Abstract
Two genes involved in protein secretion, encoding the Rab protein YPT1/YPTA and the general fusion factor NSFI/NSFA, were characterized from two filamentous fungi, Trichoderma reesei and Aspergillus niger var. awamori. The isolated genes showed a high level of conservation with their Saccharomyces cerevisiae and mammalian counterparts, and T. reesei ypt1 was shown to complement yeast Ypt1p depletion. The transcriptional regulation of the T. reesei ypt1, nsf1, and sar1 genes, involved in protein trafficking, was studied with mycelia treated with the folding inhibitor dithiothreitol (DTT) and with brefeldin A, which inhibits membrane traffic between the endoplasmic reticulum and Golgi complex. The well-known inducer of the yeast and T. reesei unfolded protein response (UPR), DTT, induced the nsf1 gene and the protein disulfide isomerase gene, pdi1, in both of the experiments, and sar1 mRNA increased in only one experiment under strong UPR induction. The ypt1 mRNA did not show a clear increase during DTT treatment. Brefeldin A strongly induced pdi1 and all of the intracellular trafficking genes studied. These results suggest the possibility that the whole secretory pathway of T. reesei could be induced at the transcriptional level by stress responses caused by protein accumulation in the secretory pathway.
Proteins destined to be secreted from eukaryotic cells pass through secretory organelles, namely, the endoplasmic reticulum (ER) and the Golgi complex, and are finally transported to the plasma membrane to be released from the cell. The proteins are transported between the organelles and to the plasma membrane in carrier vesicles. Each vesicle transport step includes packaging of the cargo proteins and budding of the vesicle from the donor compartment membrane, migration of the vesicle to the acceptor compartment, and docking and fusion of the vesicle to the acceptor compartment membrane.
In order to maintain cellular organization, each vesicle fusion must be specific for the right acceptor membrane. This specificity is ensured by protein factors that are specific only for a given fusion event. The specificity of fusion is largely due to recognition proteins in the vesicle and target membranes (v-SNAREs and t-SNAREs, respectively), but other types of stage-specific proteins that assist in vesicle docking and fusion are also important. The small GTP-binding Rab proteins belonging to the Ras superfamily associate with the vesicle membrane through lipid modification in their GTP-bound active form (19). They have a role in the formation of a large protein complex (20S complex; includes the t-SNARE and v-SNARE) that is needed for membrane fusion to take place (43). It has been discovered that during the assembly of this complex, the Saccharomyces cerevisiae Rab protein involved in ER-Golgi transport, Ypt1p, interacts transiently with the corresponding t-SNARE, Sed5p, displacing the negative regulator Sly1p and allowing interaction of the t-SNARE and v-SNARE (21, 37). According to recent evidence, Ypt1p is also involved in protein sorting upon exit from the ER (26).
In addition to proteins specific for one fusion step, general fusion factors that are functional for multiple membrane fusion steps are also needed in the secretory pathway. These include yeast Sec18p and its mammalian counterpart, NSF (11, 55, 56). Sec18p has been isolated as a member of the membrane fusion protein complex (20S complex) (53). It was believed that hydrolysis of ATP by Sec18p/NSF triggers dissociation of the fusion complex and subsequently fusion of the vesicle and target membranes (6, 44). More recent papers, however, suggest that mammalian NSF activates the SNARE proteins prior to membrane fusion (29) or that it participates in their recycling after membrane fusion (52), perhaps by acting as a chaperone that alters the conformation of the SNAREs (27, 30).
Filamentous fungi are naturally good protein secretors, and because of the high efficiency of secretion by industrially exploited fungi such as Trichoderma reesei and Aspergillus niger, the secretory pathways of these fungi are of particular interest. Knowledge at the molecular level about intracellular protein folding and trafficking in filamentous fungi is beginning to accumulate (reviewed in reference 7). Folding factors, such as Bip and protein disulfide isomerase (PDI), have been characterized at the gene level for some filamentous fungi (e.g., see references 28, 38, and 48). Of the genes involved in protein trafficking, results have hitherto been published for the ypt1 genes of Neurospora crassa (13) and Phytophthora infestans (5) and for homologs of the yeast SAR1 gene from A. niger and T. reesei (49). Punt et al. (36) have cloned several secretion-related small GTPases from A. niger, and Dumas et al. (8) have cloned a homolog of yeast SEC4 from Colletotrichum lindemuthianum. In this paper, we report the characterization of the homologs of the yeast YPT1 and SEC18 genes from the filamentous fungus T. reesei and, for comparison, from A. niger var. awamori. The regulation of the T. reesei genes in the presence of inhibitors of protein secretion was studied, and evidence is presented to explain the transcriptional regulation of the Trichoderma secretory pathway.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The temperature-sensitive sso2 mutant S. cerevisiae strain H1152 (a sso2-1 leu2-3 trp1-1 ura3-1 sso1::HIS3) (S. Keränen and M. Aalto, unpublished data) was used. The yeast strains used for YPT1 complementation studies were GFU1-6D (a GAL10-YPT1::HIS3 his3 leu2 trp1 ura3) and TSU3-5D (ypt1ts::LEU2 his3 ura3) (courtesy of Hans Dieter Schmitt). The sec4 mutant yeast strain NY774 (α sec4-8 leu2-3,112 ura3-52) (courtesy of Peter Novick) and the sec18 mutant strain mBY12-6D (α sec18-1 trp1-289 leu2-3 leu2-112 ura3-52 his−) (courtesy of Randy Shekman) were used for complementation testing. The dithiothreitol (DTT) and brefeldin A (BFA) treatments were carried out with the T. reesei strain RutC-30 (24), and a DTT treatment experiment was done with T. reesei QM9414. The plasmid host was Escherichia coli DH5α [F− endA1 hsdR17(rk mk) supE44 thi-1 λ− recA1 gyrA96 relA1 Δ(argFlacZYA)U169 φ80lacZΔM15].
