Abstract
RAD18 is an ubiquitin ligase that is involved in replication damage bypass and DNA double-strand break (DSB) repair processes in mitotic cells. Here, we investigated the testicular phenotype of Rad18-knockdown mice to determine the function of RAD18 in meiosis, and in particular, in the repair of meiotic DSBs induced by the meiosis-specific topoisomerase-like enzyme SPO11. We found that RAD18 is recruited to a specific subfraction of persistent meiotic DSBs. In addition, RAD18 is recruited to the chromatin of the XY chromosome pair, which forms the transcriptionally silent XY body. At the XY body, RAD18 mediates the chromatin association of its interaction partners, the ubiquitin-conjugating enzymes HR6A and HR6B. Moreover, RAD18 was found to regulate the level of dimethylation of histone H3 at Lys4 and maintain meiotic sex chromosome inactivation, in a manner similar to that previously observed for HR6B. Finally, we show that RAD18 and HR6B have a role in the efficient repair of a small subset of meiotic DSBs.
Key words: RAD18, HR6B, meiosis, DNA double-strand break repair, XY body
Introduction
The E3 ubiquitin ligase RAD18 is crucial for cell survival after induction of various types of DNA damage in mammalian somatic cells (Tateishi et al., 2003; Shiomi et al., 2007; Huang et al., 2009; Miyase et al., 2005; Watanabe et al., 2009). RAD18 functions in complex with HR6A (UBE2A) and HR6B (UBE2B), the two mammalian orthologs of the Saccharomyces cerevisiae E2 ubiquitin-conjugating enzyme Rad6 (Koken et al., 1991). In S. cerevisiae, Rad6 and Rad18 are most well known for their role in replication damage bypass (RDB). This pathway allows progression of DNA replication by translesion synthesis polymerases in the presence of DNA damage (reviewed by Chang and Cimprich, 2009; Guo et al., 2009; Livneh et al., 2010). Functional orthologs of proteins involved in the RDB pathway have been identified in mammals, implying that this pathway is generally well conserved. In addition to its function in the RDB pathway, RAD18 also acts at double-strand break (DSB) repair sites in mammalian mitotic cells (Huang et al., 2009; Watanabe et al., 2009). The exact function of RAD18 in DSB repair in mammalian cells is not fully clear. Recent data indicate that RAD18 facilitates homologous recombination (HR) through binding of RAD51C (Huang et al., 2009). RAD18 is expressed in multiple tissues, but the highest level is found in testis (van der Laan et al., 2000), and in S. cerevisiae, RAD18 gene expression increases during meiosis, suggesting a specific function for RAD18 in this process (Jones and Prakash, 1991). Deletion of RAD18 in yeast does not affect either meiosis or meiotic recombination (Dowling et al., 1985; Fabre et al., 1989; Game and Mortimer, 1974) but double mutants of rad18 and various genes that act during excision–repair show a drastic reduction in spore viability, compared with the single mutants (Dowling et al., 1985). This result indicates that Rad18 does perform a specific function during meiosis in S. cerevisiae.
Meiotic DSBs are induced by the meiosis-specific topoisomerase-II-like enzyme SPO11 in prophase I (Keeney et al., 1997; Keeney et al., 1999), and are required for proper homologous chromosome pairing and meiotic recombination in yeast and mammals (Cao et al., 1990; Baudat et al., 2000; Romanienko and Camerini-Otero, 2000). The chromatin surrounding mitotic and meiotic DSBs undergoes a series of orchestrated modifications. One of the first modifications is phosphorylation of the histone H2A variant H2AX (γH2AX) (Burma et al., 2001; Mahadevaiah et al., 2001), which is accompanied by the formation of foci containing the DSB-repair protein RAD51 and its meiosis-specific paralog DMC1 on chromatin (Ashley et al., 1995; Barlow et al., 1997; Tarsounas et al., 1999). In mitotic cells, DSBs can be repaired by two distinct repair pathways: HR and non-homologous DNA end-joining (NHEJ). HR is an error-free mechanism, in which a homologous sequence of the sister chromatid is used as a template to process repair. NHEJ is an error-prone mechanism, in which the two ends of the broken DNA are processed for direct ligation, with an increased chance of small deletions or insertions. In meiotic cells, this NHEJ repair mechanism is repressed (Goedecke et al., 1999), leaving HR as the only available pathway for repair. In addition, the use of the sister chromatid as a template to process repair via HR is most likely repressed (Schwacha and Kleckner, 1994; Schwacha and Kleckner, 1997). HR via one of the chromatids of the homologous chromosome might lead to the formation of crossovers, although repair of most meiotic DSBs generates noncrossovers. The repair of meiotic DSBs is accompanied by progression of synapsis, which is achieved by the formation of the synaptonemal complex (SC) between the chromosomal axes of the paired homologous chromosomes (reviewed by Inagaki et al., 2010).
During meiotic prophase, all homologous chromosomes initiate pairing and synapsis in zygotene. In pachytene nuclei, all chromosomes have completely synapsed, and these substages can be visualized using antibodies that recognize SYCP3, a major protein component of the SC, which is present in the axial and lateral elements of the SC (Heyting et al., 1987). The X and Y chromosomes are largely heterologous, and during midpachytene in mouse, synapsis is observed only along the short homologous pseudoautosomal regions. The rest of the chromosomal arms remains unsynapsed, and forms a subnuclear region called XY body (or sex body), which is first seen around early pachytene and persists into diplotene (Moses, 1977). XY body formation is associated with meiotic sex chromosome inactivation (MSCI) (Monesi, 1965). The chromatin surrounding the XY body undergoes various modifications reminiscent of DSB repair in mitotic cells, such as phosphorylation of H2AX (Mahadevaiah et al., 2001) and ubiquitylation of histone H2A at Lys119 (Baarends et al., 1999). In addition, many DSB-repair-related factors accumulate specifically on the XY body (reviewed by Inagaki et al., 2010). The RDB enzyme HR6A/B and its E3 ligase partner RAD18 also accumulate on the XY body (van der Laan et al., 2004). Single Hr6a knockout (KO) and Hr6b KO mice are viable, but double-knockout mice are embryonic lethal (Roest et al., 2004). Spermatogenesis of Hr6b KO mice is markedly affected during postmeiotic steps, leading to male infertility. In addition, Hr6b KO spermatocytes show an increased rate of apoptosis, longer synaptonemal complexes and an increased frequency of crossover formation (Baarends et al., 2003). HR6B also exerts a negative control over the level of H3K4 dimethylation on the X and Y chromosomes in diplotene, and in postmeiotic round spermatids. This function contributes to the postmeiotic maintenance of X chromosome silencing (Baarends et al., 2007; Mulugeta Achame et al., 2010). It is not known which E3 enzymes are required for the different functions of HR6B in meiotic and postmeiotic germ cell development. Here, we investigated the function of the ubiquitin ligase RAD18, a well-known HR6A and HR6B interaction partner, in mammalian meiosis, using Rad18-knockdown mice.
Results
RAD18 expression is efficiently downregulated following transgenic expression of shRNA
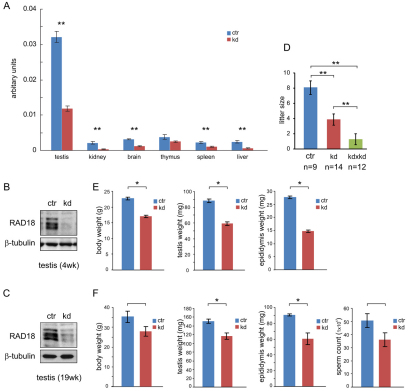
Rad18-knockdown (KD) and control mice were generated through targeted insertion of a Rad18-specific shRNA construct, and a control construct not encoding shRNA, respectively, driven by the U6 promoter in the Rosa26 locus (Seibler et al., 2005). In these animals, only one allele of the Rosa26 locus is targeted. We first investigated the expression level of Rad18 mRNA by quantitative real-time RT-PCR in testis, brain, kidney, liver, spleen and thymus from 4-week-old mice. Rad18 mRNA was most highly expressed in testis (Fig. 1A) (van der Laan et al., 2004), and the expression was significantly downregulated (to approximately 35%) in Rad18 KD mice. In most other tissues, we also found a significant reduction of Rad18 mRNA (Fig. 1A). In Rad18 KD testis, RAD18 protein expression was even more efficiently downregulated (to approximately 11% in 4-week-old mice and to 16% in 19-week-old mice) than the mRNA (Fig. 1B,C). In the rest of the tissues mentioned above, we could not detect RAD18 expression on immunoblots in either control or Rad18 KD samples (data not shown). Possible off-target genes that might be downregulated by the shRNA were searched using the BLASTN program. Three possible genes with a high query coverage (more than 70%) were detected; transmembrane protein 136 (Tmem136), slingshot homolog 3 (Ssh3) and UDP-glucose ceramide glucosyltransferase-like 2 (Ugcgl2). The mRNA expression level of these three genes was investigated in various tissues from control and Rad18 KD mice, and no significant changes in the expression of these genes were observed (supplementary material Fig. S1).
Fig. 1.
Characterization of Rad18 KD mice. (A) Rad18 mRNA expression in testis, kidney, brain, thymus, spleen and liver of three control mice and three Rad18 KD mice. Error bars indicate s.e.m. values. (B,C) RAD18 expression in total cell extracts (20 μg) from testis of 4-week-old (B) and 19-week-old (C) from control (ctr) and Rad18 KD was detected on immunoblots. β-tubulin was used as loading control. (D) Average litter size obtained from matings between control and wild type (ctr), knockdown and wild type (kd), and two knockdowns (kd×kd). Error bars indicate s.e.m. values. (E,F) Body, testis and epididymis weights (E,F) and the number of sperm (F) from 4-week-old (E) and 19-week-old (F) control and Rad18 KD mice. Error bars indicate s.e.m. *P<0.05 and **P<0.01 (Mann–Whitney U-test). Blue and red bars indicate control and Rad18 KD, respectively.
