (Handa and Guidotti, 1996), simply hydrolyze xATP more quickly than any other enzyme in the ECM.
In animals ATP is released into the ECM in a regulated manner by ATP-binding cassette transporters (Abraham et al., 1993; Reisin et al., 1994). The efflux of ATP is thought to be a way of regulating the intracellular adenylate pool involved in signaling, and this xATP would need to be hydrolyzed in part because xATP itself is a powerful elicitor of intracellular calcium signaling (Zheng et al., 1991; Suko et al., 1997). If xATP is also an elicitor in plants, apyrases may provide a governable way of inactivating xATP as a signal, an inactivation analogous to that discovered in the nervous system (Todorov et al., 1997).
We propose a role for apyrase in addition to quenching xATP signals. Apyrase may function constitutively to recoup Pi from xATP whether the xATP accumulates in the soil as it is released from various cellular sources or, as has been shown in animals, it accumulates in the ECM when cells release ATP through multidrug resistance proteins or the cystic fibrosis transmembrane conductance regulator (Abraham et al., 1993; Reisin et al., 1994). The mdr1 gene is a member of the ATP-binding cassette family and has been reported to promote the electrogenic transport of ATP across the plasma membrane into the ECM. Since a homolog of mdr1 has been found in plants (Dudler and Hertig, 1992), we hypothesize that it might have ATP-transport properties similar to channels found in animals.
Given the high catalytic efficiency of apyrases, and the fact that their ability to generate Pi is limited only by the diffusion of ATP, we speculate that apyrases could create a microenvironment of recoverable Pi in the area proximal to MDR1. Estimations of the kinetics of ATP efflux from cells overexpressing MDR1 are approximately 4 × 106 molecules cell−1 s−1 (Abraham et al., 1993), but this xATP does not accumulate. In animals a steady-state level (in femtomoles/cell) of xATP is achieved rapidly through the action of apyrase and other phosphatases. Although there is a functional linkage between this apyrase activity and nucleoside transport in eukaryotes (Che et al., 1992), the fate of the hydrolyzed Pi has yet to be reported. We believe that apyrase itself may provide a means of immediately and directly recovering Pi from xATP.
The pea apyrase seems to mobilize the γ-phosphate of ATP preferentially. This is probably because the ADPase activity of the pea apyrase is only 15% of that of the ATPase activity (Chen and Roux, 1986). The lack of responsiveness of the transgenics treated with xAMP is most likely due to the inability of apyrases to hydrolyze nucleotide monophosphates (Plesner, 1995).
Although pea apyrase does play a role in the transport of Pi, it does not facilitate the transport of the other products of ATP hydrolysis (Fig. 7). Increased extracellular apyrase activity does not result in a concomitant increase in adenylate transport, indicating that the regulation of purine uptake is independent of xATPases and Pi uptake. This exclusion could be physiologically relevant to regulating the size of the adenylate pool.
In plants considerations of scavenging xATP could also include environmental ATP. As much as 80% of the phosphorus in soil is in organic form (Schachtman et al., 1998), and much of this is thought to be unavailable to plants. Although soil flora may be in competition with plants for Pi, it seems more likely that soil flora would be the point of origin for most Pi bound in extracellular adenylate nucleotides. It has long been known that Escherichia coli secretes 99.9% of its cAMP into the extracellular fluid (Matin and Matin, 1982). Yeasts also release significant amounts of ATP into the extracellular fluid (Boyum and Guidotti, 1997). We speculate that apyrases could make the organic phosphate released by microbes in the rhizosphere available to plants.
Measurements of ATP in soil have largely focused on using the nucleotide as an indicator for soil biomass (Brookes and Jenkinson, 1989). These measurements place priority on the ratio of ATP to carbon biomass, a ratio that is typically 10 μmol of ATP g−1 carbon biomass (De Nobili et al., 1996). The procedures required for such measurements require sonication of samples prior to processing. Furthermore, most samples used for such measurements are collected from the field and have a clay content capable of chelating ATP. Although we acknowledge the benefits of current methods of correlating biomass with ATP, we would like to suggest that our estimation of aqueous ATP, the ATP fraction in soil water, may be representative of the amount of ATP available to roots in a soil rich in organic matter (Fig. 8; even though this is only a very conservative approximation of the ATP that might actually be present in the mucilaginous root-soil interface). The ATP in the soil water may originate from dead cells, efflux, or phage activity. However, regardless of the source, this ATP could be used as a Pi source by plants, in which case apyrase may function to mobilize it. Future experiments will help to delineate which xATP pools suggested in this report are physiologically relevant to apyrase and Pi transport.
