Abstract
Hermansky-Pudlak syndrome is an autosomal recessive condition characterized by a bleeding diathesis and hypopigmentation of the skin, hair and eyes. Some HPS patients develop other complications such as granulomatous colitis and/or a fatal pulmonary fibrosis. Eight genes have been associated with the condition, resulting in subtypes HPS-1 through HPS-8. The HPS gene products are involved in the biogenesis of specialized lysosome-related organelles such as melanosomes, platelet delta granules and others. HPS1 and HPS4 form a stable complex named BLOC-3, and patients with BLOC-3 or AP-3 deficiency develop pulmonary fibrosis. Therefore, it is important to subtype each HPS patient. HPS type 1 (HPS-1) occurs frequently on the island Puerto Rico due to a founder mutation. Here, we describe seven mutations, six of which are previously unreported, in the HPS1, HPS4 and HPS5 genes among patients of Mexican, Uruguayan, Honduran, Cuban, Venezuelan and Salvadoran ancestries. Our findings demonstrate that the diagnosis of HPS should be considered in Hispanic patients with oculocutaneous albinism and bleeding symptoms. Moreover, such patients should not be assumed to have the HPS-1 subtype typical of northwest Puerto Rican patients. We recommend molecular HPS subtyping in such cases, since it may have significant implications for prognosis and intervention.
Keywords: albinism, bleeding diathesis, Hermansky-Pudlak syndrome, Hispanic descent, lysosome-related organelle, pulmonary fibrosis
Introduction
Hermansky-Pudlak syndrome (HPS; OMIM 203300) is a rare multisystemic, disorder characterized by oculocutaneous albinism, and a bleeding diathesis, sometimes accompanied by immunodeficiency, granulomatous colitis and/ or fatal pulmonary fibrosis (Hermansky and Pudlak, 1959; Gahl et al., 1998; Brantly et al., 2000; Schinella et al., 1980). These clinical manifestations are due to defects in the formation or function of lysosome-related organelles such as melanosomes in melanocytes, platelet delta granules, lung lamellar bodies and/or lytic granules of cytotoxic T-cells (Wei, 2006; Huizing et al., 2008). HPS is caused by defects in one of eight genes: HPS1, AP3B1, HPS3 to HPS8 (Oh et al., 1996; Dell’Angelica et al., 1999; Anikster et al., 2001; Suzuki et al., 2002; Li et al., 2003; Zhang et al., 2003; Morgan et al., 2006). With the exception of the gene encoding a subunit of AP-3 (HPS-2) (Dell’Angelica et al., 1999), the HPS genes encode novel proteins of unknown function that have no homology to any other protein and no recognizable motif. In addition, HPS gene products have been identified as subunits of at least three novel multi-protein complexes named Biogenesis of Lysosome-related Organelles Complex (BLOC)-1 through -3. BLOC-1 is a multimeric complex composed of HPS7 and HPS8, among other subunits (Starcevic et al., 2004; Li et al., 2003; Morgan et al., 2006). BLOC-2 is composed of HPS3, HPS5 and HPS6 (Di Pietro et al., 2004), while HPS1 and HPS4 are subunits of BLOC-3 (Martina et al., 2003; Nazarian et al., 2003). Deficiency of any of these complexes can affect intracellular trafficking of proteins (Dell’Angelica, 2004).
Patients who have mutations in the same BLOC exhibit similar phenotypes (Wei, 2006; Huizing et al., 2008). BLOC-3 deficient patients exhibit a relatively more severe phenotype of hypopigmentation, frequently develop granulomatous colitis, and suffer a fatal, adult-onset pulmonary fibrosis (Huizing et al., 2008; Anderson et al., 2003; Hermos et al., 2002). Patients with deficiency in BLOC-2 manifest a milder phenotype of variable hypopigmentation and sporadic granulomatous colitis, but pulmonary fibrosis has not been found in BLOC-2 patients (Huizing et al., 2001; Huizing et al., 2004; Huizing et al. 2009). So far, only two BLOC-1 patients have been reported, one HPS-7 patient (Li et al., 2003) and one HPS-8 family (Morgan et al., 2006), but no detailed clinical features were provided apart from hypopigmentation, silvery hair (in HPS-8) and a bleeding diathesis.
HPS is described in patients worldwide (Brantly et al., 2000; Anderson et al., 2003; Witkop et al., 1990; Huizing et al., 2001; Ito et al., 2005; Merideth et al, 2009; Vincent et al., 2009), but is common on the island of Puerto Rico. One of 1,800 Puerto-Ricans in the northwest part of the island suffers from HPS type 1 (HPS-1) (Witkop et al., 1990), due to a 16 bp duplication in exon 15 of the HPS1 gene (Oh et al, 1996). In central Puerto Rico, one of 4,000 natives has HPS-3 due to another founder mutation, a 3,904-bp deletion in the HPS3 gene (Anikster et al., 2001; Santiago-Borrero et al., 2006).
Here we report six non-Puerto Rican Hispanic HPS patients. All these individuals presented with pale to light skin color, nystagmus, and bleeding problems including epistaxis and easy bruising. Molecular analysis revealed that none of these patients carried the Puerto Rican 16 bp deletion in HPS1 or the 3,904 bp deletion in HPS3 founder mutations, but instead had other mutations in the HPS1, HPS4 or HPS5 genes. These cases emphasize the molecular variability of HPS among non-Puerto Rican, Hispanic HPS patients.