Subfertility and reduced testis and body weights of Rad18 KD mice
The Rad18 KD animals appeared healthy, although the body weight was reduced by approximately 25% in both young and adult animals (Fig. 1E,F). This might be caused by general effects of the severe reduction in RAD18 levels in all tissues, in particular in kidney and liver, which showed the most severe reduction in Rad18 mRNA (to 16% and 23%, respectively). Repeated breeding experiments to obtain homozygous Rad18 KD mice were unsuccessful. The average litter size was 1.3±0.72 (mean ± s.e.m.) (Fig. 1D) and no mice were found to be homozygous for the targeted allele. Breeding experiments using the heterozygous Rad18 KD males and females in combination with wild-type C57BL/6 females and males, respectively, revealed that Rad18 KD mice are subfertile. The average litter size of Rad18 KD males and females was smaller than that of control (KD, 3.9±0.75; control, 8.1±0.9; P<0.01, Fig. 1D), and the litter size in female Rad18 KD mice (female, 3.3±0.98) was similar to that in male Rad18 KD mice (male, 4.6±1.2; P=0.42). To further analyze the possible role of RAD18 in spermatogenesis, we first compared the weights of reproductive organs in Rad18 KD and control mice. On average, testis weights were 33% reduced in 4-week-old Rad18 KD mice, and the epididymis weight was reduced by 50%. Because the body weight of the Rad18 KD mice was also reduced compared with controls, these effects might be caused by delayed testicular development (Fig. 1E). However, when we examined adult (19-week-old) mice, Rad18 KD testis, epididymis and body weight were also reduced compared with controls (Fig. 1F). In addition, the number of sperm was reduced by 29%, which corresponds to the 22% reduced testis weight in Rad18 KD males, although the differences from the control were not statistically significant (Fig. 1F).
Aberrant elongating spermatid heads in testes of in Rad18 KD mice
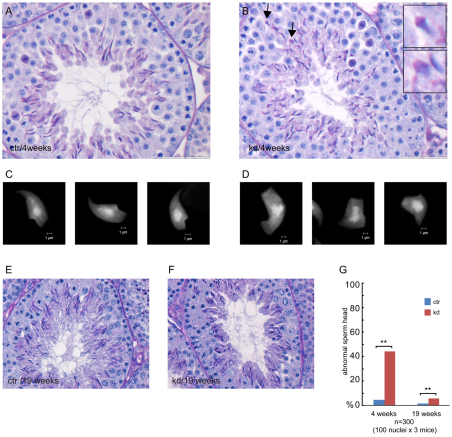
Histological analysis of cross-sections of control and Rad18 KD testes, revealed no overall differences (Fig. 2A,B). Complete spermatogenesis was apparent in both genotypes. However, in Rad18 KD mice, elongating spermatids frequently showed an aberrant head shape (Fig. 2B,C,D,G). In 19-week-old animals, the overall frequency of aberrant spermatid heads was highly reduced in testes from both control and Rad18 KD mice (Fig. 2E,F), but was still approximately fivefold increased in Rad18 KD mice versus controls (Fig. 2G). This relative increase in the number of aberrant spermatids was less than that observed at 4 weeks of age. However, the similar and very low level of persistent RAD18 protein in testes from 4-week-old and 19-week-old Rad18 KD males does not indicate that the knockdown of RAD18 becomes less efficient with age. In addition, we could not find any correlation between litter size and age of the Rad18 KD males or females (n=7 breedings for each group).
Fig. 2.
Morphological analysis of testes from Rad18 KD mice and controls. (A,B,E,F) Histological sections of 4-week-old (A,B) and 19-week-old (E,F) control (A,E) and Rad18 KD (B,F) testes were stained with hematoxylin and eosin. (B) Elongated spermatids indicated with arrows are shown in enlarged pictures in the inserts. (C,D) DAPI staining of spread nuclei from elongated spermatids of control (C) and Rad18 KD (D). (G) Percentage of elongated spermatids with aberrant shape in control and Rad18 KD mice. Blue and red bars indicate control and Rad18 KD, respectively. **P<0.01 (Chi-squared test).
Reduced XY synapsis in spermatocytes of Rad18 KD mice
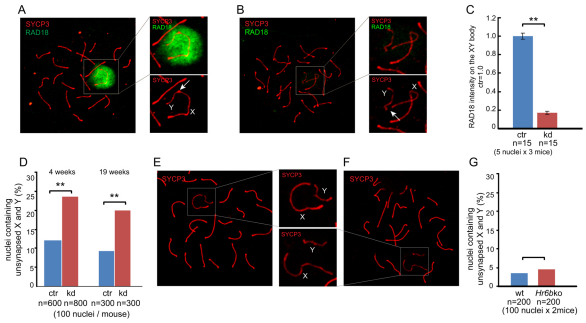
RAD18 associates with the largely unpaired X and Y chromosomes that form the transcriptionally inactive so-called XY body in pachytene and diplotene spermatocytes (Fig. 3A). In Rad18 KD spermatocytes, RAD18 expression was considerably decreased (Fig. 3B), with a 80% reduced level of RAD18 staining on the XY body compared with that in control mice (Fig. 3C). We observed an increased frequency of XY asynapsis in Rad18 KD pachytene spermatocytes (Fig. 3D). When X and Y were not synapsed, they were still adjacent to each other in 66% of these nuclei (Fig. 3E), whereas in the rest of the nuclei that displayed asynapsed X and Y chromosomes, they were at a larger distance (Fig. 3F). The presence of asynapsed X and Y chromosomes did not lead to an increased frequency of XY aneuploidies in Rad18 KD spermatids, as verified using DNA fluorescence in situ hybridization (FISH) with X and Y painting probes (data not shown). We also examined XY synapsis in Hr6b KO spermatocytes. However, the frequency of XY asynapsis in Hr6b KO pachytene spermatocytes was not different from that of controls (Fig. 3G).
Fig. 3.
Efficient knockdown of RAD18 in spermatocytes from Rad18 KD mice, and increased frequency of XY asynapsis. (A,B) Double immunostaining of pachytene spermatocyte nuclei of control (A) and Rad18 KD mice (B) with anti-SYCP3 (red) and anti-RAD18 (green). The insert shows a larger magnification of the area containing the XY body. Arrows indicate the pseudoautosomal synapsed region (A) and unsynapsed region (B). (C) RAD18 intensity on the XY body was measured with ImageJ software. The intensity of RAD18 in control nuclei was set at 1.0. Error bars indicate s.e.m. **P<0.01 (Mann–Whitney U-test). (D) The percentage of unsynapsed X and Y chromosomes in mid-pachytene of 4-week-old and 19-week-old mice. **P<0.01 (Chi-squared test). (E,F) Immunostaining of Rad18 KD pachytene spermatocyte nuclei with anti-SYCP3 (red). The insert shows a larger magnification of the area containing the XY body. X and Y chromosomes are indicated in the enlarged pictures. (G) 4-week-old wild-type and Hr6b KO mice. (C,D,G) Blue and red bars indicate control (ctr) and Rad18 KD, respectively.
RAD18-dependent localization of HR6A and HR6B at the XY body
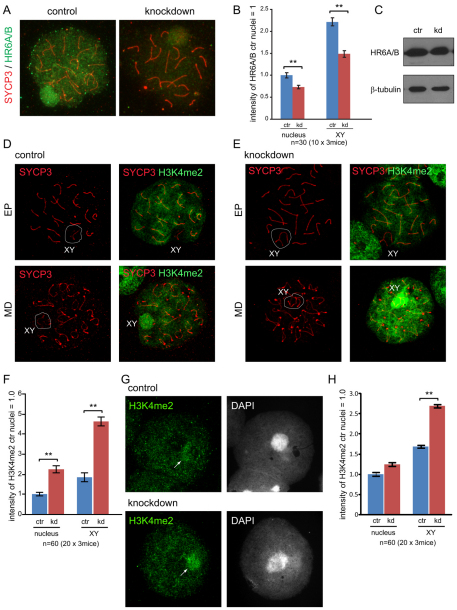
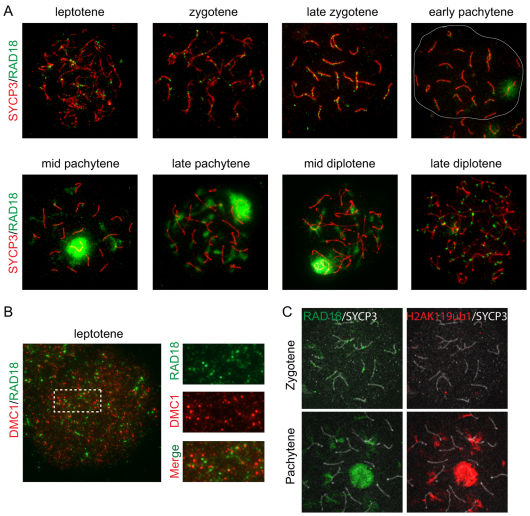
We previously found that the ubiquitin-conjugating enzymes HR6A and HR6B (HR6A/B) associate with the XY body in pachytene and diplotene spermatocytes (van der Laan et al., 2004; Mulugeta Achame et al., 2010). In mitotic cells, the mammalian RAD18-HR6A/B complex is highly stable, even under high salt conditions (1 M NaCl) (data not shown). Therefore, we examined whether the association of HR6A/B with the XY body depends on RAD18. In Rad18 KD spermatocytes, a decreased level of HR6A/B was detected in whole nuclei and on the XY body (Fig. 4A,B), although the expression level of HR6A/B in Rad18 KD testis was similar to that of the control (Fig. 4C), indicating that RAD18 mediates the chromatin association of HR6A/B in meiosis. The residual binding of HR6A/B to the XY body might result from the residual amount of RAD18 that is still present, or might be independent of RAD18.
