Abstract
Isolation and culture of both embryonic and tissue specific stem cells provide an enormous opportunity to study the molecular processes driving development. To gain insight into the initial events underpinning mammalian embryogenesis, pluripotent stem cells from each of the three distinct lineages present within the preimplantation blastocyst have been derived. Embryonic (ES), trophectoderm (TS) and extraembryonic endoderm (XEN) stem cells possess the developmental potential of their founding lineages and seemingly utilize distinct epigenetic modalities to program gene expression. However, the basis for these differing cellular identities and epigenetic properties remain poorly defined.
Quantitative reverse transcription-polymerase chain reaction (qPCR) is a powerful and efficient means of rapidly comparing patterns of gene expression between different developmental stages and experimental conditions. However, careful, empirical selection of appropriate reference genes is essential to accurately measuring transcriptional differences. Here we report the quantitation and evaluation of fourteen commonly used references genes between ES, TS and XEN stem cells. These included: Actb, B2m, Hsp70, Gapdh, Gusb, H2afz, Hk2, Hprt, Pgk1, Ppia, Rn7sk, Sdha, Tbp and Ywhaz. Utilizing three independent statistical analysis, we identify Pgk1, Sdha and Tbp as the most stable reference genes between each of these stem cell types. Furthermore, we identify Sdha, Tbp and Ywhaz as well as Ywhaz, Pgk1 and Hk2 as the three most stable reference genes through the in vitro differentiation of embryonic and trophectoderm stem cells respectively.
Understanding the transcriptional and epigenetic regulatory mechanisms controlling cellular identity within these distinct stem cell types provides essential insight into cellular processes controlling both embryogenesis and stem cell biology. Normalizing quantitative RT-PCR measurements using the geometric mean CT values obtained for the identified mRNAs, offers a reliable method to assess differing patterns of gene expression between the three founding stem cell lineages present within the mammalian preimplantation embryo.
Introduction
During mammalian pre-implantation development a series of asynchronous divisions result in the formation of the blastocyst. At this stage of development three distinct cell types have emerged: the epiblast, trophectoderm and primitive endoderm, which give rise to the fetus, placenta and extraembryonic endoderm respectively [1]–[3]. To better define the developmental and transcriptional processes unique to each of these distinct lineages, in vitro cultured progenitor stem cells have been derived [4]–[7]. Analysis of ES, TS and XEN stem cell lines have revealed much about the cellular processes controlling mammalian development and demonstrated surprising differences in the epigenetic regulation of gene expression between these three lineages [7]–[12]. Identifying the biochemical factors underlying these differences remains an essential step to understanding the molecular processes driving development and better defining crucial aspects of mammalian stem cell biology.
Quantitative reverse transcription-polymerase chain reaction (qPCR) has emerged as a powerful technique to rapidly assess transcriptional differences between cell types and differing experimental conditions. However, accurate quantitative analysis is dependent upon proper, empirical selection of a suitable reference. Using published microarray data, and a novel statistical algorithm, (geNORM) Vandesompele and colleagues demonstrated that the geometric mean of three reference genes provided the most accurate and reliable means of normalizing qPCR expression data [13]. Subsequently, this experimental strategy has been validated and additional algorithms written and utilized to identify the most suitable reference genes for a variety of experimental conditions [14]–[25].
In this study we sought to identify a list of genes most suitable for use as normalization controls in qPCR-based comparisons between ES, TS and XEN stem cells or their in vitro differentiated progeny. In order to help identify candidate genes we set two main criteria that the mRNAs would have to fulfill: 1) the transcripts needed to be expressed above background and easily detectable, and 2) candidate mRNAs needed to be stably expressed between each of the three stem cell lineages under investigation. To this end we surveyed the recent literature and compiled a short list of fourteen candidate genes, including Actb, B2m, Hsp70, Gapdh, Gusb, H2afz, Hk2, Hprt, Pgk1, Ppia, Rn7sK, Sdha, Tbp and Ywhaz [14]–[28]. These genes belong to diverse functional classes and should not be co-regulated, thus providing a non-biased method of normalizing qPCR expression data.
To evaluate the stability of our candidate genes we isolated RNA from three independent lines of varying genotypes for each of the three stem cell types. This RNA was quantified and seeded into five independent qPCR reactions measuring each of the candidate genes. Using the geNORM, NormFinder and BestKeeper algorithms, we identify the Pgk1, Sdha and Tbp transcripts as the most stably expressed reference genes between each of these stem cell types. To determine which of these candidates was most suitable for use during in vitro differentiation studies, we cultured ES and TS cells in the absence of crucial growth factors LIF and FGF4 respectively. Using three independent RNA samples isolated on Day 0 and Day 8, we identify Sdha, Tbp and Ywhaz as well as Ywhaz, Pgk1 and Hk2 as the three most stable reference genes through the in vitro differentiation of ES and TS cells. Our results suggest that normalization of qPCR data using the geometric means of the transcripts listed above will yield the most accurate quantification of gene expression between these three unique stem cell types.
Results
After a survey of the recent literature we curated a short list of fourteen commonly used reference genes and either designed new primers or pulled existing ones from references cited in the materials and methods. These genes are listed in Table 1 and represent several distinct functional classes so as to minimize the possibility of co-regulation. For each gene, a minimum of two independent primer sets were tested and of these, the primer set exhibiting the greatest efficiency was selected. To conduct an accurate survey of candidate gene expression levels between ES, TS and XEN stem cells we isolated RNA from at least three independent stem cell lines, representing at least two different genotypes. We postulate that utilizing lines derived from diverse genotypes will more accurately identify stable reference genes to be used in future studies contrasting patterns of gene expression.