For the DTT and BFA treatments, RutC-30 was grown in a minimal medium (34) with 2% lactose in conical flasks at 28°C, with shaking at 200 rpm, for 60 h, and the mycelium was diluted 1:10 into the same medium. Growth was continued for 21 h and the culture was divided into two aliquots. DTT (10 mM) or BFA (50 μg/ml) was added to one aliquot, and mycelial samples were withdrawn from both aliquots before and 15 (only BFA), 30, 60, 90, 120, 240, and 360 min after treatment. The T. reesei strain QM9414 was cultivated in minimal medium (34) with 2% sorbitol and 0.05% proteose peptone. For induction of cellulase gene expression, α-sophorose (1 mM) was added after 23 and 32 h of cultivation. Treatment of the cultures with 10 mM DTT was started after 40 h of cultivation. Mycelial samples for RNA analysis were collected during the treatment. The dry weight of the cultures was 1.1 to 1.4 g/liter at the beginning of the DTT treatment. The mycelia for Northern blots were harvested by filtration through a glass fiber filter, washed with 0.9% NaCl, and stored frozen.
Nucleic acid methods.
The yeast SEC4 (the whole coding region) and SEC18 (the region corresponding to amino acids 224 to 447) fragments were obtained by PCR from yeast chromosomal DNA. Heterologous hybridization was carried out in a hybridization mix without formamide as described previously (40). The optimal hybridization temperature was first determined with genomic Southern blots, and a T. reesei cDNA library in λZAP (46) was screened for ypt1 and nsf1 clones at 48 and 50°C, respectively. The genomic T. reesei ypt1 gene was isolated from a cosmid library and subcloned as a 6-kb BamHI fragment into pBluescript SK(−) (Stratagene). The genomic T. reesei nsf1 gene was isolated from a cosmid library (22), and its 5′ end was subcloned as a 5-kb PstI fragment into pZERO (Invitrogen). The 5′ end of the gene was sequenced from this subclone and the rest of the gene was sequenced from the cosmid DNA.
Genomic DNA from the A. niger var. awamori strains UVK143f and dgr246p2 (51) was isolated as described previously (14) and used as the template for PCR amplification of the yptA gene. A 1.2-kb PCR fragment from the 5′ end and a 1.1-kb PCR fragment from the 3′ end were obtained by the method of rapid PCR-based walking of uncloned genomic DNA (Genome Walker kit; Clontech). The PCR fragments were cloned into pCRII (Invitrogen). For cloning of the A. niger var. awamori nsfA gene, an 830-bp BamHI-XhoI fragment from the Trichoderma nsf1 cDNA was used as a probe for Southern hybridization of genomic A. niger var. awamori DNA. The hybridization mix contained 25% formamide, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 0.5% sodium dodecyl sulfate (SDS), and 0.1 mg of denatured herring sperm DNA per ml. The DNA blot was washed at 50°C for 30 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS, with 0.5× SSC-0.1% SDS, and with 0.1× SSC-0.1% SDS. The same conditions were used for screening of an A. niger (strain 2663E) genomic DNA library which was constructed from approximately 15-kb inserts generated by partial Sau3A digestion in the λDASHII vector (Clontech) (T. Fowler, personal communication). DNA fragments from one isolated λ clone were then subcloned into plasmids for sequence analysis.
Northern hybridization was carried out with 5 μg of total RNA samples on Hybond N nylon membranes (Amersham, Little Chalfont, United Kingdom) according to the manufacturer's instructions. Quantification of Northern blots was done with the Phosphorimager SI and with ImageQuant software (Molecular Dynamics). The signals obtained with the gpd1 gene, encoding glyceraldehyde phosphate dehydrogenase, were used to control the RNA loading and to normalize the quantification of signals obtained with the other probes. Primer extension analysis of the T. reesei ypt1 gene was done as described previously (40), with a 35-mer oligonucleotide binding at the translation start site (5′ AGGTAGTCGTATTCAGGGTTCATGATGGGCGGTTG 3′).
Yeast suppression and complementation.
Yeast strains were grown without plasmids in rich medium and with plasmids in synthetic complete medium without uracil (SC-Ura) (42). In an attempt to clone the homologs of yeast SSO1/2 from T. reesei and A. niger var. awamori, we transformed a T. reesei cDNA library in pAJ401 (23) and an A. niger var. awamori cDNA library in pYES2 (Invitrogen) (10) into the yeast strain H1152. This strain has a disrupted SSO1 gene and a temperature-sensitive sso2 mutation. The transformants derived from the T. reesei library were plated on SC-Ura with galactose to the restrictive temperature, 31°C. The A. niger var. awamori library transformants were first grown on glucose medium at 24°C and then replicated onto galactose medium to switch on the GAL1 promoter in pYES2 and grown at 31°C. Plasmids were rescued from clones able to grow under selection by transforming total yeast DNA into E. coli by electroporation. During testing of whether the T. reesei ypt1 cDNA can complement the Ypt1p depletion, the plasmid containing the ypt1 cDNA under the PGK1 promoter in a multicopy vector (pMS82) was transformed into the yeast strain GFU1-6D. Transformants were grown as streaks on SC-Ura medium with galactose as the only carbon source and replicated onto SC-Ura plates with glucose. The streaks were grown for 3 days and replicated again onto the same plates. During testing for complementation of the temperature-sensitive ypt1 and sec4 mutations, pMS82 was transformed into the yeast strains TSU3-5D (ypt1) and NY774 (sec4) and the transformants were grown at 24°C. They were then replicated onto three plates, which were incubated at 33, 34, and 35°C. The full-length Trichoderma nsf1 cDNA was cloned into the multicopy vector pAJ401 between the yeast PGK1 promoter and terminator. The resulting plasmid was transformed into the yeast strain mBY12-6D. The transformants were first grown at 24°C and then replicated to 31°C to test for complementation.
Nucleotide sequence accession numbers.
The sequence data presented in this article have been submitted to the DDJB/EMBL/GenBank databases under the following accession numbers: AJ277108 (T. reesei ypt1), AJ277109 (T. reesei nsf1), AF244545 (A. niger var. awamori yptA), and AF244544 (A. niger var. awamori nsfA).
RESULTS
Characterization of the T. reesei ypt1 and A. niger var. awamori yptA genes.
The T. reesei ypt1 cDNA was isolated from a λZAP cDNA library by heterologous hybridization with the yeast SEC4 whole coding region as a probe. A full-length cDNA clone had an open reading frame encoding a 202-amino-acid protein with higher identity to yeast Ypt1p than Sec4p. Based on complementation of a yeast YPT1 depletion strain described below, the new T. reesei gene was named ypt1. The genomic ypt1 gene was isolated from a cosmid library, subcloned, and sequenced. It was found to contain four introns.