Fig. 4.
Reduced HR6A/B and increased dimethylation of histone H3 at Lys4 in Rad18 KD spermatocytes and spermatids. (A) Double immunostaining of control and Rad18 KD pachytene spermatocyte nuclei with anti-SYCP3 (red) and anti-HR6A/B (green). (B) The intensities of HR6A/B in the nucleus and on the XY body were measured with ImageJ. The intensity of HR6A/B in control nuclei was set at 1.0. Error bars indicate s.e.m. (C) HR6A/B expression in total cell extracts of testis from 4-week-old control (ctr) and Rad18 KD mice was detected on immunoblots. β-tubulin was used as loading control. (D,E) Double immunostaining of spermatocyte nuclei with anti-SYCP3 (red) and anti-H3 dimethylated at Lys4 (H3K4me2) (green) in control (D) and Rad18 KD mice (E). EP, early pachytene; MD, mid diplotene. The XY body is shown in the white circle. (F) The intensities of H3K4me2 in the nucleus and on the XY body from 4-week-old and 19-week-old mice were measured using ImageJ. The intensity of H3K4me2 in control nuclei was set at 1.0. Error bars indicate s.e.m. (G) Immunostaining of spermatid nuclei with anti-H3K4me2 (green) in Rad18 KD and control mice. Arrows indicate sites of the accumulation of H3K4me2. (H) The intensities of H3K4me2 in spermatid nuclei and on the X or Y chromosomes were measured as described in F. (B,F,H) Blue and red bars indicate control (ctr) and Rad18 KD, respectively. **P<0.01 (Mann–Whitney U-test).
Overall increased di-methylation of histone H3 Lys4 in Rad18 KD mice
The reduced level of HR6A/B in Rad18 KD spermatocytes prompted us to analyze several aspects of meiotic prophase that were found to be aberrant in Hr6b KO spermatocytes, to investigate which meiotic HR6B functions depend on RAD18. In Hr6b KO mice, increased dimethylation of histone H3 at Lys4 (H3K4me2) and increased phosphorylation of H2A at Tyr120 (H2AT120p) were found on the XY body, and H3K4me2 was also increased on the X and Y in round spermatids (Baarends et al., 2007). In control mice, H3K4me2 staining patterns were similar to what was previously described (Fig. 4D) (Baarends et al., 2007). In Rad18 KD mice, the H3K4me2 signal was increased on the XY body as well as in the rest of the nucleus in diplotene nuclei, compared with controls (Fig. 4E,F). In haploid Rad18 KD round spermatids, H3K4me2 levels were also increased on the sex chromosomes (approximately 2.7-fold), compared with controls (Fig. 4G,H). In these cells, either the X or the Y chromosome is located adjacent to the chromocenter, and localization of H3K4me2 on the X or Y chromosomes has been verified previously using FISH (Baarends et al., 2007). The expression level and pattern of H2AT120p did not show any significant differences between Rad18 KD and control mice (data not shown).
De-repression of X-linked genes
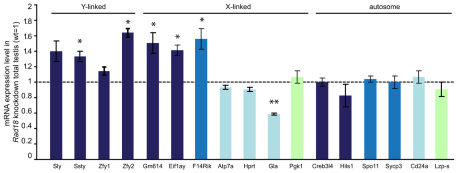
The increased level of H3K4me2 on the sex chromosomes of Hr6b KO spermatocytes and spermatids is associated with an overall increase in X-linked gene expression in spermatids, indicating that the post-meiotic maintenance of meiotic sex chromosome inactivation (MSCI) is disturbed in Hr6b KO mice (Baarends et al., 2007; Mulugeta Achame et al., 2010). To analyze whether this function of HR6B also depends on RAD18, the gene expression level of selected X-, Y- and autosome-linked genes was examined using mRNA isolated from total testes of 19-week-old control and Rad18 KD mice. We selected four Y-linked genes that are expressed selectively in postmeiotic spermatids (Sly, Ssty, Zfy1, Zfy2). In addition, we analyzed seven X-linked genes that are expressed in postmeiotic cells (Gm614 and Eif1ay), in meiotic and postmeiotic cells (4930408F14Rik), in premeiotic cells but repressed in meiotic and postmeiotic cells (Atp7a, Hprt and Gla), or in Sertoli and peritubular myoid cells (Pgk1) (Namekawa et al., 2006) [Mammalian Reproductive Genetics Database (MRGD)]. We also selected six autosomal genes that are expressed in peritubular myoid cells (Lzp-s), or at premeiotic (Cd24a), meiotic (Spo11 and Sycp3) or postmeiotic (Hils and Creb3l4) spermatogenic developmental steps (Namekawa et al., 2006) (MRGD). The Y-linked genes Sly, Ssty and Zfy2, but not Zfy1, showed increased expression in Rad18 KD testes compared with the control (Fig. 5). Three X-linked genes, Gm614, Eif1ay and F14Rik, also showed increased expression in Rad18 KD testes. However, for four other X-linked genes, Atp7a, Hrpt, Gla and Pgk1, we did not observe an effect of RAD18 depletion, except a decreased expression of Gla. Thus, in Rad18 KD total testes, only X- and Y-linked genes that are normally induced during postmeiotic germ cell development, showed increased expression. Note that for genes that are also expressed in premeiotic and somatic cells, a selective effect on the expression in meiotic and postmeiotic cells may be not apparent if the expression in premeiotic cells and somatic cells is also high, because all these cells are present in the total testis samples. For the autosomal genes, none of the five genes analyzed showed any significant differences between control and Rad18 KD mice.
Fig. 5.
Derepression of X- and Y-linked genes in Rad18 KD testis. Real-time RT-PCR quantification of mRNA levels of X-, Y- and autosome-linked genes in total testes from Rad18 KD mice. The amount of PCR products was normalized to mRNA encoding β-actin. The level of mRNA expression in control is set as 1.0, and mRNA expression levels of indicated genes in Rad18 KD testes relative to the control are shown in the graph. Error bars show s.e.m. for two independent experiments from three mice. Dark blue bars indicate genes mainly expressed in postmeiotic cells, blue bars indicate genes mainly expressed in meiotic prophase cells, light blue bars represent premeiotic genes, and the light green bars represent genes that are mainly expressed in somatic cells. *P<0.05 (Mann–Whitney U-test).
Normal formation of meiotic crossover sites in Rad18 KD mice
Another major meiotic phenotype previously observed in Hr6b KO spermatocytes is an increased number of MLH1 foci, and resultant crossovers (Baarends et al., 2003). In Rad18 KD mice, the average number of MLH1 spots was 23.5 (n=120), which was not significantly different from control mice (24.0, n=90), indicating that knockdown of RAD18 does not influence the number of crossover sites (not shown).
RAD51 localization to meiotic DSBs does not depend on RAD18
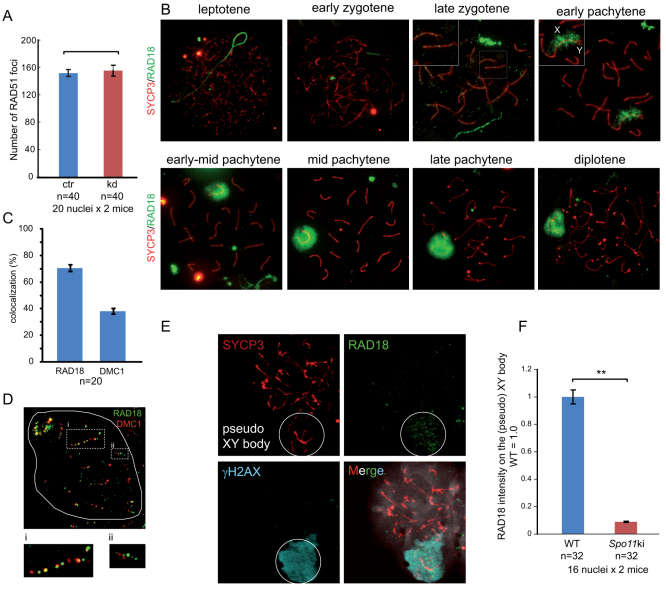
During meiotic prophase, DSBs are induced by SPO11 in leptotene nuclei (Keeney et al., 1997; Keeney et al., 1999), and these DSBs can be visualized as RAD51–DMC1 foci that form in leptotene and reduce in number as homologous chromosome synapsis proceeds (supplementary material Fig. S2A) (Moens et al., 1997; Tarsounas et al., 1999). RAD51 is an essential DSB repair protein that is involved in mitotic and meiotic HR, and DMC1 is its meiosis-specific paralog, that is required for homologous chromosome pairing during male and female meiosis in mouse (Pittman et al., 1998; Yoshida et al., 1998). In RAD18-deficient mitotic MEF cells, Huang and colleagues (Huang et al., 2009) found that the formation of radiation-induced RAD51 foci partially depends on the presence of RAD18. To examine whether RAD18 influences the number of SPO11-induced RAD51 foci in meiosis, we counted the number of RAD51 foci in Rad18 KD leptotene nuclei. We observed similar numbers of the SPO11-induced RAD51 foci in control and knockdown mice, suggesting RAD18-independent accumulation of RAD51 at SPO11-induced DSBs in meiosis (Fig. 6A and supplementary material Fig. S2B).