Table 1. Descriptions of the fourteen candidate reference genes studied.
| Symbol | Name | Accession | Brief Description |
| Rn7sk | 7SK, small nuclear RNA | NR_030687 | Small nuclear RNA that binds elongation factor P-TEFb and negatively regulates transcription. |
| Actb | Beta-Actin | NM_007393 | A highly conserved protein found in all eukaryotic cells involved in various cellular processes such as cell motility and cytokinesis. |
| B2m | beta-2 microglobulin | NM_009735 | Gene that codes for Beta-2-microglobulin, a component of the MHC |
| Gapdh | glyceraldehyde-3-phosphate dehydrogenase | NM_008084 | Enzyme involved in metabolic and non-metabolic processes such as glycolysis, transcription activation, and apoptosis. |
| Gusb | glucuronidase, beta | NM_010368 | Gene that codes for Beta-glucuronidase, which catalyzes the hydrolysis of B-D-glucuronic acid. |
| H2afz | H2A histone family, member Z | NM_016750 | Member of the H2A histone family that is required for embryonic development. |
| Hk2 | hexokinase 2 | NM_013820 | Enzyme that phosphorylates hexoses, including glucose to produce glucose-6-phosphate. |
| Hprt | hypoxanthine-phosphoribosyl transferase | NM_013556 | Transferase that aids in the generation of new purine nucleotides from degraded DNA. |
| Hsp70 | heat shock 70kD protein | NM_010478 | Heat shock protein that aids in protein folding and cellular stress response. |
| Pgk1 | phosphoglycerate kinase 1 | NM_008828 | A highly conserved transferase involved in glycolysis that catalyzes the formation of ATP. |
| Ppia | peptidylprolyl isomerase A | NM_008907 | Gene that codes for peptidylprolyl isomerase A, a protein that catalyzes the folding of proteins. |
| Sdha | Succinate dehydrogenase complex, subunit A | BC011301 | Gene that codes for a subunit of succinate dehydrogenase and is important in cellular respiration. |
| Tbp | TATA box binding protein | NM_013684 | Protein that binds to the TATA box sequence and aids in transcription initiation. |
| Ywhaz | Tyrosine 3-monooxygenase /tryptophan 5-monooxygenase activation protein, zeta polypeptide | NM_011740 | Codes for a highly conserved protein that helps mediate signal transduction. |
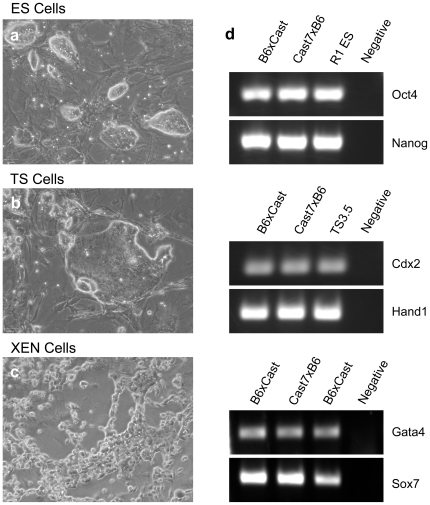
Previous studies in our laboratory have utilized stem cell lines derived from Mus musculus castaneus x mus musculus (C57Black6) F1 embryos. Polymorphisms between these genetic strains allow the examination of mono-allelic patterns of epigenetic marks and gene expression within loci regulated by genomic imprinting [29]. For ES, TS and XEN stem cell analysis we utilized lines derived from F1 embryos of reciprocal crosses between these stains (C57Black6×Castaneous and Cast7×Black6) [11], [29], [30]. For analysis of ES and TS cells we also utilized the previously described R1 ES and TS3.5 lines derived from 129 stain mice [31], [32]. Each of these different lines demonstrated cellular morphology consistent with their cell type and expressed unique cohorts of transcription factors characteristic of their lineage [7], [33] (Figure 1). Cell lines were cultured to 80% confluence, RNA isolated and seeded into five independent qRT-PCR reactions measuring our fourteen candidate genes. Results presented below are the combined analysis of all genetic backgrounds tested.
Figure 1. Characteristic cellular morphology and marker gene expression for ES, TS and XEN Stem Cells.
a–c Light micrographs of representative ES (a) TS (b) and XEN (c) stem cell lines used in this study. d) Expression of transcription factors characteristic of each of the stem cell lineages.
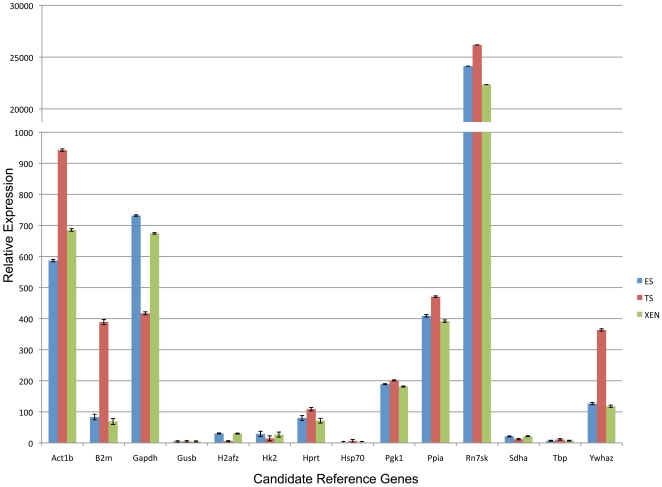
Of the candidate genes tested Rn7sk demonstrated the most robust expression averaging expression levels 125 fold higher than the remaining candidates; which were all readily detectable in each of the cell lines tested. To measure the relative stability of each of the candidate genes between the ES, TS and XEN lines, the CT values for the measured transcripts were compiled and run through the NormFinder, GENorm, and BestKeeper software packages [13]–[15]. Each of these algorithms utilize slightly different methods of estimating both the intra- and the intergroup expression variation, and allow the ranking of candidate genes based on the calculation of a “stability value”. While there was variation amongst the mid-range to least stable genes, all three software packages identified Pgk1, Sdha, Tbp and H2afz as the most consistently stable reference genes between ES, TS and XEN stem cells (Table 2). Similar to previous studies by Mamo et al., we observed the classic “housekeeping genes” Actb, Hprt and to a lesser extent Gapdh were comparatively unstable and by our analysis would not be the best choice to normalize qPCR expression levels [19].