About 600 bp of the Trichoderma ypt1 promoter region was sequenced (data not shown), and it contained two putative TATA boxes (at −314 and −431 relative to the translation start site) and two putative CCAAT elements (at −346 and −284). To find out which of the TATA boxes is more significant for transcription initiation, we carried out primer extension experiments. Multiple transcription start sites were revealed downstream from both of the putative TATA boxes (at −381, −369, and −346 and at −152, −147, and −143 from the translation start). The most intensive signals in the primer extension gel corresponded to transcription start sites between the two putative TATA elements, suggesting that the upstream TATA box is more important than the other box (data not shown).
Subsequent to the work described above, an attempt was made to clone the T. reesei and A. niger var. awamori homologs of the yeast SSO genes involved in the final stage of secretion, fusion of the secretory vesicles to the plasma membrane. Several Trichoderma cDNA library plasmids were obtained that could support growth of the sso2 temperature-sensitive mutant strain at the restrictive temperature, one of which carried the full-length T. reesei ypt1 cDNA. In a complementation screening in the same yeast mutant strain with an A. niger var. awamori cDNA library, a cDNA that is highly homologous to T. reesei ypt1 was isolated and named yptA. This cDNA lacks about 50 bp of the protein-coding region. The genomic A. niger var. awamori yptA gene was cloned by PCR, and a region of 1,920 nucleotides (nt) was sequenced. A putative TATA box (TAATA) was found in the promoter region (data not shown). Interestingly, there is a long 3′ untranslated region of 456 bp in the cDNA. The yptA gene of A. niger var. awamori encodes a putative protein of 201 amino acids, having 1 amino acid difference compared to the srgB gene cloned from A. niger by Punt et al. (36) and sharing 94% identity with the Trichoderma YPTI protein. The yptA gene has four introns at positions identical to those for introns in the T. reesei and N. crassa (13) ypt1 genes (Fig. 1). The ypt1 gene of the oomycete P. infestans has five introns (5), three of which are at conserved positions with introns in the homolog genes isolated from the other three filamentous fungi.
FIG. 1.
Amino acid sequence alignment of the T. reesei YPTI and A. niger var. awamori YPTA proteins with the yeast Ypt1p and human Rab1 proteins. Asterisks, identical amino acids; dots, similar amino acids. The regions responsible for GTP binding and hydrolysis (G1 to G5) and the prenylation signal at the C terminus are boxed. Positions of introns in the genes of the filamentous fungi are shown by arrows.
Alignment of the T. reesei YPTI and A. niger var. awamori YPTA proteins with their yeast and human counterparts (Ypt1p and Rab1, respectively) revealed the extremely high conservation level of this group of proteins (Fig. 1). Of the sequence positions in the alignment, 64% have identical and 85% have similar amino acids in all four proteins. The proteins of the Ras superfamily have five sequence regions (G1 to G5) contributing to the binding and hydrolysis of GTP (3). These regions are almost strictly conserved between the yeast, human, T. reesei, and A. niger var. awamori sequences. The region termed G2 is of particular interest, since it is conserved between Rab proteins that are functional in a given cellular location in different species but not between different Rab subclasses (17). The G2 region is strictly conserved in the alignment shown in Fig. 1. The four YPT/Rab sequences have the lowest degree of conservation at their C-terminal 50 amino acids, but even in this region the proteins from the two filamentous fungi are rather well conserved. The Rab proteins have an isoprenoid lipid modification attached to cysteines at the C terminus (16). Two cysteines that could be geranylgeranylated in the YPTI/YPTA proteins of T. reesei and A. niger var. awamori are found at their C termini (Fig. 1).
For verification of the identity of the ypt1 gene isolated from T. reesei, complementation of the corresponding yeast gene was attempted. The Trichoderma ypt1 cDNA was expressed from a multicopy plasmid with a PGK1 promoter in a yeast strain in which the only functional copy of the native YPT1 gene is under control of the GAL1 promoter. This strain is unable to grow on glucose, on which the promoter is repressed and YPT1 is not expressed. Expression of the Trichoderma YPTI protein supported growth of the strain on glucose (Fig. 2), showing that it can functionally complement the yeast YPT1 gene. Complementation was also attempted with temperature-sensitive ypt1 and sec4 mutant yeast strains in a similar way. No difference in growth at the restrictive temperature was observed for transformants with the T. reesei ypt1 expression plasmid and those having the vector alone with either of the temperature-sensitive mutants.
FIG. 2.
Complementation of the yeast Ypt1p depletion mutant by T. reesei ypt1 cDNA. The yeast strain GFU1-6D, carrying the only functional copy of YPT1 under the control of the GAL1 promoter, was transformed with the T. reesei ypt1 expression plasmid pMS82 and the vector pAJ401, and transformants were tested for growth on glucose and galactose. The pMS82 transformants grew on glucose, indicating that complementation occurred.
Characterization of the T. reesei nsf1 and A. niger var. awamori nsfA genes.
A cDNA with a high level of homology to yeast SEC18 was isolated by heterologous hybridization from a T. reesei λZAP library. The isolated Trichoderma cDNA has an open reading frame encoding a protein of 839 amino acids. Based on the amino acid similarity of the gene product to yeast Sec18p and mammalian NSF, the isolated T. reesei gene was named nsf1. The cDNA is apparently full length, as a stop codon in the same reading frame as the translation initiation codon is present in the 5′ flanking region. The corresponding genomic gene, isolated from a cosmid library, has two introns (Fig. 3). The region sequenced from the nsf1 promoter has a putative TATA element (CATAA) at −229 from the translation start site.
FIG. 3.
Amino acid sequence alignment of the T. reesei NSFI (T. r.) and A. niger var. awamori NSFA (A. a.) proteins with yeast Sec18p and human NSF. Asterisks, identical positions; dots, similar positions. The domain borders are indicated. The regions involved in ATP binding and hydrolysis are boxed. Intron positions in the T. reesei and A. niger var. awamori genes are shown by filled and open arrows, respectively. The repeat sequences close to the N terminus of T. reesei NSFI are shown in bold.