Fig. 6.
Localization of RAD18 during meiotic prophase. (A) The number of RAD51 foci in leptotene nuclei was counted in control and Rad18 KD mice. Error bars indicate s.e.m. Blue and red bars indicate control (ctr) and Rad18 KD, respectively. (B) Accumulation of RAD18 from leptotene to diplotene in wild-type spermatocytes. In early pachytene, the XY body is shown in an enlarged image, and X and Y chromosomes are indicated as X and Y. (C) The number of RAD18 and DMC1 foci in late-zygotene nuclei was counted in control mice, and the colocalization frequency of RAD18 foci with DMC1 foci (RAD18) and of DMC1 foci with RAD18 foci (DMC1) is shown. Error bars indicate s.e.m. (D) Double immunostaining of a wild-type late-zygotene spermatocyte with anti-DMC1 (red) and anti-RAD18 (green). Enlarged pictures of regions i (frequent colocalization) and ii (little colocalization) are shown below the image. (E) Triple immunostaining of Spo11 mutant spermatocyte nuclei with anti-SYCP3 (red), anti-RAD18 (green) and anti-γH2AX (light blue). The pseudo-XY body is shown in the circle. (F) Graph showing the intensity of RAD18 on the XY body in wild-type spermatocyte nuclei and on the pseudo-XY body in Spo11 mutant spermatocyte nuclei. The intensity of RAD18 on the XY body in wild type was set as 1.0. Error bars indicate s.e.m. **P<0.01 (Mann–Whitney U-test).
RAD18 localizes to a small subfraction of meiotic DSB sites
In mitotic HeLa cells, RAD18 accumulates as foci and colocalizes with both RAD51 and γH2AX at irradiation induced DSBs (Inagaki et al., 2009; Huang et al., 2009). In wild-type meiotic cells, RAD18 accumulates as RAD51-like foci on the SC only in late zygotene, but not in either leptotene or early zygotene (Fig. 6B), when DSBs are induced by the topoisomerase-like enzyme SPO11. These DSBs are essentially different from damage-induced DSBs, for several reasons. First, SPO11 remains covalently attached after forming the break, and removal of SPO11, attached to a small stretch of nucleotides, occurs by a specialized machinery (Neale et al., 2005). In addition, the presence of meiosis-specific components of the repair machinery, such as for example the RAD51 paralog DMC1, and the absence of factors involved in NHEJ (Goedecke et al., 1999), specifically regulate the repair pathways that can be chosen (reviewed by Inagaki et al., 2010). The vast majority of the meiotic RAD18 foci in late zygotene was detected on synapsed regions (93±6%; mean ± s.d.; n=40). To examine whether these RAD18 foci correspond to sites of ongoing meiotic DSB repair, we studied the colocalization of RAD18 with DMC1, which is known to be recruited to meiotic DSBs along with RAD51 (Tarsounas et al., 1999). The global pattern and dynamics of DMC1 and RAD18 accumulation during zygotene was found to be rather different, but in late zygotene nuclei, approximately 70% of the RAD18 foci colocalized with DMC1, whereas approximately 40% of the DMC1 foci colocalized with RAD18 (Fig. 6C,D). It should be noted however, that strong DMC1 foci frequently colocalized with weak RAD18 foci and vice versa. Also, the average number of RAD18 foci in late zygotene nuclei (46±7.7; mean ± s.d.) was much lower than the number of DMC1 foci (87±11). These results suggest that RAD18 does not immediately recognize the SPO11-induced DSBs, but is recruited at a later stage, to a subset of meiotic DSBs that is still present. The RAD18 foci at autosomes gradually disappeared during pachytene. Concomitantly, RAD18 started to accumulate on the chromatin surrounding the XY body (Fig. 6B). It is of interest to note that on the XY body of control early pachytene nuclei, RAD18 appears to localize first to the X chromosomal chromatin, which contains the persistent RAD51 foci, followed by a spread over the rest of the XY body (Fig. 6B). RAD51 foci on the unpaired X chromosome disappear in late pachytene or early diplotene, whereas RAD18 and γH2AX remain on the XY body until late diplotene (Fig. 6B and supplementary material Fig. S2C).
To study whether the RAD18 localization pattern depends on the presence of SPO11-induced DSBs, localization of RAD18 in Spo11 mutant mice was examined. These mice carry a null mutation at the catalytic site (D100Y), no meiotic DSBs are generated and the meiotic phenotype is morphologically indistinguishable from the Spo11 KO (Fig. 6E) (Baudat et al., 2000; Romanienko and Camerini-Otero, 2000). In Spo11 mutant mice, we could not detect RAD18 foci, whereas we still observed a very low level of RAD18 accumulation in a single large chromatin region that colocalized with the γH2AX positive area (Fig. 6E). This region has been called the pseudo-XY body (Chicheportiche et al., 2007). The intensity of RAD18 at the pseudo-XY body was reduced by 90% compared with the level on the XY body in wild-type controls (Fig. 6F), and only a subfraction of the nuclei contained RAD18-positive pseudo-XY bodies (42%), whereas the vast majority of the nuclei (80%) contained a pseudo-XY body identified by γH2AX staining. Thus, although some RAD18 accumulates on the pseudo-XY body, the RAD18 foci on synapsed chromosomes in late zygotene and early pachytene are SPO11-dependent, and thus probably represent sites of persistent meiotic DSBs.
Recruitment of RAD18 to IR-induced DSBs in spermatocytes
As described above, RAD18 is rapidly (within 20 minutes) recruited to damage-induced DSBs in somatic cells, and also to other lesions induced by DNA-damaging agents (Inagaki et al., 2009). Therefore, it is of interest that RAD18 is not recruited to SPO11-induced DSBs in leptotene nuclei. This lack of RAD18 accumulation might be caused by meiosis-specific factors that inhibit RAD18 recruitment to these sites. Alternatively, it might result from low expression of RAD18 at these stages. To distinguish between these possibilities, wild-type mice were exposed to ionizing radiation of 4 Gy, and RAD18 foci formation was analyzed after 2 hours. We have previously shown that 2 hours after irradiation, a 25% increase in the total number of RAD51 foci can be observed in wild-type leptotene nuclei, indicating the presence of irradiation-induced DSBs (Schoenmakers et al., 2008). Irradiation-induced RAD18 accumulation was observed from leptotene until late diplotene (Fig. 7A). We observed two different patterns of RAD18 accumulation. In leptotene, zygotene, and early pachytene nuclei, RAD18 accumulated as distinct foci on the axial elements or SC, whereas a more diffuse chromatin-associated accumulation pattern was observed in pachytene and early- to mid-diplotene (Fig. 7A). The time point at which RAD18 localization changed from a focus-like to a diffuse chromatin-associated pattern coincided with the RAD18 accumulation at the XY body. When RAD18 was lost from the XY body in late diplotene, RAD18 accumulated again as RAD51-like foci (Fig. 7A).
Fig. 7.
Accumulation of RAD18 at IR-induced DSBs. (A) Localization of RAD18 in spermatocyte nuclei of wild-type mice irradiated with IR at 4Gy. Two hours later, spread spermatocyte nuclei were prepared and double-stained with anti-SYCP3 (red) and anti-RAD18 (green) antibodies. The white line in the early pachytene image indicates the boundary of the nucleus. (B) Double immunostaining of irradiated (as in A) leptotene nucleus with anti-DMC1 (red) and anti-RAD18 (green). On the right, an enlarged region is shown with separate red and green images, to better visualize foci that do and do not colocalize. (C) Triple immunostaining of irradiated (as in A) zygotene and pachytene nuclei with anti-H2AK119ub1 (red), anti-RAD18 (green) and anti-SYCP3 (white).
Next, we analyzed whether the irradiation-induced RAD18 foci in leptotene represent actual DSB repair sites, or other types of damage that are also induced by ionizing radiation. Because both the RAD18 and RAD51 antibodies were raised in rabbits, we double-stained for RAD18 and DMC1. We observed an almost perfect colocalization of RAD51 and DMC1 in irradiated leptotene spermatocytes (supplementary material Fig. S2D), indicating that not only RAD51, but also DMC1 is recruited to damage-induced DSBs during leptotene. Although we did not observe a similar overall colocalization of the damage-induced RAD18 foci with DMC1 in leptotene (Fig. 7B), some foci clearly overlapped (Fig. 7B, enlargements), indicating that at least part of the RAD18 foci in irradiated leptotene nuclei represent DSB repair sites.
At DSB repair sites in mitotic cells, ubiquitylation of histone H2A at Lys119 (H2AK119ub1) by the ubiquitin E3 ligase RNF8 functions as a key modification to transmit the signal further to downstream components of the DSB recognition pathway (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007). H2AK119ub1 colocalizes with RAD18 at the XY body (supplementary material Fig. S3A) (van der Laan et al., 2004), and the accumulation of RAD18 to chromatin surrounding DSBs in mitotic cells depends on RNF8 activity and on the ubiquitin-binding zinc finger of RAD18 (Huang et al., 2009). When we analyzed the colocalization pattern of H2AK119ub1 and RAD18 throughout meiotic prophase in control nuclei and in irradiated nuclei, we found that when RAD18 displays the chromatin-associated pattern, it colocalizes with H2AK119ub1, and in nuclei that contained RAD18 foci, no specific accumulation of H2AK119ub1 was observed (supplementary material Fig. S3B and Fig. 7C).