Table 2. Candidate reference genes ranked in order of their stability.
| NormFinder | geNORM | BestKeeper | |||||||
| Rank | Gene Name | Stability Value | Rank | Gene Name | Stability Value | Rank | Gene Name | ||
| 1 | Pgk1 | 0.012 | 1 | Pgk1 | 0.121 | 1 | Pgk1 | ||
| 2 | Sdha | 0.047 | 2 | Sdha | 0.132 | 2 | Sdha | ||
| 3 | Tbp | 0.054 | 3 | Tbp | 0.138 | 3 | H2afz | ||
| 4 | H2afz | 0.057 | 4 | H2afz | 0.139 | 4 | Tbp | ||
| 5 | Gapdh | 0.061 | 5 | Gusb | 0.142 | 5 | Gusb | ||
| 6 | Ppia | 0.062 | 6 | Gapdh | 0.145 | 6 | Ppia | ||
| 7 | Gusb | 0.062 | 7 | Ppia | 0.145 | 7 | Gapdh | ||
| 8 | Hsp70 | 0.088 | 8 | Hsp70 | 0.168 | 8 | Ywhaz | ||
| 9 | Ywhaz | 0.088 | 9 | Ywhaz | 0.168 | 9 | Hsp70 | ||
| 10 | Actb | 0.099 | 10 | Actb | 0.182 | 10 | Actb | ||
| 11 | B2m | 0.107 | 11 | Hprt | 0.192 | 11 | Hprt | ||
| 12 | Hk2 | 0.109 | 12 | B2m | 0.192 | 12 | Hk2 | ||
| 13 | Hprt | 0.109 | 13 | Hk2 | 0.193 | 13 | B2m | ||
| 14 | Rn7sk | 0.128 | 14 | Rn7sk | 0.212 | 14 | Rn7sk | ||
a) Stability of the candidate genes between ES, TS and XEN stem cells ranked using the NormFinder, GENorm and BestKeepr software tools.
We next chose to make pair-wise comparisons between ES and TS, ES and XEN as well as TS and XEN to see which candidates emerged as the most stable in contrasts between any two cell types. A consensus of all three software packages can be seen in table 3. As with the comparisons between all three lines, Pgk1, Sdha, Tbp and H2afz remained in the top five most stable genes indicating no one cell type was biasing our analysis and that these five reference genes represent the best normalization controls for qPCR-based analysis of gene expression. Utilizing the geometric mean of Pgk1, Sdha, and Tbp we normalized the CT values for each of the fourteen candidates and graphed their relative expression levels as described previously [13], [34], [35]. As can bee seen in Figure 2, Rn7sk is expressed at a drastically higher level than any of the other candidates tested and therefore does not represent a viable reference gene. Similarly, analysis of Actb, B2m, Gapdh and Ywhaz all yielded significant differences in measurements of TS cell expression as compared to both ES and XEN cells eliminating their candidacy. Our results indicate normalizing quantitative RT-PCR measurements using the geometric mean CT values obtained for the Pgk1, Sdha and Tbp mRNAs, offers the most reliable method to assess differing patterns of gene expression between the three founding stem cell lineages present within the mammalian preimplantation embryo.
Table 3. Consensus of the stability rankings for pair-wise comparisons between the stem cell types.
| ES vs. TS | ES vs. XEN | TS vs. XEN | |||||||
| Rank | Gene Name | Rank | Gene Name | Rank | Gene Name | ||||
| 1 | Pgk1 | 1 | Pgk1 | 1 | Pgk1 | ||||
| 2 | Sdha | 2 | Gapdh | 2 | Sdha | ||||
| 3 | Tbp | 3 | H2afz | 3 | H2afz | ||||
| 4 | Gusb | 4 | Tbp | 4 | Tbp | ||||
| 5 | H2afz | 5 | Sdha | 5 | Ppia | ||||
| 6 | Ppia | 6 | Gusb | 6 | Gusb | ||||
| 7 | Gapdh | 7 | Ppia | 7 | Gapdh | ||||
| 8 | Ywhaz | 8 | Hsp70 | 8 | Ywhaz | ||||
| 9 | Hsp70 | 9 | Ywhaz | 9 | Hsp70 | ||||
| 10 | Actb | 10 | Actb | 10 | Actb | ||||
| 11 | Hprt | 11 | B2m | 11 | B2m | ||||
| 12 | Hk2 | 12 | Rn7sk | 12 | Hprt | ||||
| 13 | B2m | 13 | Hk2 | 13 | Hk2 | ||||
| 14 | Rn7sk | 14 | Hprt | 14 | Rn7sk |
Figure 2. Relative expression of the fourteen candidate genes between all three genetic backgrounds of ES, TS and XEN stem cells.