The existence of a SEC18/NSF homolog gene in the A. niger var. awamori genome was confirmed by Southern blot analysis of genomic DNA, with the Trichoderma nsf1 cDNA as a probe. A single band was detected for several digestion reactions of the Aspergillus genomic DNA, suggesting that only one homolog exists in Aspergillus. The Aspergillus nsfA gene was then isolated from a genomic λ library. The coding region has one intron, the position of which is not conserved in the T. reesei nsf1 gene (Fig. 3).
The identity between the T. reesei and A. niger var. awamori NSFI/NSFA proteins is 59%, their identity with yeast Sec18p is about 50%, and their identity with human NSF is about 45%. The A. niger var. awamori nsfA sequence isolated in this work has one amino acid difference when compared to the corresponding A. niger sequence submitted to the EMBL database by B. Seiboth et al. (accession number Q9C455). The alignment of the four NSF-like proteins (Fig. 3) shows that their lengths are quite variable and that their differences are mainly in the N termini. The Aspergillus sequence is the shortest and the Trichoderma protein is the longest of the four. The Trichoderma NSFI protein has an N-terminal extension of 102 amino acids compared to A. niger var. awamori NSFA. Interestingly, in the middle of this extension there is a region with four repeats of the sequence GGYGG(A/S)PQ (Fig. 3). The biological significance of the extension and of the repeats remains to be determined.
The Sec18/NSF proteins consist of three functional domains. The N-terminal domain interacts with the SNARE and SNAP proteins (54). The ATP binding domain D1 hydrolyzes ATP, which is essential for the fusion activity of NSF. The ATP binding domain D2 is responsible for the hexamerization of Sec18/NSF (12, 18). Binding of ATP by D2 is essential for its activity, but this domain does not hydrolyze ATP. In the N-terminal domain, there is hardly any homology among all four sequences in the alignment shown in Fig. 3, but the domains from Trichoderma and Aspergillus show a relatively high conservation level. The D1 domain is by far the best conserved region of the proteins, and D2 shows moderate conservation. A wide range of different ATPases have conserved Walker A and B motifs involved in ATP binding. In the D1 domains of the NSFI/NSFA proteins from filamentous fungi, these motifs follow the general consensus, whereas in the D2 domains, they show more variation when compared with the yeast and human counterparts (Fig. 3).
Complementation of the yeast temperature-sensitive sec18 mutation by T. reesei nsf1 was attempted. The nsf1 cDNA was expressed from a yeast multicopy plasmid with the PGK1 promoter in a sec18-1 temperature-sensitive mutant strain. The transformants were first grown at a permissive temperature and subsequently assayed for growth at a restrictive temperature. No difference in growth was observed between strains expressing the nsf1 cDNA and transformants carrying the vector alone; thus, complementation did not occur.
Regulation of the T. reesei genes encoding secretory pathway components.
An unfolded protein response (UPR) (33) leads to up-regulation of ER folding factors when protein folding in the ER is impaired. It has been shown previously that the UPR also regulates a subset of genes involved in other functions of the secretory pathway in S. cerevisiae (47). The T. reesei pdi1 gene, encoding protein disulfide isomerase, is regulated by the UPR (38), and thus we were interested in studying whether the genes isolated in this study and the T. reesei sar1 gene, isolated previously (49), are under the same control. Two compounds that block secretion in T. reesei, DTT (32) and BFA (T. Pakula and M. Penttilä, unpublished data), were used to induce the UPR and possibly other stress responses originating from the secretion pathway.
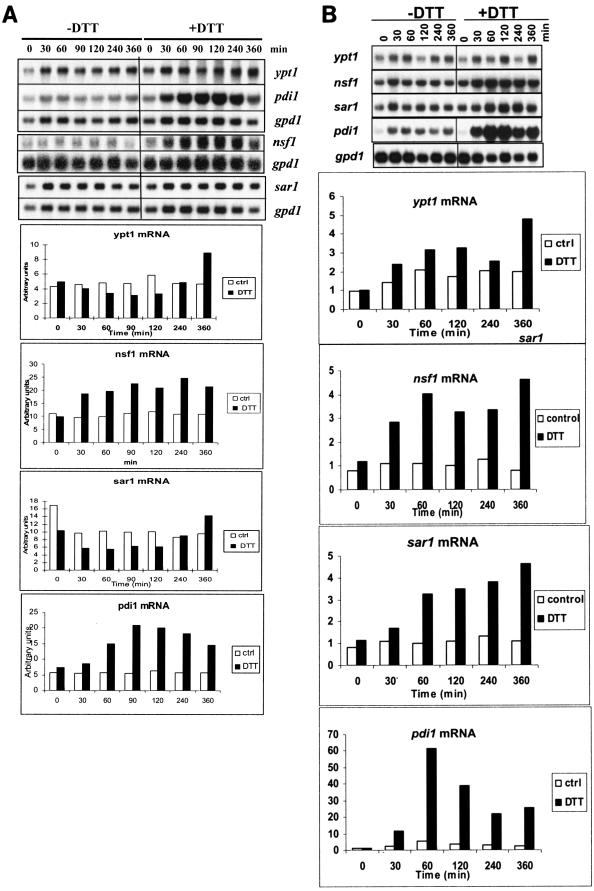
Northern hybridization done with T. reesei RutC-30 mycelia treated with DTT showed that the pdi1 mRNA level clearly increased 1 h after the addition of DTT to the culture (Fig. 4A). The mRNA levels of both ypt1 and sar1 remained constant for at least 4 h of the DTT treatment, suggesting that the UPR did not affect these genes. Interestingly, the nsf1 mRNA level increased about 2.5-fold during DTT treatment. This increase followed kinetics similar to those of pdi1 induction. To rule out the possibility that the increase of pdi1 and nsf1 mRNAs during DTT treatment was caused by some specific mutation incorporated into the strain during strain improvement by mutagenesis, a DTT treatment experiment with strain QM9414 was carried out (Fig. 4B). The pdi1 mRNA level increased dramatically after DTT addition, and the mRNAs of nsf1 and sar1 also became three- to fourfold more abundant. The ypt1 mRNA level was not significantly increased in this experiment. A possible reason for the stronger response in mRNA levels observed for this experiment than for the previous experiment (Fig. 4A) is that the DTT treatment was done in this case on a culture in which genes encoding cellulases were induced by sophorose. DTT was added to RutC-30 mycelia grown on lactose as the carbon source. It is known that sophorose is a much more potent inducer of the cellulase genes than lactose (15), and thus it can be expected that stronger secretion stress can be caused by DTT during sophorose induction than during growth on lactose.