RAD18 accumulates as foci at persistent DSBs in Sycp1 KO mice
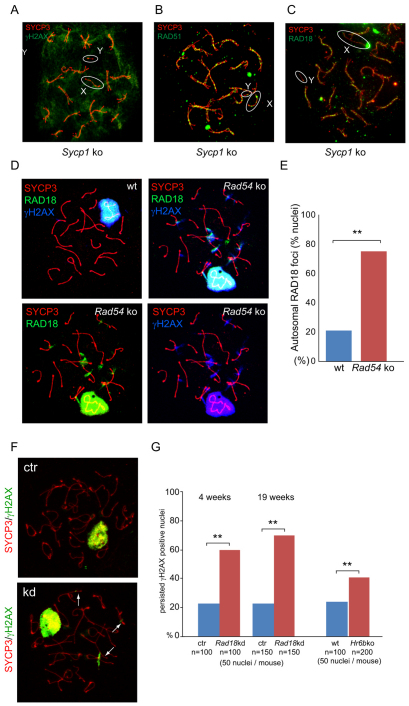
Our finding that RAD18 localizes only to a small subfraction of SPO11-induced DSBs, at a relatively late time point following the formation of these breaks and the subsequent recruitment to the chromatin surrounding the persistent DSBs on the X, indicated that the RAD18 recruitment to meiotic DSBs might only occur if these breaks persist. To study this further, we investigated RAD18 localization in spread Sycp1 KO spermatocyte nuclei. SYCP1 is a transverse filament protein that connects the lateral elements to form a SC. In Sycp1 KO mice, chromosomes align, but synapsis is not achieved. In addition, repair of meiotic DSBs is stalled, as visualized by persistent γH2AX and RAD51 staining, and no XY body is formed (Fig. 8A,B) (de Vries et al., 2005). Intriguingly, in Sycp1 KO spermatocytes, RAD18 accumulated in RAD51-like foci on all SCs in late zygotene (Fig. 8C). We also found foci-like RAD18 accumulation along the X chromosome, whereas RAD18 did not accumulate on the chromatin surrounding DSBs (Fig. 8C).
Fig. 8.
RAD18 functions in the repair of a subset of persistent DSBs in meiotic prophase. (A-C) Double immunostaining of Sycp1 KO spermatocyte nuclei with anti-SYCP3 (red) and (A) anti-γH2AX (green), (B) anti-RAD51 (green) and (C) anti-RAD18 (green) antibodies. X and Y chromosomes are indicated as X and Y, respectively. The X and Y chromosomes are shown in the white circles. (D) Triple immunostaining of wild type (only the merge is shown) and Rad54 KO (separate images are shown) in late-pachytene nuclei with anti-SYCP3 (red), anti-RAD18 (green) and anti-γH2AX (blue) antibodies. (E) The percentage of nuclei containing persistent RAD18 patches in late pachytene is shown. 100 nuclei were examined in a control and a Rad54 KO mouse. **P<0.01 (Chi-squared test). (F) Double immunostaining of control (ctr) and Rad18 KD spermatocyte nuclei with anti-SYCP3 (red) and anti-γH2AX (green). Arrows indicate γH2AX, demonstrating persistent DSBs at autosomal regions. (G) Graph showing the percentage of persistent γH2AX foci in late diplotene of 4-week-old and 19-week-old control and Rad18 KD mice, and in wild-type and Hr6b KO mice. Blue bars indicate control or wild type, and red bars indicate Rad18 KD or Hr6b KO. **P<0.01 (Chi-squared test).
RAD18 at persistent meiotic DSBs in Rad54 KO mice
RAD54 is a HR-repair protein that might also function in meiosis. In yeast meiosis, meiosis-specific proteins appear to attenuate Rad54 functions, to facilitate interhomolog repair (Niu et al., 2009). In mammals, two yeast Rad54 orthologs, RAD54 and RAD54B, have been identified (Hiramoto et al., 1999). Both Rad54 single-knockout (Essers et al., 1997) and Rad54,Rad54b double-knockout mice (Wesoly et al., 2006) are viable, fertile and show normal development. However, persistent RAD51 foci and formation of RAD51 aggregates has been reported in diplotene spermatocyte of both Rad54 single- and double-knockout mice (Wesoly et al., 2006). To study whether RAD18 also shows aberrant localization in spermatocytes on this somewhat HR-deficient background, we examined accumulation of RAD18 in Rad54 KO spermatocytes. In late pachytene or early diplotene spermatocytes, more-frequent RAD18 accumulation at several areas of autosomal chromatin was found in Rad54 KO mice, compared with the wild type (Fig. 8D,E). Here, RAD18 did not accumulate as foci, but displayed a more diffuse pattern on the chromatin, which was similar to the pattern observed at radiation-induced breaks at this stage (late pachytene or early diplotene) (Fig. 7A). At these RAD18-positive sites in Rad54 KO late pachytene or early diplotene spermatocytes, γH2AX accumulation was also observed (Fig. 8D). These RAD18- and γH2AX-positive sites were still present in mid to late diplotene (not shown). These data indicate that RAD18 is recruited to sites of persisting DNA damage because of the lack of RAD54.
Persistent γH2AX on autosomal chromatin in late diplotene spermatocytes of Rad18 KD and Hr6b KO mice
Similarly to DSB repair sites in mitotic cells, the surrounding chromatin of SPO11-induced DSBs was marked by γH2AX in leptotene and zygotene nuclei (supplementary material Fig. S2C) (Mahadevaiah et al., 2001). In addition, γH2AX is known as the earliest marker of the XY body (Mahadevaiah et al., 2001). As pachytene progresses, the γH2AX intensity decreases at autosomes but not on the XY body, and γH2AX has disappeared from autosomes in diplotene (supplementary material Fig. S2C) (Mahadevaiah et al., 2001). We observed that γH2AX was still present on the XY body just before entering metaphase in control nuclei (supplementary material Fig. S2C). In young as well as adult Rad18 KD mice, the γH2AX staining pattern was similar to that of control mice from leptotene to pachytene. However, γH2AX-positive sites still marked autosomes in 60% of late diplotene nuclei in Rad18 KD mice (Fig. 8F,G). This was much less frequently observed in control mice. To examine whether this function depends on the presence of a functional RAD18–HR6A/B complex, we examined γH2AX staining in Hr6b KO mice. Similarly to Rad18 KD mice, approximately 40% of late diplotene nuclei showed accumulation of γH2AX at autosomal chromatin, compared with approximately 20% of the nuclei in wild-type testis (Fig. 8G).
Discussion
RAD18 and HR6A/B
RAD18 is most well known for its role in RDB that allows progression of DNA replication in the presence of DNA damage in mitotic cells (reviewed in Chang and Cimprich, 2009; Guo et al., 2009; Livneh et al., 2010). In addition, RAD18 is also required for DSB repair (Huang et al., 2009; Watanabe et al., 2009). Herein, we analyzed Rad18 KD mice to study the role of RAD18 in meiotic DSB repair. RAD18 and HR6A/B function as a complex. RAD18 binds to DNA directly via its SAP domain, or indirectly via its zinc finger, which might interact with ubiquitylated chromatin components such as ubiquitylated histones (Huang et al., 2009; Notenboom et al., 2007). HR6A/B does not have any known direct DNA or chromatin binding properties. We found that the localization of HR6A/B to chromatin, including the XY body, is at least partially dependent on RAD18. The intimate functional and physical relation between the two proteins would suggest that loss of the proteins might lead to similar phenotypes, and identification of overlapping phenotypic characteristics could help to distinguish between RAD18-dependent and RAD18-independent functions of HR6A/B. Here, we show that the regulation of the H3K4me2 level at the XY body by HR6B is RAD18 dependent. None of the other meiotic phenotypic characteristics of the Hr6b KO mice were observed in the Rad18 KD mice. In addition to RAD18, HR6A/B also functions with other E3 ligases, such as RNF20 and RNF40 to ubiquitylate histone H2B at Lys120 (H2BK120ubi) (Kim et al., 2009). HR6A/B also functions with members of the UBR family of E3 ligases (Kwon et al., 2003; Kwon et al., 2001; Tasaki and Kwon, 2007). Thus, most likely the other functions of HR6B in meiosis depend on these, or as yet unknown, E3 ligases.
HR6B also functions during postmeiotic spermatid differentiation, because condensing spermatids of Hr6b KO mice showed severe morphological disturbances (Roest et al., 1996). In addition, the expression of many genes (approximately 25% of the annotated genes) becomes disturbed in Hr6b KO round spermatids (Mulugeta Achame et al., 2010). In Rad18 KD mice, we noted a milder disturbance of postmeiotic spermatid development. It is not excluded that a low level of remaining RAD18 expression in Rad18 KD spermatids generates a partial phenotype. However, the postmeiotic expression of X- and Y-linked genes might be affected in a similar way as in Hr6b KO mice. Because we used total testis material to analyze mRNA expression in this analysis, it is not possible to pinpoint the cell types that show increased expression of X- and Y-linked genes. Recently, Royo and colleagues (Royo et al., 2010), elegantly showed that disruption of MSCI in pachytene spermatocytes causes cell death at a stage corresponding to Stage IV of the spermatogenic cycle. In addition, it was found that inappropriate expression of either Zfy1 or Zfy2 (both Y chromosome genes) in pachytene spermatocytes was sufficient to trigger apoptosis. Here, we also observe upregulation of Zfy2 (not Zfy1), but no pachytene arrest or cell death. Most likely, upregulation of Zfy2 in Rad18 KD spermatocytes does not start before the diplotene stage, when increased H3K4me2 is first detected. Thus, we suggest that derepression of X- and Y-chromosomal genes starts in Rad18 KD (and Hr6b KO) diplotene spermatocytes, or thereafter. At this phase in their development, they are probably no longer sensitive to Zfy2.