CT values for each measured transcript were normalized to the geometric mean of Pgk1, Sdha and Tbp, and then graphed as relative values using methods described [13], [34], [35]. Error bars represent the standard error of the mean.
We next sought to determine which of the candidate genes remained the most stable throughout the process of differentiation. Therefore we chose to differentiate our ES and TS cell lines by removal of the key growth factors LIF and FGF4 respectively [6], [36], [37]. To this end ES cells were cultured in LIF - ES cell medium, allowed to form embryoid bodies on untreated plastic dishes and then plated on regular tissue culture plastic to differentiate into fibroblast like cells. Similarly, TS cells were plated on tissue culture treated plastic at low density in FGF4- medium which promoted the formation of TS giant-like cells. We chose not to investigate the process of XEN cell differentiation as reliable protocols for the induction of differentiation have not yet been established. In contrast to both ES and TS cell lines, when XEN cells are plated on plastic many cells simply senesce, while the remainder do not uniformly differentiate into one cell type, thus complicating our analysis.
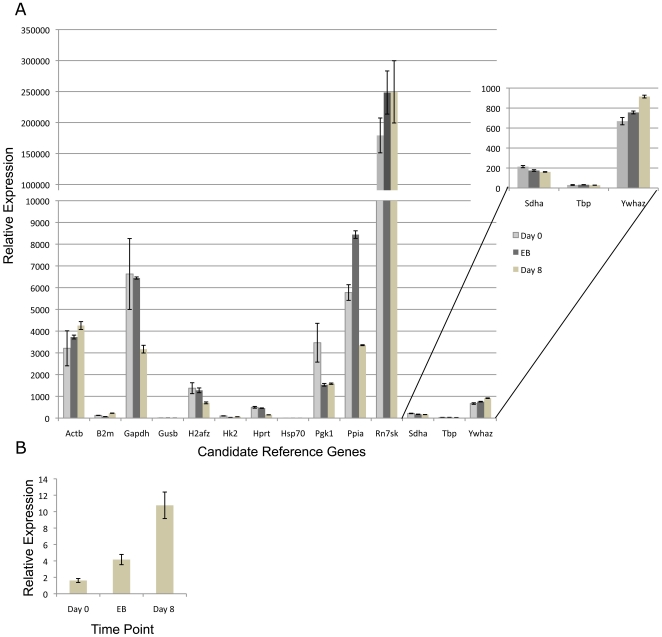
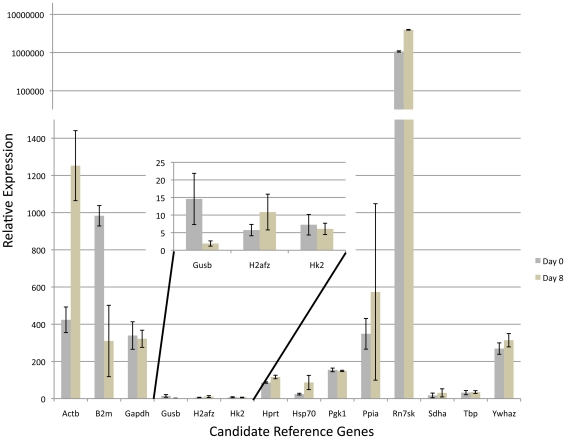
RNA samples were collected from ES cells on Day 0, Day 4 (embryoid body) and Day 8 and RNA seeded into five independent qPCR reactions measuring each of the fourteen candidate genes. Using a similar experimental design as described above, we identify Sdha, Tbp and Ywhaz as the three most stable transcripts (Table 4). To examine relative changes in gene expression, we utilized the geometric mean of these three most stable candidates to normalize CT values and graphed the relative expression of all fourteen candidate genes though ES cell differentiation (Figure 3a). We then chose to examine the expression of the cell lineage marker fibroblast-specific protein-1 (FSP-1) which is active in fibroblasts but not in epithelium, mesangial cells or embryonic endoderm [38]. In accordance with previous studies this maker demonstrated increasing expression in differentiating cell cultures, indicating our three candidate genes provided a valid reference point (Figure 3b) [39], [40]. In contrast, transcripts encoding Pgk1, H2afz, Ppia (Cyclophillin) and Gapdh all demonstrate a significant down-regulation and therefore are not suitable reference genes for this experimental time course. Similar to results reported by Willems et al., examining ES cell differentiation induced by both DMSO and Retinoic acid, we also identify B2m and Hprt as among the most unstable transcripts [18]. Using similar methodologies, we identified the Ywhaz, Pgk1 and Hk2 transcripts as the most stable during TS cell differentiation (Table 5). After applying the geometric mean of these three candidates to normalize CT values we observed massive changes in transcripts encoding Actb, B2m and Rn7sk (Figure 4). Previous studies have identified increased actin mobilization as a key feature of trophectoderm stem cell differentiation, validating our identified reference genes [41]. Taken together our data indicate Sdha, Tbp and Ywhaz and Ywhaz, Pgk1 and Hk2 represent the most stable of our fourteen candidate reference genes for use as qPCR normalization controls during ES and TS cell differentiation respectively.