Results and Discussion
Clinical Findings
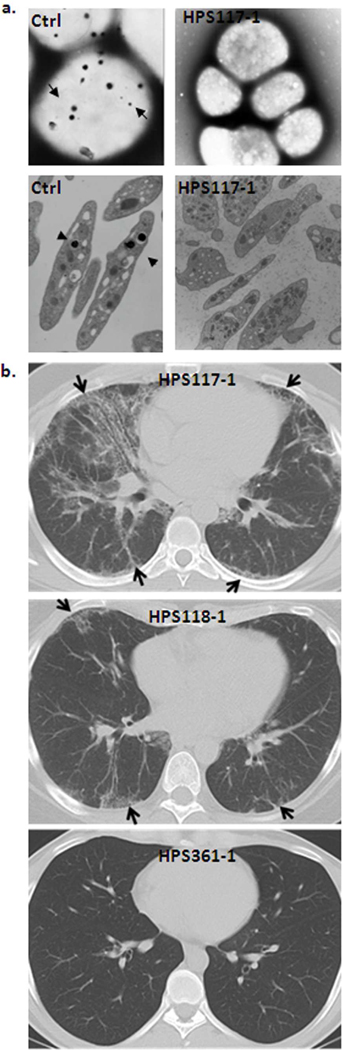
Patient HPS117-1 is a 29-year-old Mexican man who was seen for advanced pulmonary fibrosis and symptoms of inflammatory bowel disease. The diagnosis of HPS was suspected because he had albinism, nystagmus, and pulmonary fibrosis on CT scan and lung biopsy. The patient was referred to the NIH, where DNA analysis revealed a heterozygous mutation in exon 11 of HPS1, i.e., c.972delC; p. M325WfsX6 (Figure 1a and Figure S1a). On electron microscopic examination, the platelets had no dense bodies (Figure 2a). The patient reported relatively severe exertional dyspnea, fatigue, and a progressive cough present for more than a year. Pulmonary function tests revealed severe restriction and a severe reduction in gas exchange; the forced vital capacity (FVC) was 44% of predicted, total lung capacity (TLC) was 52% of predicted and diffusion capacity for carbon monoxide (DLCO) was 43% of predicted. Conventional and high-resolution chest computerized axial tomography (HRCT) scans showed diffuse bilateral interstitial infiltrates (Figure 2b). Biopsies from a colonoscopy revealed evidence of colitis in the sigmoid colon and rectum. The patient died from complications of end-stage pulmonary fibrosis at age 30.
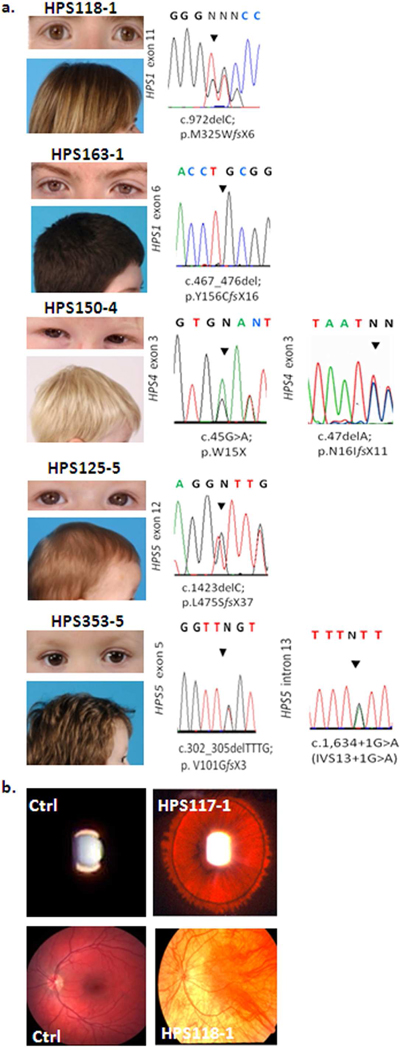
Figure 1. Clinical and molecular findings among non-Puerto Rican Hispanic HPS patients.
(a) Pigmentation and sequencing chromatograms of all six patients reported in this study. All patients showed hypopigmented skin with hair colors ranging from blond to brown. (b) Representative images of iris transillumination (in HPS117-1) and pale fundus (in patient HPS118-1), compared to controls (Ctrl).
Figure 2. Hallmarks of HPS patients.
(a) Representative whole mount (top panel) and transmission (bottom panel) EM images of selected patient compared to controls (Ctrl), showing absence of platelet delta granules (arrows in control images). (b) Conventional chest CT of HPS117-1 (top image) shows diffuse bilateral peripheral interstitial pulmonary infiltrates (arrows). A CT scan from HPS118-1 (middle image) shows mild bilateral interstitial opacities (arrows) predominantly in subpleural areas. In contrast, no pulmonary infiltrates were found in a CT scan from HPS361-1 (bottom image), who is a 22 year-old male with HPS-1 and no lung disease.
Patient HPS118-1, the sister of HPS117-1, is a 39-year-old Mexican woman with mild pulmonary fibrosis, essential hypertension, and rheumatoid arthritis. She had 3 unaffected siblings, but her paternal great grandfather reportedly had albinism. Her initial NIH evaluation at age 32 revealed that she had a heterozygous mutation in HPS1: c.972delC; p. M325WfsX6 (exon 11) (Figure 1a); platelet electron microscopy confirmed the absence of dense bodies. She had no symptoms of lung disease, but a CT scan showed mild bilateral basilar peripheral interstitial lung opacities (Figure 2b). Pulmonary function testing showed mild restriction with a TLC of 70% of predicted and a mild reduction in diffusion capacity. At age 33, she was diagnosed with rheumatoid arthritis and essential hypertension, both of which are well controlled with medications.