FIG. 4.
Effect of DTT on ypt1, nsf1, sar1, and pdi1 mRNA levels in T. reesei RutC-30 (A) and QM9414 (B). Northern hybridization was carried out with total RNA isolated from mycelia treated with 10 mM DTT and from control mycelia. The time points after DTT addition are shown. The signal intensities were quantified with a phosphorimager and normalized with respect to gpd1 signals.
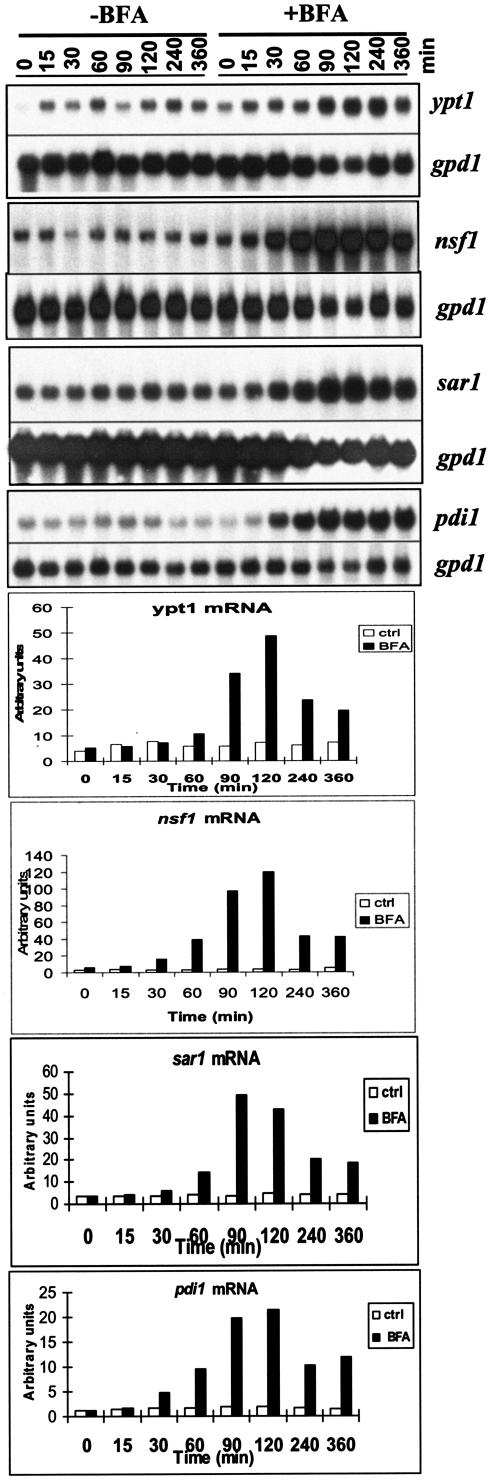
To further elucidate the mechanisms regulating the T. reesei secretory pathway, the effect of BFA, an inhibitor of intracellular membrane traffic (e.g., see references 35, 41, and 45), was tested. Northern hybridization of RNA from T. reesei mycelia treated with BFA showed that the mRNA levels of all the genes studied were strongly increased by BFA treatment (Fig. 5). The increase in mRNA levels was evident 1 h after BFA addition, and the maximal extent of induction was about 7-fold for ypt1, about 20-fold for nsf1, about 13-fold for sar1, and about 11-fold for pdi1.
FIG.5.
Effect of BFA on expression of T. reesei ypt1, nsf1, sar1, and pdi1 genes. Northern hybridization was carried out with total RNA derived from mycelia treated with BFA and from control mycelia. The gpd1 signal intensities were used to normalize the other signals.
DISCUSSION
In order to gather information on the secretory machinery of filamentous fungi, we isolated two genes from T. reesei and A. niger var. awamori, namely ypt1/yptA, involved in ER-Golgi transport, and nsf1/nsfA, involved in multiple membrane fusion steps. Interestingly, both ypt1 of T. reesei and yptA of A. niger var. awamori were identified as multicopy suppressors of the temperature-sensitive sso2 mutation in yeast. The Sso1 and Sso2 proteins are t-SNAREs residing in the plasma membrane, recognizing the v-SNARE Snc1/2 on the secretory vesicle, and giving specificity to the docking of the secretory vesicles on the plasma membrane (1). Since yeast Ypt1p is functional at the ER-Golgi and possibly at the cis-medial Golgi secretion stages (17), it was unexpected to find its close homologs as suppressors of the sso2 mutation. The explanation might be that a heterologous Ypt1p-like protein, heavily overexpressed from the strong PGK1 promoter in a multicopy plasmid, functions partially in the final stage of secretion as well, playing the role of Sec4p. Yeast Sec4p and Ypt1p are highly conserved, and it has been observed that when a nine-amino-acid segment of Ypt1p is replaced with the corresponding Sec4p sequence, the protein can fulfill the functions of both proteins (4, 9). Alternatively, the observed suppression of sso2 by the fungal ypt1/yptA cDNAs could be due to an indirect effect, e.g., the overexpression of the Ypt1p-like fungal protein could enhance the transport of a yeast secretory component that causes the actual suppression.
The identity of T. reesei ypt1 was verified in the experiment in which it clearly complemented a yeast Ypt1p depletion strain in which the only functional YPT1 gene is under the control of the GAL1 promoter (Fig. 2). No complementation, however, was detected with either the ypt1 or sec4 temperature-sensitive mutant. An explanation for this could be that in a temperature-sensitive mutant, the native protein is probably present in a misfolded state and can perhaps exclude the foreign protein from the necessary interactions even though it is unable to fulfill its task completely. Complementation of a temperature-sensitive sec18 mutant strain was attempted with the T. reesei nsf1 cDNA, but that experiment gave a negative result, perhaps for reasons similar to those causing the failure of the complementation of the yeast ypt1 temperature-sensitive mutation by T. reesei ypt1. It has been reported that the A. niger SEC4 homolog srgA is not able to complement the temperature-sensitive yeast sec4 mutant (36). Yeast SEC4 and SEC18 depletion strains were not available to us.