Table 4. Candidate reference genes ranked in order of their stability throughout ES cell differentiation using the NormFinder, GENorm and BestKeepr software tools.
| NormFinder | geNORM | BestKeeper | |||||||
| Rank | Gene Name | Stability Value | Rank | Gene Name | Stability Value | Rank | Gene Name | ||
| 1 | Sdha | 0.033 | 1 | Sdha | 0.005 | 1 | Sdha | ||
| 2 | Tbp | 0.033 | 2 | Tbp | 0.005 | 2 | Tbp | ||
| 3 | Ywhaz | 0.038 | 3 | Ywhaz | 0.015 | 3 | Ywhaz | ||
| 4 | H2afz | 0.039 | 4 | Gusb | 0.017 | 4 | Gusb | ||
| 5 | Gusb | 0.039 | 5 | Actb | 0.017 | 5 | H2afz | ||
| 6 | Actb | 0.04 | 6 | H2afz | 0.017 | 6 | Hsp70 | ||
| 7 | Hsp70 | 0.04 | 7 | Hsp70 | 0.018 | 7 | Actb | ||
| 8 | Gapdh | 0.045 | 8 | Gapdh | 0.024 | 8 | Gapdh | ||
| 9 | Ppia | 0.047 | 9 | Ppia | 0.026 | 9 | Ppia | ||
| 10 | Hk2 | 0.05 | 10 | Hk2 | 0.027 | 10 | Hk2 | ||
| 11 | Pgk1 | 0.051 | 11 | Pgk1 | 0.028 | 11 | Hprt | ||
| 12 | Hprt | 0.054 | 12 | Hprt | 0.033 | 12 | Pgk1 | ||
| 13 | Rn7sk | 0.057 | 13 | Rn7sk | 0.034 | 13 | Rn7sk | ||
| 14 | B2m | 0.058 | 14 | B2m | 0.036 | 14 | B2m | ||
Figure 3. A. Relative expression of the fourteen candidate genes throughout differentiation of all three genetic backgrounds of ES cells examined.
CT values for each transcript were measure on Day 0, Day 4 (embryoid body) and Day 8 and were then normalized to the geometric mean of Sdha Tbp and Ywhaz. Relative values were determined using methods described previously [13], [34], [35] and graphed. Error bars represent the standard error of the mean. B. Increased expression of fibroblast specific protein 1 throughout ES cell differentiation. Error bars represent the standard error of the mean for three independent replicates.
Table 5. Candidate reference genes ranked in order of their stability throughout TS cell differentiation using the NormFinder, GENorm and BestKeepr software tools.
| NormFinder | geNORM | BestKeeper | |||||||
| Rank | Gene Name | Stability Value | Rank | Gene Name | Stability Value | Rank | Gene Name | ||
| 1 | Ywhaz | 0.095 | 1 | Ywhaz | 0.018 | 1 | Ywhaz | ||
| 2 | Pgk1 | 0.096 | 2 | Pgk1 | 0.029 | 2 | Pgk1 | ||
| 3 | H2afz | 0.102 | 3 | Hk2 | 0.036 | 3 | Hk2 | ||
| 4 | Hk2 | 0.104 | 4 | H2afz | 0.04 | 4 | H2afz | ||
| 5 | Hprt | 0.11 | 5 | Hprt | 0.042 | 5 | Hprt | ||
| 6 | Tbp | 0.111 | 6 | Tbp | 0.044 | 6 | Tbp | ||
| 7 | Actb | 0.112 | 7 | Actb | 0.044 | 7 | Sdha | ||
| 8 | Sdha | 0.122 | 8 | Ppia | 0.048 | 8 | Actb | ||
| 9 | Hsp70 | 0.127 | 9 | Sdha | 0.06 | 9 | Ppia | ||
| 10 | Ppia | 0.128 | 10 | Hsp70 | 0.061 | 10 | Hsp70 | ||
| 11 | Gusb | 0.133 | 11 | Gusb | 0.065 | 11 | Gusb | ||
| 12 | Gapdh | 0.153 | 12 | Gapdh | 0.093 | 12 | Gapdh | ||
| 13 | B2m | 0.183 | 13 | B2m | 0.12 | 13 | B2m | ||
| 14 | Rn7sk | 0.202 | 14 | Rn7sk | 0.24 | 14 | Rn7sk | ||
Figure 4. Relative expression of the fourteen candidate genes throughout differentiation of all three genetic backgrounds of TS cells.
CT values for each transcript were measured on Day 0 and Day 8 and were then normalized to the geometric mean of Ywhaz, Pgk1 and Hk2. Relative values were determined using methods described previously [13], [34], [35] and graphed. Error bars represent the standard error of the mean. Note that the top third of the graph is in an exponential scale.
Discussion
Analysis of gene expression using qPCR has become the corner stone to nearly every facet of the biological sciences. However, despite numerous studies demonstrating the importance of careful selection and validation of appropriate reference genes, several studies continue to emerge utilizing inappropriate methods of qPCR normalization [13]–[15], [18], [20], [21], [42]. A recent survey of the literature identified the single use of either Actb or Gapdh to normalize expression data in the vast majority of qPCR based studies without any form of validation to ensure their experimental stability [13]. In this study we sought to identify the most stable and appropriate reference genes for studies contrasting patterns of gene expression between the three founding stem cell lineages present within the mammalian preimplantation embryo. From a list of fourteen commonly utilized reference genes we identify Pgk1, Sdha and Tbp as the most suitable reference genes and further find compelling evidence to suggest that both Actb and Gapdh are not suitable normalization controls for these experiments.