Her recent evaluation at age 39 revealed some progression of her lung disease, but she remains asymptomatic. HRCT of the chest showed a mild increase in fibrosis in the lung bases, especially on the right, and continued involvement in the right middle lobe. Pulmonary function test results were stable, with an FVC 69% of predicted, TLC 70% of predicted, and an adjusted DLCO 64% of predicted.
Patient HPS125-5 is an 8-month-old boy of Mexican ancestry with brown hair (Figure 1a) and irides, decreased retinal pigmentation, and nystagmus. He had easy bruising (Figure S1c) and epistaxis, which led to the discovery of absent platelet dense bodies (Figure S1c). He has no signs of colitis. DNA analysis showed a heterozygous mutation in HPS5: c.1423delC; p.L475SfsX37 (exon 12) (Figure 1a).
Patient HPS150-4 is a 2-year-old boy of Uruguayan ancestry with HPS-4. He has blond hair, grey irides (Figure 1a), iris transillumination, nystagmus and pale fundi. He presented with easy bruising (Figure S1c). Bloody stools began at 18 months, along with epistaxis and prolonged gingival bleeding after trauma. His platelets showed no dense bodies. Genomic DNA revealed two compound heterozygous mutations in HPS4: c.45 G>A; p.W15X (exon 3) and c.47delA; p.N16IfsX11 (exon3) (Figure 1a).
Patient HPS163-1 is a 16-year-old male of Honduran-Salvadoran ancestry. He has pale skin and his blond hair darkened considerably since birth (Figure 1a); his irides are brown (Figure 1a), and nystagmus is present. Bruising was noted at age 13; his platelets have no dense bodies. Episodes of abdominal pain, bloody diarrhea, and anal fissures prompted treatment for Crohn’s disease. Nausea caused a 20 pound weight loss; syncopal episodes and anemia let to a colectomy with end ileostomy. Repair of a perforated bowel required 18 units of platelets. There were no pulmonary symptoms. DNA analysis showed a homozygous mutation in HPS1, i.e., c.467_476del; p.Y156CfsX16 (exon 6) (Figure 1a).
Patient HPS353-5 is a 3 and 9/12 year-old boy of Cuban-Venezuelan ancestry. He has medium brown hair, brown irides (Figure 1a), and nystagmus with photosensitivity. Ocular albinism was considered but genetic testing for X-linked ocular albinism was negative. Although there was no excessive bleeding with previous surgeries (i.e. circumcision, strabismus or dental surgery), the patient presented with easy bruising (Figure S1c) of his lower extremities and two minor episodes of spontaneous epistaxis. On electron microscopic examination, his platelets had no dense bodies (Figure S1b). He had no manifestations of colitis. His visual acuity was OD 20/100 and OS 20/125 with pendular horizontal nystagmus, diffuse iris transillumination, optic disc pallor, and blond periphery of the retinae with no foveal reflex. DNA analysis revealed compound heterozygous mutations in HPS5, i.e., c.302_305delTTTG; p.V101GfsX3 (exon 5) and c.1,634+1G>A (intron 13) (Figure 1a).
Molecular and Cellular Analysis
DNA sequencing of all coding exons and intronic boundaries of the HPS1, HPS3, HPS4, HPS5, and HPS6 genes (Wei, 2006; Huizing et al., 2008) revealed the mutations listed in Table 1.
Table 1.
Summary of clinical and molecular features in non-Puerto Rican Hispanic patients.*
| Patient | Age (years) |
Sex | Ancestry | Gene | Allele 1 | Allele 2 | Bleeding | Lung fibrosis |
GI |
|---|---|---|---|---|---|---|---|---|---|
| HPS117-1 | 29 | M | Mex | HPS1 | c.972delC (ex 11) | ND | Br | + | − |
| p.M325WfsX6 | (FVC 44%) | ||||||||
| HPS118-1 | 39 | F | Mex | HPS1 | c.972delC (ex 11) | ND | Br | + | − |
| p.M325WfsX6 | (FVC 69%) | ||||||||
| HPS125-5 | 8 mo | M | Mex | HPS5 | c.1423delC (ex 12) | ND | E, Br | − | − |
| p.L475SfsX37 | |||||||||
| HPS150-4 | 2 | M | Uru | HPS4 | c.45G>A (ex 3) | c.47delA | E, Br | − | − |
| p.W15X | p.N16IfsX11 | ||||||||
| HPS163-1 | 16 | M | Hon/Sal | HPS1 | c.467_476del | c.467_476del | Br | − | + |
| p.Y156CfsX16 (ex6) | p.Y156CfsX16 (ex6) | ||||||||
| HPS353-5 | 3 | M | Cub/Ven | HPS5 | c.302_305delTTTG | c.1,634+1G>A (in13) | E, Br | − | − |
| p.V101GfsX3 (ex 5) |
Abbreviations: +, present; −, absent; Br, easy bruising; Cub, Cuban; E, epistaxis; F, female; FVC, Forced Vital Capacity; GI, gastrointestinal symptoms; Hon, Honduran; M, male; mo, months; ND, not determined; Mex, Mexican; Sal, Salvadoran; Uru, Uruguayan; Ven, Venezuelan
In addition, the patients’ fibroblast proteins were electrophoresed and immunoblotted using antibodies against HPS4 to evaluate the protein expression of BLOC-3, consisting of HPS1 and HPS4, and against HPS5 to evaluate BLOC-2, consisting of HPS3,HPS5, and HPS6 (Figure 3). Mutation in one member of a BLOC complex destabilizes the entire complex, causing degradation of the other proteins in the complex. Therefore, utilization of a specific antibody for one member of BLOC-3 (HPS4) or BLOC-2 (HPS5) assists in demonstrating deficiency of other members of the same complex (Nazarian et al., 2008).