The production of extracellular proteins by T. reesei is strongly regulated at the transcriptional level according to the carbon source (15). We reported earlier that the protein disulfide isomerase gene pdi1 of T. reesei is also regulated according to the carbon source, largely in a manner similar to that for the cellulase genes (38). Thus, it would appear that T. reesei can adjust the level of the PDII protein according to the protein load in the secretory pathway. It has also been reported that the homolog of yeast SEC4 in the fungus C. lindemuthianum is regulated according to the available carbon source after expression of the secreted pectinase enzymes (8). We tested by Northern hybridization whether the T. reesei ypt1, nsf1, and sar1 genes are regulated by the carbon source similarly to pdi1, but clear differences in the expression levels between different carbon sources were not observed (data not shown). A similar observation has been made with A. niger, in which six genes encoding secretion-related small GTPases were not regulated according to the carbon source (36).
It has been reported that genes encoding components of the secretory pathway of the yeast S. cerevisiae are not regulated at the transcriptional level (50). More recently, however, it was found that this is not the case. The UPR reacts to accumulation of unfolded proteins in the ER (33), and foremost it regulates the genes involved in protein folding and quality control in the ER. In a transcriptional profiling study, researchers observed that the UPR affects 381 genes, including some genes involved in vesicle trafficking between the ER and the Golgi and between the Golgi and the plasma membrane, in vacuolar protein sorting, and in protein glycosylation (47). Our Northern hybridization data for T. reesei mycelia treated with DTT to induce the UPR indicate that for this fungus, also, a part of the secretory genes are under UPR control. The ypt1 and sar1 genes were not affected by DTT in the strain RutC-30, and nsf1 was induced by DTT (Fig. 4A). In strain QM9414, in which a stronger UPR induction apparently took place (based on the larger increase in pdi1 mRNA level), nsf1 and sar1 mRNAs responded to DTT but ypt1 did not (Fig. 4B). The T. reesei snc1 gene, encoding a v-SNARE protein involved in secretion vesicle fusion with the plasma membrane, has also been shown to be induced by DTT (M. Valkonen, M. Penttilä, and M. Saloheimo, submitted). In S. cerevisiae, SEC18, which is the T. reesei nsf1 homolog, and the yeast SNC1/2 genes are induced about twofold by the UPR pathway and the YPT1 and SAR1 genes are not induced (47; http://www.cell.com/cgi/content/full/101/3/249/DC1).
We have characterized the transcription factor responsible for UPR induction, HACI, from T. reesei (39). Our band shift experiments (unpublished results) with the HACI protein and putative UPR elements (UPRE) from the T. reesei pdi1 and bip1 (encodes the major ER chaperone [unpublished]) promoters have shown that the binding specificity of this transcription factor closely resembles the UPRE consensus sequence of yeast genes under UPR control (GNCAGNGTGNC) (25). Analysis of a 1.1-kb region of the T. reesei nsf1 promoter revealed a sequence closely resembling the UPRE sequences of the T. reesei pdi1 and bip1 and yeast promoters (ATCAGTCGTGAC; base pairs that are conserved with the consensus sequence are underlined). This is consistent with the fact that nsf1 was induced in cells treated with DTT.
In contrast to the DTT treatment results, all the T. reesei secretory genes studied for this work were strongly induced by BFA treatment (Fig. 5). The nsf1 and pdi1 genes, which were affected by both drugs, were much more strongly induced by BFA than by DTT. This, together with the fact that ypt1 and sar1 were only induced by BFA in strain RutC-30, indicates the possibility that BFA can induce a second regulatory pathway in addition to the UPR. DTT is an inhibitor of protein folding, and it induces the UPR by causing an accumulation of unfolded protein in the ER. BFA, on the other hand, is an inhibitor of membrane traffic between the ER and the Golgi complex and can cause disruption of the Golgi complex in mammalian cells (20). In the filamentous fungus Magnaporthe grisea, BFA appears to have somewhat different effects, as it blocks protein secretion and reduces the amount of apical vesicles but apparently does not disrupt the Golgi equivalents (2). Our in vivo labeling studies have shown that BFA causes a high intracellular accumulation of secreted proteins in T. reesei which is higher than that caused by DTT (T. Pakula and M. Penttilä, submitted). Therefore, it is possible that a regulation system reacting to a large amount of protein in the secretory pathway, perhaps in a later compartment than the ER, is responsible for the induction of the secretory genes in the presence of BFA. A regulatory system responding to the amount of folded protein in the secretory pathway and affecting the mammalian GRP78 gene encoding the ER chaperone Bip has been described previously (31). Another possible explanation for the results of our DTT and BFA treatment experiments is that the UPR pathway is responsible for the induction of secretory genes in both cases. The different genes could react with different intensities to the UPR, perhaps due to different affinities of the regulatory protein(s) for the promoters, and only at the highest level of protein accumulation in the secretory pathway during BFA treatment would they all respond to this regulatory pathway.
In conclusion, our results suggest that the secretory pathway of T. reesei is regulated at the transcriptional level. Our BFA treatment experiment indicates the possibility of an interesting novel stress response pathway from secretion to gene expression. This regulation could affect the secretory pathway as a whole, since all the secretory genes tested so far have been induced. The novel regulatory pathway would be induced by the accumulation of proteins, possibly folded ones, in the secretory pathway. Alternatively, it may be that all of the secretory genes studied can react to the UPR pathway under conditions of utmost secretion stress, in contrast to results obtained with S. cerevisiae, in which only a small subset of the secretory pathway genes have been suggested to respond to the UPR.
Acknowledgments
We thank Riitta Nurmi and Aili Grundström for excellent technical assistance. We are grateful to Sirkka Keränen for useful discussions and for providing yeast strains and to Randy Shekman, Peter Novick, and Hans Dieter Schmitt for yeast strains.
This work was supported by the Finnish National Technology Agency (Tekes) and the Academy of Finland in the research program “VTT Industrial Biotechnology” (Finnish Centre of Excellence Program 2000-2005; project 64330).