Of the top three candidates to emerge from our analysis two are components of pathways controlling cellular respiration. Pgk1 - phosphoglycerate kinase 1 is the seventh step of glycolysis and Sdha - Succinate dehydrogenase or succinate-coenzyme Q reductase is an enzyme complex that binds to the inner mitochondrial membrane and is an essential component of both the citric acid cycle and electron transport chain [43], [44]. One potential weakness of our top three candidates is that although Pgk1 and Sdha are components of distinct pathways, they are both components of cellular respiration leaving the possibility that an experimental condition that impacts metabolic processes would significantly alter these normalization controls. The third and fourth candidates to emerge from our analysis were Tbp and H2afz respectively. Tbp is a central component of the RNA polymerase II pre-initiation complex and H2afz is an essential component of chromatin structure which is hypothesized to play a role in chromosome organization and stability [45]–[48]. The third and fourth candidates are truly functionally distinct from both each-other and from pathways controlling cellular respiration. As such, where experimental design permits we would recommend normalizing CT values to the geometric mean of all four of these reference genes to improve experimental rigor. However, when we incorporated this strategy we did not observe any meaningful changes in relative gene expression (data not shown).
The first differentiation event during mammalian embryogenesis is the formation of the epiblast, trophectoderm and primitive endoderm which go on to give rise to the three founding embryonic lineages. Stem cells derived from each of these lineages represent an excellent model system to study mammalian development and understand crucial aspects of stem cell biology necessary in developing regenerative therapies. Analysis of gene expression using qPCR will undoubtedly play a pivotal role in deciphering the cellular and molecular properties that define these different cell types. In these analysis, the identification of stable reference genes is an essential prerequisite to accurately interpreting experimental data. Using three independent, highly referenced and validated statistical methods, our analysis of fourteen potential candidate reference genes identify Pgk1, Sdha and Tbp as the most stable reference genes with which to normalize qPCR data. We believe these three genes will serve as excellent reference controls examining the basis for the differing developmental and epigenetic properties unique to embryonic, trophectoderm and extraembryonic endoderm stem cells.
Materials and Methods
Stem Cell Culture
Primary ES cells, TS cells, and XEN cells were derived from either 129 strain (R1 ES cells, [31] TS 3.5) [32] B6×CAST or CAST7×B6 F1 embryos [11] as previously described [5]–[7], [11]. Briefly, ES cultures were maintained in DMEM (Sigma, St. Lousi MO. Cat# D5671) supplemented with 50 µg/ml Penicillin/Streptomycin (Invitrogen, Carlsbad CA.), 100 µm β-mercaptoethanol, 1X LIF, (Sigma, St. Lousi MO.) 2 mM L-Glutamine, 1X MEM non-essential amino acids (Invitrogen, Carlsbad CA.) and 15% Hyclone ES grade fetal bovine serum (Fisher Scientific, Pittsburgh PA.). TS and XEN cell cultures were maintained as described (Kunath et al., 2005; Tanaka et al., 1998) using RPMI (Sigma, St. Lousi MI. Cat# R0883) supplemented with 50 µg/ml Penicillin/Streptomycin, 1 mM Sodium Pyruvate (Invitrogen, Carlsbad CA.), 100 µm β -mercaptoethanol, 1 µg/ml Heparin (Sigma, St. Lousi MO), 2 mM L-Glutamine, 1X FGF basic, 1X FGF4 (R&D Systems) and 20% Hyclone ES grade FBS. Cells were grown on a Mytomycin C (Sigma) treated feeder mouse fibroblast layer. For studies examining ES cell differentiation, sub-confluent cultures were dissociated with 1X trypsin (Accutase - Millipore Billerica, MA) and plated on non-tissue culture treated petri dishes in ES cell medium lacking LIF for four days and subsequently plated on 10 cm tissue culture treated dishes to differentiate into fibroblast like cells. To differentiate TS cells we followed methods described previously [32].
RNA Isolation and Reverse Transcription
Cultured cells were grown to 80% confluence, washed twice in warm PBS, and dissociated with 1X trypsin (Accutase - Millipore Billerica, MA). Cells were spun down, washed once in cold PBS, then RNA isolated using Trizol (Invitrogen, Carlsbad CA.) according to the manufacturer's protocol. One µg of purified total RNA was treated with amplification grade DNaseI (Invitrogen) according to the manufacturer's protocol, and 250 ng RNA seeded into a reverse transcription reaction using the SuperScriptII system (Invitrogen) by combining 1 µl random hexamer oligonucleotides (Invitrogen), 1 µl 10 mM dNTP (Invitrogen), 11 µl RNA plus water. This mixture was brought to 70°C for 5 minutes then cooled to room temperature. SuperScriptII reaction buffer, DTT (Invitrogen) and SuperScriptII were then added according to manufacturer's protocol and the mixture was brought to 25°C for 5 minutes, 42°C for 50 minutes, 45°C for 20 minutes, 50°C for 15 minutes then 70°C for five minutes.
Real-Time PCR Amplification
Real-time PCR analysis of mRNA levels was carried out using the DyNAmo Flash SYBR Green qPCR Mastermix (Fisher Scientific, Pittsburgh PA.) following the manufacturer's instructions. Reactions were performed on a StepOnePlus Real Time PCR system (Applied Biosystems, Foster City CA.). DNA primer information is available in Table 6. Analysis of Real Time PCR Data
Table 6. Description and sequences of the primers used in the in the analysis of both the candidate reference genes and lineage specific transcription factors.