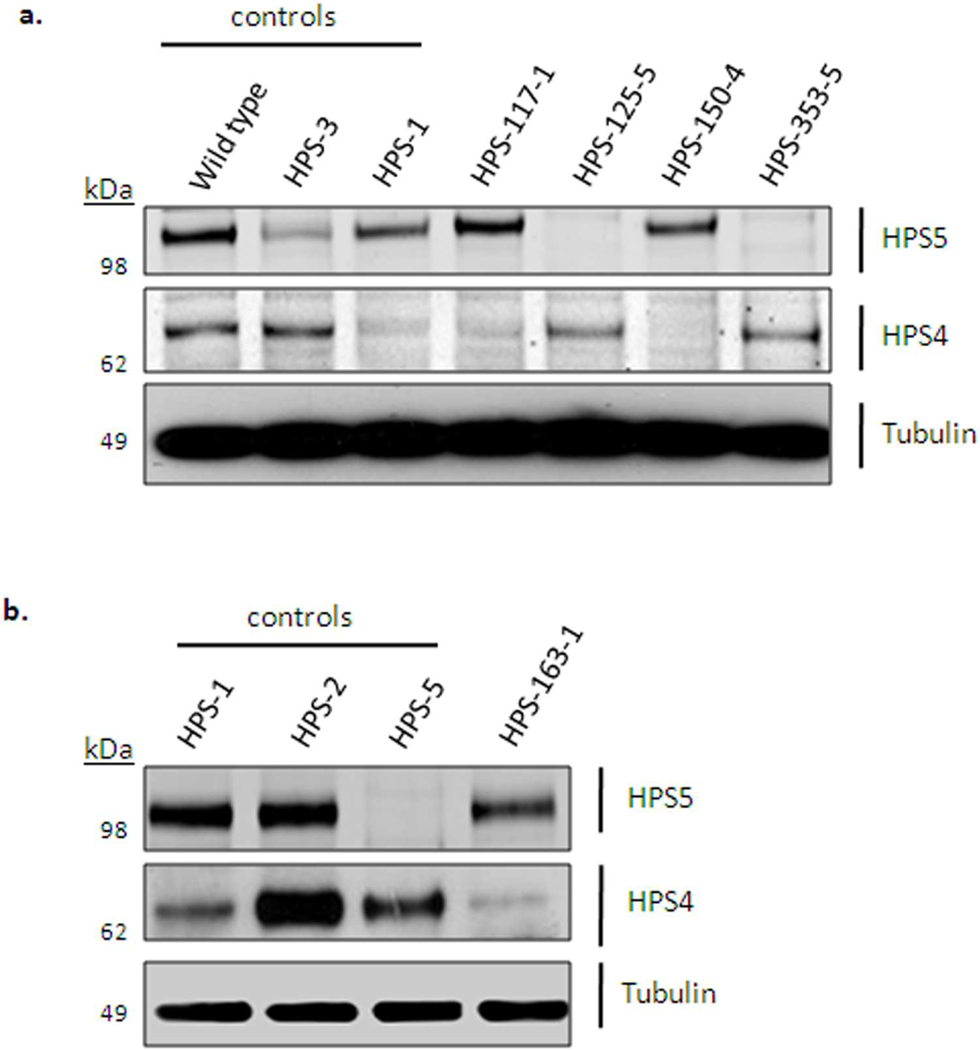
Figure 3. Immunoblot analysis of patients fibroblast extracts.
Immunoblotting was performed with antibodies against HPS4 (to detect BLOC-3 defects), HPS5 (to detect BLOC-2 defects), and α-tubulin (loading control). (a) Normal, HPS-3, and HPS-1 fibroblast extracts were loaded as controls (lanes 1–3) and compared to the patients’ protein expression. Patient HPS117-1 showed decreased HPS4 expression, similar to that of control HPS-1 cells (lane 3), suggesting an HPS1 defect. Patients HPS125-5 and HPS353-5 expressed no HPS5 protein, confirming an HPS5 defect. Patient HPS150-4 expressed no HPS4 protein, confirming an HPS4 defect. (b) Patient HPS163-1 showed reduced levels of HPS4 protein, similar to the HPS-1 loading control (lane 1), suggesting HPS-1 disease in this patient.