REFERENCES
- 1.Aalto, M. K., H. Ronne, and S. Keränen. 1993. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 12:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourett, T. M., and R. J. Howard. 1996. Brefeldin A-induced structural changes in the endomembrane system of a filamentous fungus, Magnaporthe grisea. Protoplasma 190:151-163. [Google Scholar]
- 3.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 4.Brennwald, P., and P. Novick. 1993. Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature 362:560-563. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., and R. Roxby. 1996. Characterisation of a Phytophthora infestans gene involved in vesicle transport. Gene 181:89-94. [DOI] [PubMed] [Google Scholar]
- 6.Colombo, M. I., S. C. Gelberman, S. W. Whiteheart, and P. D. Stahl. 1998. N-Ethylmaleimide-sensitive factor-dependent α-SNAP release, an early event in the docking/fusion process, is not regulated by Rab GTPases. J. Biol. Chem. 273:1334-1338. [DOI] [PubMed] [Google Scholar]
- 7.Conesa, A., P. J. Punt, N. van Luijk, and C. A. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 8.Dumas, B., C. Borel, C. Herbert, J. Maury, C. Jacquet, R. Balsse, and M.-T. Esquerre-Tugaye. 2001. Molecular characterization of CLPT1, a SEC4-like Rab/GTPase of the phytopathogenic fungus Colletotrichum lindemuthianum which is regulated by the carbon source. Gene 272:219-225. [DOI] [PubMed] [Google Scholar]
- 9.Dunn, B., T. Stearns, and D. Botstein. 1993. Specificity domains distinguish the Ras-related GTPases Ypt1 and Sec4. Nature 362:563-565. [DOI] [PubMed] [Google Scholar]
- 10.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas, A. 1998. NSF—fusion and beyond. Trends Cell Biol. 8:471-473. [DOI] [PubMed] [Google Scholar]
- 12.Hanson, P. I., R. Roth, H. Morisaki, R. Jahn, and J. E. Heuser. 1997. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch microscopy. Cell 90:523-535. [DOI] [PubMed] [Google Scholar]
- 13.Heintz, K., K. Palme, T. Diefenthal, and V. F. A. Russo. 1992. The Ncypt1 gene from Neurospora crassa is located on chromosome 2: molecular cloning and structural analysis. Mol. Gen. Genet. 235:413-421. [DOI] [PubMed] [Google Scholar]
- 14.Hynes, M. J., C. M. Corrick, and J. A. King. 1983. Isolation of genomic clones containing the amdS gene of Aspergillus nidulans and their use in the analysis of structural and regulatory mutations. Mol. Cell. Biol. 3:1430-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilmén, M., A. Saloheimo, M.-L. Onnela, and M. Penttilä. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, Y., G. Rossi, and S. Ferro-Novick. 1993. Bet2p and Mad2p are components of a prenyltransferase that adds geranylgeranyl onto Ypt1p and Sec4p. Nature 366:84-86. [DOI] [PubMed] [Google Scholar]
- 17.Lazar, T., M. Götte, and D. Gallwitz. 1997. Vesicular transport: how many Ypt/Rab-GTPases make a eukaryotic cell? Trends Biochem. Sci. 22:468-472. [DOI] [PubMed] [Google Scholar]
- 18.Lenzen, C. U., D. Steimann, S. W. Whiteheart, and W. I. Weis. 1998. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell 94:525-536. [DOI] [PubMed] [Google Scholar]
- 19.Lian, J. P., and S. Ferro-Novick. 1993. Bos1p, an integral membrane protein of the endoplasmic reticulum to Golgi transport vesicles, is required for their fusion competence. Cell 73:735-745. [DOI] [PubMed] [Google Scholar]
- 20.Lippincott-Schwartz, J., L. C. Yuan, J. S. Bonifacino, and R. D. Klausner. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56:801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupashin, V. V., and M. G. Waters. 1997. t-SNARE activation through transient interaction with a Rab-like guanosine triphosphatase. Science 276:1255-1258. [DOI] [PubMed] [Google Scholar]
- 22.Mäntylä, A., K. H. Rossi, S. Vanhanen, M. Penttilä, P. Suominen, and H. Nevalainen. 1992. Electrophoretic karyotyping of wild type and mutant Trichoderma longibrachiatum (reesei) strains. Curr. Genet. 21:471-477. [DOI] [PubMed] [Google Scholar]
- 23.Margolles-Clark, E., M. Ilmén, and M. Penttilä. 1997. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J. Biotechnol. 57:167-179. [Google Scholar]
- 24.Montenecourt, B. S., and D. E. Eveleigh. 1979. Selective screening methods for the isolation of high yielding cellulase mutants of Trichoderma reesei. Adv. Chem. Ser. 181:289-301. [Google Scholar]
- 25.Mori, K., N. Ogawa, T. Kawahara, H. Yanagi, and T. Yura. 1998. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 273:9912-9920. [DOI] [PubMed] [Google Scholar]
- 26.Morsomme, P., and H. Riezman. 2002. The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev. Cell 2:307-317. [DOI] [PubMed] [Google Scholar]
- 27.Neuwald, A. F. 1999. The hexamerization domain of N-ethylmaleimide sensitive factor: structural clues to chaperone function. Structure 7:19-23. [DOI] [PubMed] [Google Scholar]
- 28.Ngiam, C., D. J. Jeenes, P. J. Punt, C. A. M. J. J. van den Hondel, and D. B. Archer. 2000. Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl. Environ. Microbiol. 66:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nichols, B. J., C. Ungermann, H. R. B. Pelham, W. T. Wickner, and A. Haas. 1997. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature 387:199-202. [DOI] [PubMed] [Google Scholar]
- 30.Nishimune, A., J. T. Isaac, E. Molnar, J. Noel, S. R. Nash, M. Tagaya, G. L. Collingridge, S. Nakanishi, and J. M. Henley. 1998. NSF binding to GluR2 regulates synaptic transmission. Neuron 21:87-97. [DOI] [PubMed] [Google Scholar]