We observed an increased frequency of XY asynapsis in Rad18 KD mice. This phenotype is not present in Hr6b KO mice. It cannot be excluded that RAD18 carries out some HR6B-independent functions. However, we presume that it is more likely that this phenotype is related to systemic dysregulation in Rad18 KD mice that is evidenced by the overall decrease in body weight. In Atm KO mice, a similar effect on XY synapsis was described (Barchi et al., 2008). Atm-deficient mice also display reduced body weight compared with their littermate controls (Xu et al., 1996). In these mice, overall meiotic recombination was increased (Barchi et al., 2008), indicating that the XY asynapsis could not be caused by decreased meiotic recombination. The observed asynapsis could result from premature desynapsis or from a complete lack of synapsis. If the latter process occurs, we would expect to find a relative reduction in the frequency of MLH1 focus formation of the XY pair, and an increase in the number of spermatids with aberrant sex chromosome constitution. Because we did not observe any aberration in the frequency of MLH1 focus formation on the XY pair, and found normal frequencies of X and Y bearing spermatids, we suggest that premature XY desynapsis occurs in the Rad18 KD spermatocytes.
Sun and co-workers (Sun et al., 2009) described the testicular phenotype of Rad18 KO mice. They reported normal spermatogenesis in young mice, and progressive loss of stem cells in old (>12 months) Rad18 KO mice. Because they did not study meiotic prophase in detail in their mouse model, and the meiotic defects that we observed in the Rad18 KD mice are rather subtle, it is not excluded that similar defects will be present in these males. Conversely, we do not exclude that upon aging, Rad18 KD mice will show a similar loss of stem cells as was described for the Rad18 KO mice. The average litter size of the control mice in their study (Sun et al., 2009) is much lower compared to what we observed, indicating that the genetic background might also be different. Finally, the Rad18 KO allele was generated through insertional mutagenesis, and it cannot be excluded that (truncated) RAD18 is expressed in some tissues in the knockout mouse model, leading to a hypomorphic phenotype.
Recruitment of RAD18 to persistent meiotic DSBs and chromatin of the XY body
In the present study, we found that RAD18 accumulates only at a small subset of meiotic DSBs in late zygotene to early pachytene, but not in leptotene, in a SPO11-dependent manner. This subset of meiotic DSBs might represent persistent DSBs, because in Sycp1 KO mice, in which many DSBs remain unrepaired owing to the failure of complete chromosome synapsis, many RAD18 foci appear along the chromosomal axes. Our finding that RAD18 is able to associate with radiation-induced DSBs in leptotene indicates that the amount of RAD18 is not limiting at this stage. Thus, it appears that factors involved in recruiting RAD18 to damage-induced DSBs are absent or masked at SPO11-induced DSBs in leptotene. We suggest that the two known RAD18 interaction partners RPA (Davies et al., 2008) and RAD51C (Huang et al., 2009) are somehow involved in the recruitment of RAD18 only to the persistent meiotic DSBs in late zygotene. The recruitment of RAD18 to the chromatin surrounding persistent meiotic (in Rad18 KD and Rad54 KO spermatocytes) or damage-induced DSBs is most likely mediated by H2AK119 ubiquitylation, which provides a binding platform for the ubiquitin-binding zinc finger of RAD18.
Chromosomes or chromosomal regions without a pairing partner can only repair their meiotic DSBs in the unpaired regions through recombination with the sister chromatid or by NHEJ. Because both these pathways appear to be repressed during early meiotic prophase, meiotic DSBs in such regions might remain unrepaired. Indeed, persistent RAD51 foci are observed along the unsynapsed arm of the X chromosome in pachytene (Moens et al., 1997). Surprisingly, these are only rarely observed along the unsynapsed part of the Y, suggesting that SPO11 generates only a few breaks in this region of the Y, or that these breaks are repaired via an alternative, rapid pathway. Corresponding to these persistent meiotic DSBs on the X chromosome in pachytene, RAD18 accumulates first on the chromatin regions of the X chromosome, and subsequently spreads to the synapsed chromatin and the region of the Y chromosome. This suggests that the persistent DSBs on the X might stimulate recruitment of RAD18 to the X chromosome, followed by spreading to the Y chromosome.
Possible role of RAD18 in inter-sister-chromatid-mediated repair of persistent meiotic DSBs
The repair mechanism for DSBs on the X and Y chromosomes remains unclear. Recent analyses of radiation-induced DSB repair in spermatocytes revealed that the NHEJ pathway is reactivated in late pachytene spermatocytes, and most damage-induced DSBs appear to be repaired via this pathway, although the HR pathway takes over when NHEJ is compromised (Ahmed et al., 2010). For the persistent breaks at the XY body, it is not known whether NHEJ or HR (using the sister chromatid as a template for repair) is the favoured pathway. Because we did not observe a change in the dynamics of RAD51 foci on the XY body in Rad18 KD mice, it is unlikely that RAD18 is required for the repair of these breaks. Moreover, the fact that RAD51 foci disappear from the XY body before γH2AX, suggests that the NHEJ pathway mediates repair of persistent DSBs on the unsynapsed XY axes. This would fit with the idea that inter-sister-chromatid-mediated DSB repair should be inhibited as long as synapsis is not achieved, which maintains the interhomolog bias to stimulate chromosome pairing. In yeast, a protein named Hop1 triggers dimerization and activation of a kinase Mek1 (Carballo et al., 2008; Niu et al., 2005). Mek1 subsequently mediates phosphorylation of Rad54, which inhibits its activity, and is involved in mediating the interhomolog bias (Niu et al., 2009). In mammals, two possible Hop1 homologs, Hormad1 and Hormad2 have been identified (Wojtasz et al., 2009). HORMAD1 and HORMAD2 preferentially accumulate on unsynapsed chromosome axes in spermatocytes, and are removed upon synapsis. This is consistent with the idea that the interhomolog bias could be relieved upon synapsis. Thus, at synapsed autosomal sites, on paired chromosomes that repaired most of the breaks and formed at least a single crossover, the inhibition to repair by the sister chromatid might be relieved. At such DSB sites, RAD18 and RAD54 might function together to mediate inter-sister chromatid repair.
Taken together, our data indicate that RAD18, together with HR6B, might ubiquitylate an unknown substrate to facilitate repair of persisting meiotic DSBs at synapsed regions, possibly using the sister chromatid as a template for repair in late zygotene and early pachytene. In the absence of RAD18, these DSB sites persist until the NHEJ pathway is reactivated in diplotene. At the XY body, the function of the RAD18–HR6B complex might be different. At the DSB sites along the unsynapsed X, it might try to stimulate HR, but fails to succeed because of the inhibition of inter-sister-chromatid-mediated repair. On the surrounding chromatin RAD18 might be involved in regulating chromatin structure and thereby help to maintain MSCI.
Materials and Methods
Generation of Rad18 KD and control mice
The recombinase mediated cassette exchange (RMCE) for the efficient generation of targeted transgenes has been previously described (Seibler et al., 2005). Into the exchange vector for RMCE (pRMCE-U6) the following oligonucleotides representing the shRNA sequence against the Rad18 mRNA were cloned using the restriction endonucleases BbsI and AscI, (20-1 s: 5′-ACCGCTGAAACAGTATGGCTTATTCAAGAGATAAGCCATACTGTTTCAGCTTTTTGG-3′) and (20-1 as, 5′-CGCGCCAAAAAGCTGAAACAGTATGGCTTATCTCTTGAATAAGCCATACTGTTTCAG-3′). The resulting vector is called pshRad18 and contains the following elements in 5′ to 3′ direction: a synthetic polyA signal, an F3 site, a neomycin-resistance gene lacking the start ATG, the Pgk polyadenylation signal, the human U6 promoter, shRNA 20-1 and an FRT site.
A empty vector control plasmid contained the following elements in 5′ to 3′ direction: a synthetic polyA signal, an F3 site, a neomycin-resistance gene lacking the start ATG, the Pgk polyadenylation signal and an FRT site. ES cell culture was carried out as described previously (Hogan et al., 1994). The generation of ES cells carrying the RMCE configuration at the Rosa26 locus with the Rad18 shRNA expression vector or the control vector was performed as described (Seibler et al., 2005).
All animal experiments were approved by the local animal experiments committee. Mice heterozygote for the targeted allele with the Rad18 shRNA and mice heterozygote for the control vector at the Rosa26 locus were generated via tetraploid embryo complementation as described (Seibler et al., 2005). Founder mice were backcrossed to C57BL/6 mice. Mice were genotyped using specific primers for the targeted control sequence and Rad18 shRNA; forward, 5′-CATCAGAAGCTGACTCTAGATGGC-3′; reverse, 5′-CTTGTCCCTCCAATTTTACACC-3′ with conditions of 95°C 5 minutes, 35 cycles of 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 1 second, and then 72°C for 5 minutes. Amplification of DNA from Rad18 shRNA mice resulted in a 688 bp PCR fragment and from the targeted control in a 250 bp PCR fragment
Spo11 mutant mice, and Sycp1 and Rad54 KO mice
We used a Spo11-knock-in mouse model in which the catalytically active Tyr100 residue is replaced by a Phe. Identically to the Spo11 knockout (Bellani et al., 2005), male and female double-knock-in mice (KI) are infertile, and meiotic prophase is blocked, with spermatocytes and oocytes reaching a zygotene-like stage with variable degrees of (heterologous) synapsis (data not shown). Sycp1 and Rad54 KO mice were described previously (de Vries et al., 2005; Essers et al., 1997).
Analyses of fertility and sperm count
Adult heterozygous male and female control and Rad18 KD mice were bred with control females and males, respectively for a maximum of 6 weeks. Litter number and litter size were recorded. The number of spermatozoa was counted as described (Roest et al., 1996). At least 200 sperm in two different samples from three animals were counted.