These immunoblotting techniques also demonstrated the pathogenicity of certain mutations, in cases where we found only one mutation in an HPS gene. For example, HPS117-1 and HPS118-1 carried a single copy of c.972delC in HPS1. This frame-shift mutation generates a premature termination codon, and causes nonsense-mediated RNA decay (Shotelersuk et al., 1998). It was previously found in Puerto Rican patients (Carmona-Rivera et al., 2010), but was also described in other Caucasians (Oh et al., 1998) and in an African-American (Merideth et al., 2009). In fact, the C-nucleotide at position 972 was recognized as an HPS1 mutation ‘hotspot’ (Oh et al., 1998). However, no second mutation could be found in our patient’s coding region of HPS1, so we used immunoblotting of HPS117-1 fibroblast extracts to illustrate a dramatic reduction in the levels of the HPS4 antibody signal, similar to that of HPS-1 control fibroblast extracts. This suggested lack of functional HPS1 (Figure 3a); the patients’ second HPS1 mutation probably involves a non-coding region of HPS1.
DNA analysis of patient HPS125-5 showed a previously unreported single copy mutation in HPS5, c.1423delC (Figure 1a), causing a frame-shift and generating a premature termination codon. No second mutation was found, but protein analysis showed complete absence of the HPS5 protein in this patient’s fibroblasts (Figure 3a), supporting the diagnosis of HPS-5.
Sequence analysis of HPS150-4 showed two unreported compound heterozygous mutations in exon 3 of the HPS4 gene, a nonsense mutation and a 1-bp deletion leading to a premature termination codon (Figure 1a). RNA transcripts from both alleles are likely to be degraded by nonsense mediated RNA decay, a prediction supported by complete absence of the HPS4 protein in the patient’s fibroblasts (Figure 3a).
Similarly, an unreported homozygous 10-bp deletion in exon 6 of HPS1, found in HPS163-1 (Figure 1a), is predicted to result in nonsense-mediated RNA decay. We used qPCR (Griffin et al., 2005) to rule out hemizygosity in this patient; qPCR demonstrated the presence of two HPS1 alleles, confirming homozygosity of the 10-bp deletion (data not shown). In addition, protein expression levels of HPS4 were reduced (Figure 3b), suggesting degradation of HPS4 in the absence of the HPS1 protein.
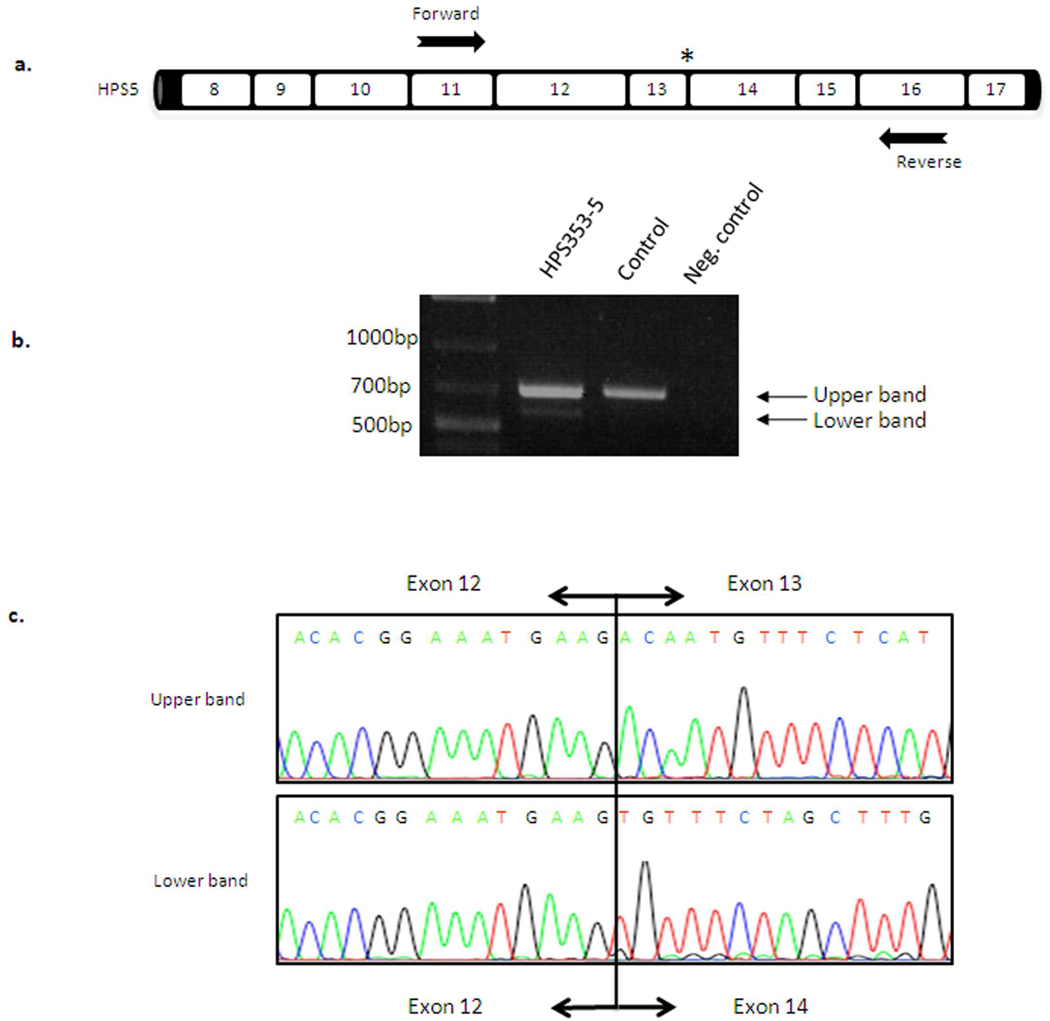
Patient HPS353-5 was compound heterozygous for two unreported mutations in HPS5, including a 4-bp deletion (TTTG) in exon 5 leading to a premature termination codon. The second mutation was a splice site variant in intron 13 (c.1,634+1G>A). To demonstrate aberration in splicing, we amplified HPS5 cDNA using primers located in exon 11 and exon 16 (Figure 4a). In addition to the expected 694-bp PCR product, we found a band of approximately 570-bp (Figure 4b). Sequence analysis of the additional band demonstrated skipping of exon 13 (124-bp) (Figure 4c), altering the reading frame. Immunoblot analysis showed total absence of the HPS5 protein in fibroblasts of patient HPS353-5 compared to normal (Figure 3a), indicating that these two mutations have pathogenic implications for the HPS5 gene product.