- 31.Pahl, H. L., and P. A. Bäuerle. 1995. A novel signal transduction pathway from the endoplasmic reticulum to the nucleus is mediated by transcription factor NF-κB. EMBO J. 14:2580-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pakula, T. M., J. Uusitalo, M. Saloheimo, K. Salonen, R. J. Aarts, and M. Penttilä. 2000. Monitoring the kinetics of glycoprotein synthesis and secretion in the filamentous fungus Trichoderma reesei: cellobiohydrolase I (CBHI) as a model protein. Microbiology 146:223-232. [DOI] [PubMed] [Google Scholar]
- 33.Patil, C., and P. Walther. 2001. Intracellular signalling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349-356. [DOI] [PubMed] [Google Scholar]
- 34.Penttilä, M., H. Nevalainen, M. Rättö, E. Salminen, and J. K. C. Knowles. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155-164. [DOI] [PubMed] [Google Scholar]
- 35.Peyroche, A., B. Antonny, S. Robineau, J. Acker, J. Cherfils, and C. L. Jackson. 1999. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3:275-285. [DOI] [PubMed] [Google Scholar]
- 36.Punt, P. J., B. Seiboth, X. O. Weenink, C. van Zeijl, M. Lenders, C. Konetschny, A. F. J. Ram, and C. A. M. J. J. van den Hondel. 2001. Identification and characterisation of a family of secretion-related small GTPase-encoding genes from the filamentous fungus Aspergillus niger: a putative SEC4 homologue is not essential for growth. Mol. Microbiol. 41:513-525. [DOI] [PubMed] [Google Scholar]
- 37.Rothman, J. E., and T. H. Söllner. 1997. Throttles and dampers: controlling the engine of membrane fusion. Science 276:1212-1213. [DOI] [PubMed] [Google Scholar]
- 38.Saloheimo, M., M. Lund, and M. Penttilä. 1999. The protein disulphide isomerase gene of the fungus Trichoderma reesei is induced by endoplasmic reticulum stress and regulated by the carbon source. Mol. Gen. Genet. 262:35-45. [DOI] [PubMed] [Google Scholar]
- 39.Saloheimo, M., M. Valkonen, and M. Penttilä. Induction mechanisms of the HACI-mediated unfolded protein response in filamentous fungi. Mol. Microbiol., in press. [DOI] [PubMed]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd. ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Satiat-Jeunemaitre, B., L. Cole, C. T. Bourett, R. Howard, and C. Hawes. 1996. Brefeldin A effects in plant and fungal cells: something new about vesicle trafficking? J. Microsc. 181:162-177. [DOI] [PubMed] [Google Scholar]
- 42.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 43.Sogaard, M., K. Tani, R. R. Ye, S. Geromanos, P. Tempst, T. Kirchhausen, J. E. Rothman, and T. Söllner. 1994. A Rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell 78:937-948. [DOI] [PubMed] [Google Scholar]
- 44.Söllner, T., M. K. Bennett, S. W. Whiteheart, R. H. Scheller, and J. E. Rothman. 1993. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation and fusion. Cell 75:409-418. [DOI] [PubMed] [Google Scholar]
- 45.Spanfo, S., M. G. Silletta, A. Colanzi, S. Alberti, G. Fiucci, C. Valente, A. Fusella, M. Salmonas, A. Mironov, A. Luini, and D. Corda. 1999. Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. J. Biol. Chem. 274:17705-17710. [DOI] [PubMed] [Google Scholar]
- 46.Stålbrand, H., A. Saloheimo, J. Vehmaanperä, B. Henrissat, and M. Penttilä. 1995. Cloning and expression in Saccharomyces cerevisiae of a Trichoderma reesei β-mannanase gene containing a cellulose binding domain. Appl. Environ. Microbiol. 61:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walther. 2000. Functional and genomic analyses reveal an essential co-ordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 48.van Gemeren, I. A., P. J. Punt, A. Drint-Kuyvenhoven, M. P. Broekhuijsen, A. van't Hoog, A. Beijersbergen, C. T. Verrips, and C. A. M. J. J. van den Hondel. 1997. The ER chaperone encoding bipA gene of the black Aspergilli is induced by heat shock and unfolded proteins. Gene 198:43-52. [DOI] [PubMed] [Google Scholar]
- 49.Veldhuisen, G., M. Saloheimo, M. A. Fiers, P. J. Punt, R. Contreras, M. Penttilä, and C. A. M. J. J. van den Hondel. 1997. Isolation and analysis of functional homologues of the secretion-related SAR1 gene of Saccharomyces cerevisiae from Aspergillus niger and Trichoderma reesei. Mol. Gen. Genet. 256:446-455. [DOI] [PubMed] [Google Scholar]
- 50.Wahlensieck, Y., H. Riezman, and B. Meyhack. 1995. Transcriptional studies on yeast SEC genes provide no evidence for regulation at the transcriptional level. Yeast 11:901-911. [DOI] [PubMed] [Google Scholar]
- 51.Ward, M., L. J. Wilson, and K. H. Kodama. 1993. Use of Aspergillus overproducing mutants, cured for integrated plasmid, to overproduce heterologous proteins. Appl. Microbiol. Biotechnol. 39:738-743. [DOI] [PubMed] [Google Scholar]
- 52.Weber, T., B. V. Zemelman, J. A. McNew, B. Westerman, M. Gmalch, F. Parlati, T. H. Söllner, and J. E. Rothman. 1998. SNAREpins: minimal machinery for membrane fusion. Cell 92:759-772. [DOI] [PubMed] [Google Scholar]
- 53.Whiteheart, S. W., M. Brunner, D. W. Wilson, M. Wiedman, and J. E. Rothman. 1992. Soluble N-ethylmaleimide-sensitive fusion attachment proteins (SNAPs) bind to a multi-SNAP receptor complex in Golgi membranes. J. Biol. Chem. 267:12239-12243. [PubMed] [Google Scholar]
- 54.Whiteheart, S. W., K. Rossnagel, S. A. Buhrow, M. Brunner, R. Jaenicke, and J. E. Rothman. 1994. N-Ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol. 126:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whiteheart, S. W., and E. W. Kubalek. 1995. SNAPs and NSF: general members of the fusion apparatus. Trends Cell Biol. 5:64-68. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, D. W., C. A. Wilcox, G. C. Flynn, E. Chen, W. J. Kuang, W. J. Henzel, M. R. Block, A. Ullrich, and J. E. Rothman. 1989. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature 339:355-359. [DOI] [PubMed] [Google Scholar]