ACKNOWLEDGMENTS
We thank Dr. G. Thompson, A. Rajagopal, and B. Windsor for valuable suggestions that aided in the preparation of this manuscript, and Dr. M. Harrison, who helped us to obtain the yeast strains.
Abbreviations:
- ECM
extracellular matrix
- NTP
nucleotide triphosphate
- xATP
extracellular ATP
Footnotes
This work was supported by grants from the National Science Foundation and the National Aeronautics and Space Administration and by a National Science Foundation graduate fellowship to C.T.
LITERATURE CITED
- Abraham E, Prat A, Gerweck L, Seneveratne T, Arceci R, Kramer R, Guidotti G, Cantiello H. The multidrug resistance (mdr1) gene product functions as an ATP channel. Proc Natl Acad Sci USA. 1993;90:312–316. doi: 10.1073/pnas.90.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyum R, Guidotti G. Glucose-dependent, cAMP-mediated ATP efflux from Saccharomyces cerevisiae. Microbiology. 1997;143:1901–1908. doi: 10.1099/00221287-143-6-1901. [DOI] [PubMed] [Google Scholar]
- Brookes P, Jenkinson D. ATP and adenylate energy charge levels in soil microbial biomass. In: Stanley P, McCarthy B, Smither R, editors. ATP Luminescence: Rapid Methods in Microbiology. Oxford, UK: Blackwell Scientific Publications; 1989. pp. 119–127. [Google Scholar]
- Bun-ya M, Nishimura M, Harashima S, Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991;11:3229–3238. doi: 10.1128/mcb.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che M, Nishida T, Gatmaitan Z, Arias I. A nucleoside transporter is functionally linked to ectonucleotidases in rat liver canalicular membrane. J Biol Chem. 1992;267:9684–9688. [PubMed] [Google Scholar]
- Chen Y-R, Datta N, Roux S. Purification and partial characterization of a calmodulin-stimulated nucleoside triphosphatase from pea nuclei. J Biol Chem. 1987;262:10689–10694. [PubMed] [Google Scholar]
- Chen Y, Roux S. Characterization of nucleoside triphosphatase activity in isolated pea nuclei and its photoreversible regulation by light. Plant Physiol. 1986;81:609–613. doi: 10.1104/pp.81.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall P. Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 1995;107:207–213. doi: 10.1104/pp.107.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nobili M, Diaz-Ravina M, Brookes P, Jenkinson D. Adenosine 5′-triphosphate measurements in soils containing recently added glucose. Soil Biol Biochem. 1996;28:1099–1104. [Google Scholar]
- Dudler R, Hertig C. Structure of an mdr-like gene from Arabidopsis thaliana. J Biol Chem. 1992;267:5882–5888. [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Handa M, Guidotti G. Purification and cloning of a soluble ATP- diphosphohydrolase (apyrase) from potato tubers (Solanum tuberosum) Biochem Biophys Res Commun. 1996;218:916–923. doi: 10.1006/bbrc.1996.0162. [DOI] [PubMed] [Google Scholar]
- Harrison M, van Buuren M. A phosphate transporter from the mycorrhizal fungus Glomus versiforme. Nature. 1995;378:626–629. doi: 10.1038/378626a0. [DOI] [PubMed] [Google Scholar]
- Hsieh H-L, Tong C-G, Thomas C, Roux S. Light-modulated abundance of an mRNA encoding a calmodulin-regulated, chromatin-associated NTPase in pea. Plant Mol Biol. 1996;30:135–147. doi: 10.1007/BF00017808. [DOI] [PubMed] [Google Scholar]
- Komoszynski M, Wojtczak A. Apyrases (ATP diphosphohydrolases, EC 3.6.1.5) function and relationship to ATPases. Biochim Biophys Acta. 1996;1310:233–241. doi: 10.1016/0167-4889(95)00135-2. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Leggecie G, Willmitzer L, Riesmeier J. Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: identification of phosphate transporters from higher plants. Plant Cell. 1997;9:381–392. doi: 10.1105/tpc.9.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus A, Broekman J, Drosopoulos J, Islam N, Alyonycheva T, Safier L, Hajjar K, Posnett D, Schoenborn M, Schooley K, and others (1997) Control of platelet reactivity by an ecto-ADPase on human endothelial cells. In L Plesner, T Kirley, Knowles A, eds, Ecto-ATPases: Recent Progress on Structure and Function. Plenum Press, New York, pp 167–170