Figure 4. cDNA analysis of patient HPS353-5.
(a) Schematic representation of the HPS5 gene and location of the primers used for PCR analysis to detect splice-site alteration in patient HPS353-5. The patient’s splice site variant c.1,634+1G>A (intron 13) is located one bp intronic from the exon12-exon13 boundary (asterisk). (b) cDNA amplification of exons 11–16 of the HPS5 cDNA transcript showing the expected band of 694-bp and a lower molecular weight band around 570-bp. (c) Sequence analysis of the lower molecular weight band revealed skipping of exon 13 (124-bp) in patient HPS353-5, confirming the pathogenicity of the novel HPS5 splice site variant in this patient.
In summary, we report six non-Puerto-Rican Hispanic HPS patients with Mexican, Uruguayan, Honduran, Cuban, Venezuelan and Salvadoran ancestries, identifying mutations in the HPS1, HPS4 and HPS5 genes (Table 1). Importantly, we did not identify the two common Puerto-Rican HPS founder mutations, i.e., the 16-bp duplication in HPS1 and the 3,904-bp deletion in HPS3, in our cohort. Hispanic patients with oculocutaneous albinism and bleeding symptoms should not be assumed to have the HPS-1 subtype typical of northwest Puerto Rican patients. We recommend molecular HPS subtyping in such cases, since a diagnosis of the type of HPS allows anticipation of clinical complications, appropriate management and genetic counseling (Huizing et al., 2008; Gahl et al., 2002).
Materials and Methods
Patients and cells
All patients in this study were enrolled in the clinical protocol “Clinical and Basic Investigations into Hermansky-Pudlak Syndrome” (NCT00001456, www.clinicaltrials.gov), approved by the NHGRI Institutional Review Board, and adhered to Helsinki guidelines. All patients or their parents provided written, informed consent. Patients were diagnosed with HPS based on the presence of oculocutaneous albinism (i.e., decreased visual acuity, nystagmus, and some degree of hypopigmentation relative to family members) and the absence of platelet delta granules on whole-mount electron microscopy.
Primary cultures of skin fibroblasts, obtained from a 4-mm punch biopsy, were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum containing 100 U/ml penicillin and 0.1 mg/ml streptomycin.
Electron microscopy of platelet delta granules
Presence of platelet delta granules in platelet-rich plasma was analyzed using whole mount and transmission electron microscopy as previously described (Witkop et al., 1987; Huizing et al., 2007).
Molecular analysis
Genomic DNA was isolated from patients’ peripheral blood mononuclear cells using the Gentra Puregene Blood Kit (Qiagen, Valencia, CA). All HPS1, HPS3, HPS4, HPS5 and HPS6 coding exons and flanking intronic boundaries were amplified by polymerase chain reaction (PCR) amplification, and subjected to bi-directional sequencing. RNA was extracted from fibroblast using the RNeasy Mini kit (Qiagen) and transcribed into cDNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). Primer sequences for amplifying exon 11–16 of HPS5 (Figure 3) were: forward 5’- GACTGCAGATAAATTGGAGCATTT-3’ and reverse 5’-GTAGCTTGGTCATTGCTTCTGCTG-3’.
Sequencing was performed using ABI BigDye Terminator chemistry (Applied Biosystems, Foster City, CA) with detection on an ABI 3130×l genetic analyzer (Applied Biosystems). Results were analyzed using Sequencher v4.9 software (Gene Codes Corporation, Ann Arbor, MI).
Pulmonary function testing and lung computed tomography scanning
Pulmonary function testing measurements were made using standard equipment according to American Thoracic Society recommendations as previously described (SensorMedics, Yorba Linda, CA) (Rouhani et al., 2009). Conventional and high-resolution computed tomography scans of the chest were performed as previously described, without intravenous contrast during end-inspiration in the supine and prone positions, respectively (General Electric Medical Systems, Milwaukee, WI) (Gochuico et al., 2008).