Irradiation of mice
Adult wild-type mice were subjected to whole-body γ-irradiation with Elekta linear accelerator (Crawley). Mice received a total dose of 4 Gy and were sacrificed at 2 hours after irradiation. Testes were collected and used to prepare spread nuclei preparations as described below.
Antibodies
For primary antibodies, we used mouse monoclonal antibodies anti-phosphorylated H2AX (Upstate), anti-ubiquitylated histone H2A (Millipore, #05-678), anti-DMC1 (Abcam) and anti-MLH1 (Becton and Dickinson), rabbit polyclonal antibodies anti-RAD18 (Inagaki et al., 2009), anti-RAD51 (Essers et al., 2002), anti-HR6A/B (Baarends et al., 2003), anti-SYCP3 (gift from C. Heyting, Wageningen University, Wageningen, The Netherlands), and anti-H3K4me2 (Upstate), and rat polyclonal antibody anti-SYCP3 (Baarends et al., 2007). For secondary antibodies, we used a goat anti-IgG-peroxidase (rabbit/mouse) (Sigma), goat anti-IgG (rabbit) Alexa Fluor 488, 564 or 633, goat anti-IgG (mouse) Alexa Fluor 488, 564 or 633, goat antibody against rat IgG Alexa Fluor 488, 564 or 633, or goat antibodies against mouse IgM Alexa Fluor 564 (Molecular Probes).
Protein isolation and immunoblot analysis
Cell lysates from testes were prepared in 2 ml lysis buffer [50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 0.2% v/v Nonidet P-40, 10% v/v glycerol, 0.5 mM dithiothreitol, and protease inhibitors (Roche)]. Testis tissue was homogenized and sonicated. The expression level of protein from whole-cell lysates was analyzed by SDS-PAGE and immunoblotting was carried out as described (Inagaki et al., 2009).
Meiotic spread nuclei preparations and immunocytochemistry
Testis tissues were processed to obtain spread nuclei for immunocytochemistry as described (Peters et al., 1997). Spread nuclei of spermatocytes were stained as described previously (Baarends et al., 2007) with the antibodies mentioned above.
Histological analysis
Testes were isolated from control and heterozygous Rad18 KD mice that were 4 and 19 weeks old. Testes were fixed in Bouins' fixative for morphological analysis, and embedded in paraffin according to standard procedures. Mounted sections were deparaffinized, rehydrated and stained with hematoxylin and eosin.
Confocal microscopy
Images of cells were obtained using a Zeiss LSM510NLO microscope (Carl Zeiss) as described previously (Inagaki et al., 2009). For quantification of immunofluorescent signal, slides were analyzed on the same day. Fluorescent images were taken under identical conditions for all slides, and images were analyzed using ImageJ software (National Institutes of Health, Bethesda, MD). Nuclei were selected and the mean (μ) and standard deviation (σ) was calculated using ImageJ software, and the threshold was determined (van Royen et al., 2007). To select the HR6A/B or H3K4me2 positive area on the XY body, threshold was set by using the formula: μ + 1.5*σ. To count the number of RAD51 or DMC1 foci, threshold was set by using the formula, μ + 2.5*σ. To further prevent counting non-specific background as RAD51 or DMC1 foci, areas with a signal above the threshold that was less than 2 pixels in size were not counted. The intensity of selected area and the number of foci were counted using ImageJ. For colocalization analysis of RAD18 with DMC1, the AIM software (Carl Zeiss, Jena) was used for data analysis. When pachytene nuclei were analyzed, the XY body area was excluded.
Real-time RT-PCR
For real-time RT-PCR, RNA was prepared from testes using Trizol, treated with DNase and reverse transcribed using random hexamer primers and Superscript II reverse transcriptase (Invitrogen). PCR was carried out with the iQ SYBR green PCR mastermix (Applied Biosystems) in the DNA engine Opticon 2 real-time PCR detection system (Bio-Rad). The names of examined genes, primer sequences, and annealing temperatures are shown in supplementary material Table S1. PCR of mRNA encoding β-actin was included in each reaction and used to normalize the data. Three independent experiments were performed and each real-time PCR was performed in duplicate. All -RT reactions were negative.
Statistics
The Mann–Whitney U-test or Chi-squared test was used for the significance tests.
Supplementary Material
Acknowledgments
We thank Hein te Riele and Sandra de Vries (The Netherlands Cancer Institute, Amsterdam, The Netherlands) for Spo11 mutant mice, Albert Pastink (Leiden University Medical Center, Leiden, The Netherlands) for Sycp1 KO mice and Roland Kanaar (Erasmus MC, Rotterdam, The Netherlands) for Rad54 KO mice. This work was supported by the Netherlands Organization for Scientific Research (NWO) through ALW (VIDI 864.05.003). Deposited in PMC for immediate release.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/16/2837/DC1
References
- Ahmed E. A., Philippens M. E., Kal H. B., de Rooij D. G., de Boer P. (2010). Genetic probing of homologous recombination and non-homologous end joining during meiotic prophase in irradiated mouse spermatocytes. Mutat. Res. 688, 12-18 [DOI] [PubMed] [Google Scholar]
- Ashley T., Plug A. W., Xu J., Solari A. J., Reddy G., Golub E. I., Ward D. C. (1995). Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma 104, 19-28 [DOI] [PubMed] [Google Scholar]
- Baarends W. M., Hoogerbrugge J. W., Roest H. P., Ooms M., Vreeburg J., Hoeijmakers J. H., Grootegoed J. A. (1999). Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207, 322-333 [DOI] [PubMed] [Google Scholar]
- Baarends W. M., Wassenaar E., Hoogerbrugge J. W., van Cappellen G., Roest H. P., Vreeburg J., Ooms M., Hoeijmakers J. H., Grootegoed J. A. (2003). Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol. Cell. Biol. 23, 1151-1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baarends W. M., Wassenaar E., Hoogerbrugge J. W., Schoenmakers S., Sun Z. W., Grootegoed J. A. (2007). Increased phosphorylation and dimethylation of XY body histones in the Hr6b-knockout mouse is associated with derepression of the X chromosome. J. Cell Sci. 120, 1841-1851 [DOI] [PubMed] [Google Scholar]
- Barchi M., Roig I., Di Giacomo M., de Rooij D. G., Keeney S., Jasin M. (2008). ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 4, e1000076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow A. L., Benson F. E., West S. C., Hulten M. A. (1997). Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 16, 5207-5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Manova K., Yuen J. P., Jasin M., Keeney S. (2000). Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989-998 [DOI] [PubMed] [Google Scholar]
- Bellani M. A., Romanienko P. J., Cairatti D. A., Camerini-Otero R. D. (2005). SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J. Cell Sci. 118, 3233-3245 [DOI] [PubMed] [Google Scholar]
- Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. (2001). ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276, 42462-42467 [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. (1990). A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61, 1089-1101 [DOI] [PubMed] [Google Scholar]
- Carballo J. A., Johnson A. L., Sedgwick S. G., Cha R. S. (2008). Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132, 758-770 [DOI] [PubMed] [Google Scholar]
- Chang D. J., Cimprich K. A. (2009). DNA damage tolerance: when it's OK to make mistakes. Nat. Chem. Biol. 5, 82-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicheportiche A., Bernardino-Sgherri J., de Massy B., Dutrillaux B. (2007). Characterization of Spo11-dependent and independent phospho-H2AX foci during meiotic prophase I in the male mouse. J. Cell Sci. 120, 1733-1742 [DOI] [PubMed] [Google Scholar]
- Davies A. A., Huttner D., Daigaku Y., Chen S., Ulrich H. D. (2008). Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol. Cell 29, 625-636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries F. A., de Boer E., van den Bosch M., Baarends W. M., Ooms M., Yuan L., Liu J. G., van Zeeland A. A., Heyting C., Pastink A. (2005). Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376-1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling E. L., Maloney D. H., Fogel S. (1985). Meiotic recombination and sporulation in repair-deficient strains of yeast. Genetics 109, 283-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., Hendriks R. W., Swagemakers S. M., Troelstra C., de Wit J., Bootsma D., Hoeijmakers J. H., Kanaar R. (1997). Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell 89, 195-204 [DOI] [PubMed] [Google Scholar]
- Essers J., Hendriks R. W., Wesoly J., Beerens C. E., Smit B., Hoeijmakers J. H., Wyman C., Dronkert M. L., Kanaar R. (2002). Analysis of mouse Rad54 expression and its implications for homologous recombination. DNA Repair (Amst.) 1, 779-793 [DOI] [PubMed] [Google Scholar]
- Fabre F., Magana-Schwencke N., Chanet R. (1989). Isolation of the RAD18 gene of Saccharomyces cerevisiae and construction of rad18 deletion mutants. Mol. Gen. Genet. 215, 425-430 [DOI] [PubMed] [Google Scholar]
- Game J. C., Mortimer R. K. (1974). A genetic study of x-ray sensitive mutants in yeast. Mutat. Res. 24, 281-292 [DOI] [PubMed] [Google Scholar]
- Goedecke W., Eijpe M., Offenberg H. H., van Aalderen M., Heyting C. (1999). Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat. Genet. 23, 194-198 [DOI] [PubMed] [Google Scholar]
- Guo C., Kosarek-Stancel J. N., Tang T. S., Friedberg E. C. (2009). Y-family DNA polymerases in mammalian cells. Cell. Mol. Life Sci. 66, 2363-2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyting C., Moens P. B., van Raamsdonk W., Dietrich A. J., Vink A. C., Redeker E. J. (1987). Identification of two major components of the lateral elements of synaptonemal complexes of the rat. Eur. J. Cell Biol. 43, 148-154 [PubMed] [Google Scholar]
- Hiramoto T., Nakanishi T., Sumiyoshi T., Fukuda T., Matsuura S., Tauchi H., Komatsu K., Shibasaki Y., Inui H., Watatani M., et al. (1999). Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene 18, 3422-3426 [DOI] [PubMed] [Google Scholar]
- Hogan B. L., Blessing M., Winnier G. E., Suzuki N., Jones C. M. (1994). Growth factors in development: the role of TGF-beta related polypeptide signalling molecules in embryogenesis. Dev. Suppl. 1994, 53-60 [PubMed] [Google Scholar]