- Mass E, Ogata G, Finkel M. Salt-induced inhibition of phosphate transport and release of membrane proteins from barley roots. Plant Physiol. 1979;64:139–143. doi: 10.1104/pp.64.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matin A, Matin M. Cellular levels, excretion, and synthesis rates of cyclic AMP in Escherichia coil grown in continuous culture. J Bacteriol. 1982;149:801–807. doi: 10.1128/jb.149.3.801-807.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchal U, Pardo J, Raghothama K. Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA. 1996;93:10519–10523. doi: 10.1073/pnas.93.19.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesner L. Ecto-ATPases: indentities and functions. Int Rev Cytol. 1995;158:141–214. doi: 10.1016/s0074-7696(08)62487-0. [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J. A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991;97:1087–1093. doi: 10.1104/pp.97.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid M, Bieleski R. Changes in phosphatase activity in phosphorus-deficient Spirodela. Planta. 1970;94:273–281. doi: 10.1007/BF00385759. [DOI] [PubMed] [Google Scholar]
- Reisin I, Prat A, Abraham E, Amara J, Gregory R, Ausiello D, Cantiello H. The cystic fibrosis transmembrane conductance regulator is a dual ATP and chloride channel. J Biol Chem. 1994;269:20584–20591. [PubMed] [Google Scholar]
- Ros Barceló A, Muñoz R, Sabater F. Lupin peroxidases. I. Isolation and characterization of cell wall-bound isoperoxidase activity. Physiol Plant. 1987;71:448–454. [Google Scholar]
- Schachtman D, Reid R, Ayling S. Phosphorus uptake by plants: from soil to cell. Plant Physiol. 1998;116:447–453. doi: 10.1104/pp.116.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl C, Byrd A, Benzion G, Altschuler M, Hildebrand D, Hunt A. Design and construction of a versatile system for the expression of foreign genes in plants. Gene. 1987;61:1–11. doi: 10.1016/0378-1119(87)90359-3. [DOI] [PubMed] [Google Scholar]
- Sedaa K, Bjur R, Shinozuka K, Westfall D. Nerve and drug induced release of adenine nucleosides and nucleotides from rabbit aorta. J Pharmacol Exp Ther. 1990;252:1060–1067. [PubMed] [Google Scholar]
- Somerville C, Ogren W (1982) Isolation of photorespiration mutants in Arabidopsis thaliana. In M Edelman, R Hallick, NH Chua, eds, Methods in Chloroplast Biology. Elsevier Biomedical Press, Amsterdam, The Netherlands, pp 129–138
- Suko Y, Kawahara K, Fukuda Y, Masuda Y. Nuclear and cytosolic calcium signaling induced by extracellular ATP in rat kidney inner medullary collecting duct cells. Biochem Biophys Res Commun. 1997;234:224–229. doi: 10.1006/bbrc.1997.6488. [DOI] [PubMed] [Google Scholar]
- Todorov L, Mihaylova-Todorova S, Westfall T, Sneddon P, Kennedy C, Bjur R, Westfall D. Neuronal release of soluble nucleotidases and their role in neurotransmitter inactivation. Nature. 1997;387:76–79. doi: 10.1038/387076a0. [DOI] [PubMed] [Google Scholar]
- Toh-E A, Oshima Y. Characterization of a dominant, constitutive mutation, PHO0, for the repressible acid phosphatase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1974;120:608–617. doi: 10.1128/jb.120.2.608-617.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C-G, Dauwalder M, Clawson G, Hatem C, Roux S. The major nucleoside triphosphatase in pea (Pisum sativum L.) nuclei and rat liver share common epitopes also present on nuclear lamins. Plant Physiol. 1993;101:1005–1011. doi: 10.1104/pp.101.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1992;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Guidotti G. CD39 is an Ecto-(Ca 2+,Mg 2+)-apyrase. J Biol Chem. 1996;271:9898–9901. [PubMed] [Google Scholar]
- Wang T-F, Ou Y, Guidotti G. The transmembrane domains of ectoapyrase (CD39) affect its enzymatic activity and quaternary structure. J Biol Chem. 1998;273:24814–24821. doi: 10.1074/jbc.273.38.24814. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Ogawa N, Oshima Y. Function of the PHO regulatory genes for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. Mol Gen Genet. 1989;217:40–46. doi: 10.1007/BF00330940. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zychlinsky A, Liu C, Ojcius D, Young J. Extracellular ATP as a trigger for apoptosis or programmed cell death. J Cell Biol. 1991;112:279–288. doi: 10.1083/jcb.112.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]