Immunoblotting
Fibroblast pellets were lysed by incubation in ice-cold lysis buffer [50 mM Tris-HCl pH 7.4, 300 mM NaCl, 0.5% w/v Triton X-100, 5mM EDTA, 1× protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN)] for 30 min at 4°C, followed by centrifugation at 16,000 ×g for 15 min. Protein concentrations were determined using the BCA Protein Assay (Pierce, Rockford, IL). Total protein samples (20 µg) were separated on a 4–12% gradient NuPAGE Bis-Tris Gel (Invitrogen, Carlsbad, CA), and blotted to nitrocellulose membranes. Membranes were blocked with 10% bovine serum albumin (BSA) for 30 min at room temperature, followed by incubation with one of three primary antibodies, i.e., rabbit polyclonal antibodies against HPS4 (1:500) (H-150; Santa Cruz Biotechnology Inc, Santa Cruz, CA), rabbit polyclonal antibody against HPS5 (1:500) (Proteintech Group Inc, Chicago, IL), or mouse monoclonal antibody against α-tubulin (1:10,000) (Sigma, St. Louis, MO). Subsequently, the membranes were probed with either secondary donkey anti-rabbit or anti-mouse antibodies, Horseradish peroxidase linked (1:10,000; GE Healthcare, UK) followed by detection with enhanced chemiluminescence ( ECL) Western Blotting substrate from Pierce (Rockford, IL), or with secondary IRDye® 800CW goat anti-rabbit or anti-mouse antibodies (1:10,000) for 1 hour at room temperature and detected using the Li-Cor Odyssey® Infrared Imaging System per the manufacturer’s instructions (Li-Cor Biosciences, Lincoln, NE).
Supplementary Material
Acknowledgments
We thank the patients for participating in the NIH protocol. We also thank Roxanne Fischer for excellent laboratory support. This work was supported by the Intramural Research program of the National Human Genome Research Institute, National Institutes of Health, Bethesda, Maryland, USA.
Footnotes
Conflict of interest
All authors declare no conflict of interest.
References
- Anderson PD, Huizing M, Claassen DA, et al. Hermansky-Pudlak syndrome type 4 (HPS-4): clinical and molecular characteristics. Hum Genet. 2003;113:10–17. doi: 10.1007/s00439-003-0933-5. [DOI] [PubMed] [Google Scholar]
- Anikster Y, Huizing M, White J, et al. Mutation of a new gene causes a unique form of Hermansky-Pudlak syndrome in a genetic isolate of central Puerto Rico. Nat Genet. 2001;28:376–380. doi: 10.1038/ng576. [DOI] [PubMed] [Google Scholar]
- Brantly M, Avila NA, Shotelersuk V, et al. Pulmonary function and high-resolution CT finding in patients with an inherited form of pulmonary fibrosis, Hermansky-Pudlak syndrome, due to mutations in HPS-1. Chest. 2000;117:129–136. doi: 10.1378/chest.117.1.129. [DOI] [PubMed] [Google Scholar]
- Carmona-Rivera C, Hess R, O’Brien K, et al. Novel mutations in the HPS1 gene among Puerto Rican patients. Clin Genet Epub ahead of print 2010 Jun 28. 2010 doi: 10.1111/j.1399-0004.2010.01491.x. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica EC. The building BLOC(k)s of lysosomes and related organelles. Curr Opin Cell Biol. 2004;16:458–464. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Dell’Angelica EC, Shotelersuk V, Aguilar RC, et al. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the b3A subunit of the AP-3 adaptor. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Di Pietro SM, Falcon-Perez JM, Dell’Angelica EC. Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic. 2004;5:276–283. doi: 10.1111/j.1600-0854.2004.0171.x. [DOI] [PubMed] [Google Scholar]
- Gahl WA, Brantly M, Kaiser-Kupfer M, et al. Genetic defect and clinical characteristics of patients with a form of oculocutaneous albinism (Hermansky Pudlak Syndrome) N Engl J Med. 1998;338:1258–1265. doi: 10.1056/NEJM199804303381803. [DOI] [PubMed] [Google Scholar]
- Gahl WA, Brantly M, Troendle J, et al. Effect of pirfenidone on the pulmonary fibrosis of Hermansky-Pudlak syndrome. Mol Genet Metab. 2002;76:234–242. doi: 10.1016/s1096-7192(02)00044-6. [DOI] [PubMed] [Google Scholar]
- Gochuico BR, Avila NA, Chow CK, et al. Progressive preclinical interstitial lung disease in rheumatoid arthritis. Arch Intern Med. 2008;168:159–166. doi: 10.1001/archinternmed.2007.59. [DOI] [PubMed] [Google Scholar]
- Griffin AE, Cobb BR, Anderson PD, et al. Detection of hemizyocity in Hermansky-Pudlak syndrome by quantitative real-time PCR. Clin Genet. 2005;68:23–30. doi: 10.1111/j.1399-0004.2005.00461.x. [DOI] [PubMed] [Google Scholar]
- Hermos CR, Huizing M, Kaiser-Kupfer, et al. Hermansky-Pudlak Syndrome type 1: Gene organization, Novel mutations, and clinical-molecular review of Non-Puerto Rican cases. Hum Mutat. 2002;20:482. doi: 10.1002/humu.9097. [DOI] [PubMed] [Google Scholar]
- Hermansky F, Pudlak P. Albinism associated with hemorrhagic diathesis and unusual pigmented reticular cells in the bone marrow: Report of two cases with histochemical studies. Blood. 1959;14:162–169. [PubMed] [Google Scholar]