- Huang J., Huen M. S., Kim H., Leung C. C., Glover J. N., Yu X., Chen J. (2009). RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 11, 592-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007). RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901-914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki A., van Cappellen W. A., van der Laan R., Houtsmuller A. B., Hoeijmakers J. H., Grootegoed J. A., Baarends W. M. (2009). Dynamic localization of human RAD18 during the cell cycle and a functional connection with DNA double-strand break repair. DNA Repair (Amst.) 8, 190-201 [DOI] [PubMed] [Google Scholar]
- Inagaki A., Schoenmakers S., Baarends W. M. (2010). DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics 5, 255-266 [DOI] [PubMed] [Google Scholar]
- Jones J. S., Prakash L. (1991). Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD18 increase in UV irradiated cells and during meiosis but not during the mitotic cell cycle. Nucleic Acids Res. 19, 893-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., Kleckner N. (1997). Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375-384 [DOI] [PubMed] [Google Scholar]
- Keeney S., Baudat F., Angeles M., Zhou Z. H., Copeland N. G., Jenkins N. A., Manova K., Jasin M. (1999). A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics 61, 170-182 [DOI] [PubMed] [Google Scholar]
- Kim J., Guermah M., McGinty R. K., Lee J. S., Tang Z., Milne T. A., Shilatifard A., Muir T. W., Roeder R. G. (2009). RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137, 459-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken M. H., Reynolds P., Jaspers-Dekker I., Prakash L., Prakash S., Bootsma D., Hoeijmakers J. H. (1991). Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. USA 88, 8865-8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., et al. (2007). Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637-1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. T., Xia Z., Davydov I. V., Lecker S. H., Varshavsky A. (2001). Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol. Cell. Biol. 21, 8007-8021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. T., Xia Z., An J. Y., Tasaki T., Davydov I. V., Seo J. W., Sheng J., Xie Y., Varshavsky A. (2003). Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 23, 8255-8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh Z., Ziv O., Shachar S. (2010). Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle 9, 729-735 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah S. K., Turner J. M., Baudat F., Rogakou E. P., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W. M., Burgoyne P. S. (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27, 271-276 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007). RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887-900 [DOI] [PubMed] [Google Scholar]
- Miyase S., Tateishi S., Watanabe K., Tomita K., Suzuki K., Inoue H., Yamaizumi M. (2005). Differential regulation of Rad18 through Rad6-dependent mono- and polyubiquitination. J. Biol. Chem. 280, 515-524 [DOI] [PubMed] [Google Scholar]
- Moens P. B., Chen D. J., Shen Z., Kolas N., Tarsounas M., Heng H. H., Spyropoulos B. (1997). Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106, 207-215 [DOI] [PubMed] [Google Scholar]
- Monesi V. (1965). Differential rate of ribonucleic acid synthesis in the autosomes and sex chromosomes during male meiosis in the mouse. Chromosoma 17, 11-21 [DOI] [PubMed] [Google Scholar]
- Moses M. J. (1977). Synaptonemal complex karyotyping in spermatocytes of the Chinese hamster (Cricetulus griseus). II. Morphology of the XY pair in spread preparations. Chromosoma 60, 127-137 [DOI] [PubMed] [Google Scholar]
- Mulugeta Achame E., Wassenaar E., Hoogerbrugge J. W., Sleddens-Linkels E., Ooms M., Sun Z. W., van Ijcken W. F., Grootegoed J. A., Baarends W. M. (2010). The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids. BMC Genomics 11, 367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa S. H., Park P. J., Zhang L. F., Shima J. E., McCarrey J. R., Griswold M. D., Lee J. T. (2006). Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16, 660-667 [DOI] [PubMed] [Google Scholar]
- Neale M. J., Pan J., Keeney S. (2005). Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Baumgartner B., Schaefer D., Loidl J., Hollingsworth N. M. (2005). Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 16, 5804-5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Busygina V., Kwon Y., Allen J. A., Li X., Kunz R. C., Kubota K., Wang B., Sung P., et al. (2009). Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell 36, 393-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notenboom V., Hibbert R. G., van Rossum-Fikkert S. E., Olsen J. V., Mann M., Sixma T. K. (2007). Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 35, 5819-5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. H., Plug A. W., van Vugt M. J., de Boer P. (1997). A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66-68 [DOI] [PubMed] [Google Scholar]
- Pittman D. L., Cobb J., Schimenti K. J., Wilson L. A., Cooper D. M., Brignull E., Handel M. A., Schimenti J. C. (1998). Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1, 697-705 [DOI] [PubMed] [Google Scholar]
- Roest H. P., van Klaveren J., de Wit J., van Gurp C. G., Koken M. H. M., Vermey M., van Roijen J. H., Vreeburg J. T. M., Baarends W. M., Bootsma D., et al. (1996). Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes a defect in spermatogenesis associated with chromatin modification. Cell 86, 799-810 [DOI] [PubMed] [Google Scholar]
- Roest H. P., Baarends W. M., de Wit J., van Klaveren J. W., Wassenaar E., Hoogerbrugge J. W., van Cappellen W. A., Hoeijmakers J. H., Grootegoed J. A. (2004). The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol. Cell. Biol. 24, 5485-5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko P. J., Camerini-Otero R. D. (2000). The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975-987 [DOI] [PubMed] [Google Scholar]
- Royo H., Polikiewicz G., Mahadevaiah S. K., Prosser H., Mitchell M., Bradley A., de Rooij D. G., Burgoyne P. S., Turner J. M. (2010). Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 20, 2117-2123 [DOI] [PubMed] [Google Scholar]
- Schoenmakers S., Wassenaar E., van Cappellen W. A., Derijck A. A., de Boer P., Laven J. S., Grootegoed J. A., Baarends W. M. (2008). Increased frequency of asynapsis and associated meiotic silencing of heterologous chromatin in the presence of irradiation-induced extra DNA double strand breaks. Dev. Biol. 317, 270-281 [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. (1994). Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 76, 51-63 [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. (1997). Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90, 1123-1135 [DOI] [PubMed] [Google Scholar]
- Seibler J., Kuter-Luks B., Kern H., Streu S., Plum L., Mauer J., Kuhn R., Bruning J. C., Schwenk F. (2005). Single copy shRNA configuration for ubiquitous gene knockdown in mice. Nucleic Acids Res. 33, e67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi N., Mori M., Tsuji H., Imai T., Inoue H., Tateishi S., Yamaizumi M., Shiomi T. (2007). Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 35, e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Yomogida K., Sakao S., Yamamoto H., Yoshida K., Watanabe K., Morita T., Araki K., Yamamura K., Tateishi S. (2009). Rad18 is required for long-term maintenance of spermatogenesis in mouse testes. Mech. Dev. 126, 173-183 [DOI] [PubMed] [Google Scholar]
- Tarsounas M., Morita T., Pearlman R. E., Moens P. B. (1999). RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 147, 207-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T., Kwon Y. T. (2007). The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem. Sci. 32, 520-528 [DOI] [PubMed] [Google Scholar]
- Tateishi S., Niwa H., Miyazaki J., Fujimoto S., Inoue H., Yamaizumi M. (2003). Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 23, 474-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan R., Roest H., Hoogerbrugge J., Smit E., Slater R., Baarends W., Hoeijmakers J., Grootegoed J. (2000). Characterization of mRAD18Sc, a mouse homolog of the yeast post-replication repair gen RAD18. Genomics 69, 86-94 [DOI] [PubMed] [Google Scholar]
- van der Laan R., Uringa E. J., Wassenaar E., Hoogerbrugge J. W., Sleddens E., Odijk H., Roest H. P., de Boer P., Hoeijmakers J. H., Grootegoed J. A., et al. (2004). Ubiquitin ligase Rad18Sc localizes to the XY body and to other chromosomal regions that are unpaired and transcriptionally silenced during male meiotic prophase. J. Cell Sci. 117, 5023-5033 [DOI] [PubMed] [Google Scholar]
- van Royen M. E., Cunha S. M., Brink M. C., Mattern K. A., Nigg A. L., Dubbink H. J., Verschure P. J., Trapman J., Houtsmuller A. B. (2007). Compartmentalization of androgen receptor protein-protein interactions in living cells. J. Cell Biol. 177, 63-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Iwabuchi K., Sun J., Tsuji Y., Tani T., Tokunaga K., Date T., Hashimoto M., Yamaizumi M., Tateishi S. (2009). RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res. 37, 2176-2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesoly J., Agarwal S., Sigurdsson S., Bussen W., Van Komen S., Qin J., van Steeg H., van Benthem J., Wassenaar E., Baarends W. M., et al. (2006). Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell. Biol. 26, 976-989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L., Daniel K., Roig I., Bolcun-Filas E., Xu H., Boonsanay V., Eckmann C. R., Cooke H. J., Jasin M., Keeney S., et al. (2009). Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 5, e1000702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ashley T., Brainerd E. E., Bronson R. T., Meyn M. S., Baltimore D. (1996). Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 10, 2411-2422 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Kondoh G., Matsuda Y., Habu T., Nishimune Y., Morita T. (1998). The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell 1, 707-718 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.