| Primer Name | Sequence | Tm | Amplicon Size | Reference |
| mH2afz Sen | GCGCAGCCATCCTGGAGTA | 60 | Mamo et al. 2007 | |
| m2afz Asen | CCGATCAGCGATTTGTGGA | 60 | 202 | |
| mGAPDH Sen | TGACGTGCCGCCTGGAGAAA | 60 | Mamo et al. 2007 | |
| mGAPDH Asen | AGTGTAGCCCAAGATGCCCTTCAG | 60 | 98 | |
| mHPRT1 Sen | CTGGTGAAAAGGACCTCTCGAA | 60 | Mamo et al. 2007 | |
| mHPRT1 Asen | CTGAAGTACTCATTATAGTCAAGGGCAT | 60 | 117 | |
| mPPIA Sen | CGCGTCTCCTTCGAGCTGTTTG | 60 | Mamo et al. 2007 | |
| mPPIA Asen | TGTAAAGTCACCACCCTGGCACAT | 60 | 150 | |
| mYWHAZ Sen | TTGATCCCCAATGCTTCGC | 60 | This study | |
| mYWHAZ Asen | CAGCAACCTCGGCCAAGTAA | 60 | 88 | |
| mSDHA Sen 2 | GCTCCTGCCTCTGTGGTTGA | 60 | This study | |
| mSDHA Asen 2 | AGCAACACCGATGAGCCTG | 60 | 134 | |
| mB2M Sen | CCGCCTCACATTGAAATCCA | 60 | This study | |
| mB2M Asen | TCGATCCCAGTAGACGGTCTTG | 60 | 199 | |
| mGUSB Sen 2 | GGCTGGTGACCTACTGGATTT | 60 | This study | |
| mGUSB Asen 2 | TTGGCACTGGGAACCTGAAGT | 60 | 134 | |
| mTBP Sen | GAAGAACAATCCAGACTAGCAGCA | 60 | This study | |
| mTBP Asen | CCTTATAGGGAACTTCACATCACAG | 60 | 127 | |
| mPGK1 Sen | CTGACTTTGGACAAGCTGGACG | 60 | This study | |
| mPGK1 Asen | GCAGCCTTGATCCTTTGGTTG | 60 | 110 | |
| mHexokinase II Fwd | CCCTGTGAAGATGTTGCCCAC | 60 | Allen et al.,2004 | |
| mHexocinase II Rev | TGCCCATGTACTCAAGGAAGT | 60 | 251 | |
| mActb Fwd | TGGTGGGTATGGGTCAGAAG | 57 | Allen et al.,2004 | |
| mActb Rev | GGTCATCTTTTCACGGTTGG | 57 | 269 | |
| mhsp70 Fwd | ACGTGGCCTTCACCGACACC | 57 | Allen et al.,2004 | |
| mhsp70 Rev | CGATCTCCTTCATCTTCGTC | 57 | 270 | |
| mRn7sk Fwd | ATTGATCGCCAGGGTTGATT | 60 | Allen et al.,2004 | |
| mRn7sk Rev | CGGGGAAGGTCGTCCTCTTC | 60 | 123 | |
| mOct4 Fwd | ATGGCTGGACACCTGGCTTC | 60 | Golding et al., 2010 | |
| mOct4 Rev | GGTCGGCACAGGGCTCAGA | 60 | 337 | |
| mCDX2 Fwd | GCAGTCCCTAGGAAGCCAAGTGA | 60 | Strumpf et al., 2005 | |
| mCDX2 Rev | CTCTCGGAGAGCCCAAGTGTG | 60 | 162 | |
| mGATA4 Fwd | CCTCTATCACAAGATGAACGGC | 60 | Golding et al., 2010 | |
| mGATA4 Rev | CACTGCTGCTGCTGCTGCTA | 60 | 356 | |
| mNanog Fwd | GCACTCAAGGACAGGTTTCAGA | 60 | Golding et al., 2010 | |
| mNanog Rev | GGTGGAGTCACAGAGTAGTTCAGG | 60 | 578 | |
| mSox7 Fwd | CGCCGCCCGCCGTCCCCCGA | 60 | Golding et al., 2010 | |
| mSox7 Rev | CACCCCTGTCCTCCTTCTCC | 60 | 399 | |
| mHand 1 Fwd | GATGCTGCCCCAGATTTCCCT | 60 | Golding et al., 2010 | |
| mHand 1 Rev | CCCTTTTCCGCTTGCTTTCG | 60 | 388 | |
| mFSP1 Fwd | GGCAAGACCCTTGGAGGAG | 60 | This study | |
| mFSP1 Rev | CCTTTTCCCCAGGAAGCTAG | 60 | 212 |
The measured CT (Cycle Threshold) values for each sample were complied and the stability of each of the fourteen reference genes analyzed using the GENorm, NORMFinder and BESTKeeper software tools; which have been described in detail elsewhere [13]–[15]. Once suitable reference genes were identified, the geometirc mean CT values of the best three candidate genes were calculated for each individual sample and used to normalize expression levels using the ▵▵CT method described previously [13], [34], [35]. These normalized values were averaged and the standard error of the mean calculated and graphed using Excel.