- Huizing M, Anikster Y, Fitzpatrick DL, et al. Hermansky-Pudlak Syndrome type 3 in Ashkenazi Jews and other non-Puerto Rican patients with hypopigmentation and platelet storage-pool deficiency. Am J Hum Genet. 2001;69:1022–1032. doi: 10.1086/324168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Helip-Wooley A, Westbroek W, et al. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genomics Hum Genet. 2008;9:359–386. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Hess R, Dorward H, et al. Cellular, molecular and clinical characterization of patients with Hermansky-Pudlak syndrome type 5. Traffic. 2004;5:711–722. doi: 10.1111/j.1600-0854.2004.00208.x. [DOI] [PubMed] [Google Scholar]
- Huizing M, Parkes JM, Helip-Wooley A, et al. Platelet alpha granules in BLOC-2 and BLOC-3 subtypes of Hermansky-Pudlak syndrome. Platelets. 2007;18:150–157. doi: 10.1080/13576500600936039. [DOI] [PubMed] [Google Scholar]
- Huizing M, Pederson B, Hess RA, et al. Clinical and cellular charaterisation of Hermansky-Pudlak syndrome type 6. J Med Genet. 2009;46:803–810. doi: 10.1136/jmg.2008.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Suzuki T, Inagaki K, et al. High frequency of Hermansky-Pudlak syndrome type 1 (HPS1) among Japanese albinism patients and functional analysis of HPS1 mutant protein. J Invest Dermatol. 2005;125:715–720. doi: 10.1111/j.0022-202X.2005.23884.x. [DOI] [PubMed] [Google Scholar]
- Li W, Zhang Q, Oiso N, et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nat Genet. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Moriyama K, Bonifacino JS. BLOC-3, a protein complex containing the Hermansky-Pudlak syndrome gene products HPS1 and HPS4. J Biol Chem. 2003;278:29376–29384. doi: 10.1074/jbc.M301294200. [DOI] [PubMed] [Google Scholar]
- Merideth MA, Vincent LM, Sparks SE, et al. Hermansky-Pudlak syndrome in two African-American brothers. Am J Med Genet A. 2009;149:987–992. doi: 10.1002/ajmg.a.32757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan NV, Pasha S, Johnson CA, et al. A germline mutation in BLOC1S3/reduced pigmentation causes a novel variant of Hermansky-Pudlak syndrome (HPS8) Am J Hum Genet. 2006;78:160–166. doi: 10.1086/499338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Falcón-Pérez JM, Dell'Angelica EC. Biogenesis of lysosome-related organelles complex 3 (BLOC-3): a complex containing the Hermansky-Pudlak syndrome (HPS) protein HPS1 and HPS4. Proc Natl Acad Sci U S A. 2003;100:8770–8775. doi: 10.1073/pnas.1532040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, Huizing M, Helip-Wooley A, et al. An immnunoblotting assay to facilitate the molecular diagnosis of Hermansky-Pudlak syndrome. Mol Genet Metab. 2008;93:134–144. doi: 10.1016/j.ymgme.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Bailin T, Fukai K, Feng GH, et al. Positional cloning of a gene for Hermansky-Pudlak syndrome, a disorder of cytoplasmic organelles. Nat Genet. 1996;14:300–306. doi: 10.1038/ng1196-300. [DOI] [PubMed] [Google Scholar]
- Oh J, Ho L, Ala-Mello S, et al. Mutation analysis of patients with Hermansky-Pudlak syndrome: a frameshift hot spot in the HPS gene and apparent locus heterogeneity. Am J Hum Genet. 1998;62:593–598. doi: 10.1086/301757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhani FN, Brantly ML, Markello TC, et al. Alveolar macrophage dysregulation in Hermansky-Pudlak syndrome type 1. Am J Respir Crit Care Med. 2009;180:1114–1121. doi: 10.1164/rccm.200901-0023OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Borrero PJ, Rodríguez-Pérez Y, Renta JY, et al. Genetic testing for oculocutaneous albinism type 1 and 2 and Hermansky-Pudlak syndrome type 1 and 3 mutations in Puerto Rico. J Invest Dermatol. 2006;126:85–90. doi: 10.1038/sj.jid.5700034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinella RA, Greco MA, Cobert BL, et al. Hermansky-Pudlak Syndrome with granulomatous colitis. Ann Inter Med. 1980;1:20–23. doi: 10.7326/0003-4819-92-1-20. [DOI] [PubMed] [Google Scholar]
- Shotelersuk V, Hazelwood S, Larson D, et al. Three new mutations in a gene causing Hermansky-Pudlak syndrome: clinical correlations. Mol Genet Metab. 1998;64:99–107. doi: 10.1006/mgme.1998.2679. [DOI] [PubMed] [Google Scholar]
- Starcevic M, Dell’Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Li W, Zhang Q, et al. Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat Genet. 2002;30:321–324. doi: 10.1038/ng835. [DOI] [PubMed] [Google Scholar]
- Vincent LM, Adams D, Hess RA, et al. Hermansky-Pudlak syndrome type 1 in patients of Indian descent. Mol Genet Metab. 2009;97:227–233. doi: 10.1016/j.ymgme.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei ML. Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res. 2006;19:19–42. doi: 10.1111/j.1600-0749.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- Witkop CJ, Krumwiede M, Sedano H, et al. Reliability of absent platelet dense bodies as a diagnostic criterion for Hermansky-Pudlak syndrome. Am J Hematol. 1987;26:305–311. doi: 10.1002/ajh.2830260403. [DOI] [PubMed] [Google Scholar]
- Witkop CJ, Nunez BM, Rao GH, et al. Albinism and Hermansky-Pudlak syndrome in Puerto Rico. Biol Asoc Med PR. 1990;82:333–339. [PubMed] [Google Scholar]
- Zhang Q, Zhao B, Li W, et al. Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat Genet. 2003;33:1–9. doi: 10.1038/ng1087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.