Acknowledgments
The authors would like to thank Dr. C.R. Long and R.D. Landry for critical review of the manuscript. K.J.V. was supported through the Texas A&M University College of Veterinary Medicine & Biomedical Sciences Graduate Student Research Trainee Award.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Institutes of Health - NIAA grant 1R03AA020129-01. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rossant J. Investigation of the determinative state of the mouse inner cell mass. II. The fate of isolated inner cell masses transferred to the oviduct. J Embryol Exp Morphol. 1975;33:991–1001. [PubMed] [Google Scholar]
- 2.Rossant J, Tam PP. Emerging asymmetry and embryonic patterning in early mouse development. Dev Cell. 2004;7:155–164. doi: 10.1016/j.devcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- 4.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 7.Kunath T, Arnaud D, Uy GD, Okamoto I, Chureau C, et al. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development. 2005;132:1649–1661. doi: 10.1242/dev.01715. [DOI] [PubMed] [Google Scholar]
- 8.Cherry SR, Biniszkiewicz D, van Parijs L, Baltimore D, Jaenisch R. Retroviral expression in embryonic stem cells and hematopoietic stem cells. Mol Cell Biol. 2000;20:7419–7426. doi: 10.1128/mcb.20.20.7419-7426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, et al. Selective loss of imprinting in the placenta following preimplantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- 10.Fortier AL, Lopes FL, Darricarrère N, Martel J, Trasler JM. Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet. 2008;17:1653–1665. doi: 10.1093/hmg/ddn055. [DOI] [PubMed] [Google Scholar]
- 11.Golding MC, Zhang L, Mann MR. Multiple epigenetic modifiers induce aggressive viral extinction in extraembryonic endoderm stem cells. Cell Stem Cell. 2010;6:457–467. doi: 10.1016/j.stem.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 15.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 16.Goossens K, Van Poucke M, Van Soom A, Vandesompele J, Van Zeveren A, Peelman LJ. Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev Biol. 2005;5:27. doi: 10.1186/1471-213X-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilsbach R, Kouta M, Bönisch H, Brüss M. Comparison of in vitro and in vivo reference genes for internal standardization of real-time PCR data. Biotechniques. 2006;40:173–177. doi: 10.2144/000112052. [DOI] [PubMed] [Google Scholar]
- 18.Willems E, Mateizel I, Kemp C, Cauffman G, Sermon K, Leyns L. Selection of reference genes in mouse embryos and in differentiating human and mouse ES cells. Int J Dev Biol. 2006;50:627–635. doi: 10.1387/ijdb.052130ew. [DOI] [PubMed] [Google Scholar]
- 19.Mamo S, Gal AB, Bodo S, Dinnyes A. Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev Biol. 2007;7:14. doi: 10.1186/1471-213X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J. 2008;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- 21.Tatsumi K, Ohashi K, Taminishi S, Okano T, Yoshioka A, et al. Reference gene selection for real-time RT-PCR in regenerating mouse livers. Biochem Biophys Res Commun. 2008;374:106–110. doi: 10.1016/j.bbrc.2008.06.103. [DOI] [PubMed] [Google Scholar]
- 22.Boda E, Pini A, Hoxha E, Parolisi R, Tempia F. Selection of reference genes for quantitative real-time RT-PCR studies in mouse brain. J Mol Neurosci. 2009;37:238–253. doi: 10.1007/s12031-008-9128-9. [DOI] [PubMed] [Google Scholar]
- 23.Suter DM, Tirefort D, Julien S, Krause KH. A Sox1 to Pax6 switch drives neuroectoderm to radial glia progression during differentiation of mouse embryonic stem cells. Stem Cells. 2009;27:49–58. doi: 10.1634/stemcells.2008-0319. [DOI] [PubMed] [Google Scholar]
- 24.van den Bergen JA, Miles DC, Sinclair AH, Western PS. Normalizing gene expression levels in mouse fetal germ cells. Biol Reprod. 2009;81:362–370. doi: 10.1095/biolreprod.109.076224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galiveti CR, Rozhdestvensky TS, Brosius J, Lehrach H, Konthur Z. Application of housekeeping npcRNAs for quantitative expression analysis of human transcriptome by real-time PCR. RNA. 2010;16:450–461. doi: 10.1261/rna.1755810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 27.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 28.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 29.Golding MC, Magri LS, Zhang L, Lalone SA, Higgins MJ, et al. Depletion of Kcnq1ot1 non-coding RNA does not affect imprinting maintenance in stem cells. Development. 2011;138(17):3667–78. doi: 10.1242/dev.057778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19:36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- 31.Nagy A, Gócza E, Diaz EM, Prideaux VR, Iványi E, et al. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 33.Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 36.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 38.Strutz F, Okada H, Lo CW, Danoff T, Carone RL, et al. Identification and characterization of a fibroblast marker: FSP1. J Cell Biol. 1995;130:393–405. doi: 10.1083/jcb.130.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee CH, Shah B, Moioli EK, Mao JJ. CTGF directs fibroblast differentiation from human mesenchymal stem/stromal cells and defines connective tissue healing in a rodent injury model. J Clin Invest. 2010;120:3340–3349. doi: 10.1172/JCI43230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernandez L, Kozlov S, Piras G, Stewart CL. Paternal and maternal genomes confer opposite effects on proliferation, cell-cycle length, senescence, and tumor formation. Proc Natl Acad Sci U S A. 2003;100:13344–13349. doi: 10.1073/pnas.2234026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vong QP, Liu Z, Yoo JG, Chen R, Xie W, et al. A role for borg5 during trophectoderm differentiation. Stem Cells. 2010;28:1030–1038. doi: 10.1002/stem.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida A, Tani K. Phosphoglycerate kinase abnormalities: functional, structural and genomic aspects. Biomed Biochim Acta. 1983;42:S263–S267. [PubMed] [Google Scholar]
- 44.Oyedotun KS, Lemire BD. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies. J Biol Chem. 2004;279:9424–9431. doi: 10.1074/jbc.M311876200. [DOI] [PubMed] [Google Scholar]
- 45.Kornberg RD. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci U S A. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rangasamy D, Greaves I, Tremethick DJ. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat Struct Mol Biol. 2004;11:650–655. doi: 10.1038/nsmb786. [DOI] [PubMed] [Google Scholar]
- 47.Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Greaves IK, Rangasamy D, Ridgway P, Tremethick DJ. H2A.Z contributes to the unique 3D structure of the centromere. Proc Natl Acad Sci U S A. 2007;104:525–530. doi: 10.1073/pnas.0607870104. [DOI] [PMC free article] [PubMed] [Google Scholar]