Abstract
Mucin 1 (MUC1) is a diagnostic factor and therapy target in lung adenocarcinoma. MUC1 C-terminal intracellular domain (CD) interacts with estrogen receptor α (ERα) and increases gene transcription in breast cancer cells. Because lung adenocarcinoma cells express functional ERα and estrogen receptor β (ERβ) we examined MUC1 expression and MUC1-ER interaction. Since blocking MUC1 CD with an inhibitory peptide (PMIP) inhibited breast tumor growth, we tested whether PMIP would inhibit lung adenocarcinoma cell proliferation. We report that MUC1 interacts with ERα and ERβ within the nucleus of H1793 lung adenocarcinoma cells in accordance with MUC1 expression. PMIP was taken up by H23 and H1793 cells and inhibited the proliferation of H1793, but not H23 cells, concordant with higher MUC1 in H1793 cells. Lower MUC1 protein in H23 does not correspond to miR-125b and miR-145 that have been reported to reduce MUC1 expression. PMIP had no effect on the viability of normal human bronchial epithelial cells, which lack MUC1 expression. PMIP inhibited estradiol (E2) –activated reporter gene transcription and endogenous cyclin D1 and nuclear respiratory factor-1 (NRF-1) gene transcription in H1793 cells. These results indicate MUC1-ER functional interaction in lung adenocarcinoma cells and that inhibiting MUC1 inhibits lung adenocarcinoma cell viability.
Keywords: MUC1, lung adenocarcinoma, estrogen receptor, miRNA, NSCLC
Introduction
Although the incidence of lung adenocarcinoma has increased in never and former smokers, women, and young adults, the role of gender in risk remains unresolved (1). Interestingly, breast cancer patients receiving antiestrogens for breast cancer had lower lung cancer mortality, indicating a possible role for estrogens in disease progression (2). We reported that lung adenocarcinoma cell lines from females respond proliferatively to estradiol (E2) and were inhibited by estrogen receptor (ER) antagonists 4-hydroxytamoxifen (4-OHT) or ICI 182,780 (ICI, Fulvestrant) (3). In contrast, lung adenocarcinoma cells from males were neither stimulated by E2 nor blocked by 4-OHT or ICI. Similar responses were detected when examining E2-induced cyclin D1 (CCND1) transcription (3). ERα and ERβ expression was similar among cell lines from males and females, indicating that the observed phenotype was not due to lower ER in cells from males. Other investigators detected E2-induced proliferation and activation of intracellular MAPK and PI3K by ERα, ERβ, GPR30, and epidermal growth factor receptor (EGFR) in lung adenocarcinoma cell lines in a gender-independent manner (4, 5).
Mucin1 (MUC1) is a high MW plasma membrane (PM)-bound protein with a heavily O-glycosylated N-terminal domain (NTD) that protrudes from the apical surface of glandular epithelial cells in lung, breast, and colon (6). MUC1 has a single transmembrane domain and a 72 aa C-terminal domain (CD) that is cleaved and regulated by tyrosine phosphorylation (7).
MUC1 is a marker of type II pneumocyte progenitor cell lineage (8). MUC1 surface expression is critical for the protective function of the airway epithelium (9). A MUC1 gene polymorphism for the large MUC1 allele was associated with lung adenocarcinoma but not with squamous cell carcinoma (10). MUC1 is an oncogene (11), increased in lung cancer (12), and is a clinical marker for lung adenocarcinoma (13). During dedifferentiation in lung adenocarcinoma progression, MUC1 is reduced and mislocalizes to the entire PM where it blocks cell-cell and cell-matrix adhesion, allowing tumor cell invasion (14). The NTD is secreted by tracheobronchial epithelial cells and is a mucus component (15).
Within cells, MUC1-CD interacts with proteins including PI3K, Shc, PLC1, c-Src, Grb-2, p53, IKKβ, IKKγ, β-catenin, hsp90, hsp70, and ERα (7). MUC1 overexpression in cancer cells results in MUC1-CD nuclear and mitochondrial localization (16). MUC1-CD interacts with ERα in the nucleus of breast cancer cells where it increased ERα-transcriptional activity (17).
Since MUC1 is an oncogene, it makes a tempting therapeutic target (18). MUC1 knockdown by siRNA in A549 lung adenocarcinoma cells increased sensitivity to cisplatin (19). Similarly, stable transfection of siMUC1 inhibited H358 non-small cell lung cancer (NSCLC) tumor formation in mice (20). Treatment of breast cancer cells with a MUC1 inhibitory peptide (MIP) cloned adjacent to the protein transduction domain (PTD4) created a peptide called PMIP that is taken up by cells without transfection reagents (21). PMIP acts as a decoy for MUC1 binding partners. PMIP inhibited MUC1/β-catenin and MUC1/EGFR interactions and inhibited MDA-MB-231 breast cancer cell proliferation, migration, and invasion in vitro and tumor growth in mice (21). Similarly, a MUC1 inhibitor called GO-201 bound MUC1-CD, blocked MUC1 oligomerization and induced necrosis in MCF-7, ZR-75-1, and MDA-MB-231 breast cancer cells (16). GO-201 was recently reported to inhibit the proliferation of lung adenocarcinoma cell lines (22).
This study tested the hypotheses that ERα and ERβ interact functionally with MUC1 in lung adenocarcinoma cells and that PMIP selectively inhibits lung adenocarcinoma, not normal human bronchial epithelial cells (HBECs), proliferation and inhibits ER-responses.
Materials and Methods
Chemicals
17-β-estradiol (E2) and 4-hydroxytamoxifen (4-OHT) were from Sigma. ICI 182,780 was from Tocris. Sequences of the control peptide (CP: NH2- YARAAARQARATNPAVAATSANL-COOH) and PMIP (MUC1 inhibitory peptide (MIP) adjacent to the protein transduction domain (PTD4)): NH2-YARAAARQARARYEKVSAGNGGSSLS-COOH, as reported in (21). FITC-PMIP and PIMP were purchased from New England Peptide.
Antibodies
Antibodies were purchased: HC-20 for ERα from Santa Cruz Biotechnology, ERβ from Upstate (cat #06-629), α-tubulin from LabVision (Fisher Scientific), β-actin from Sigma, Armenian hamster anti-MUC1-CD (Ab-5, MUC1; CT2) from Thermo Scientific; anti-MUC1 NTD (DF3) from Abcam. The secondary antibody for CT2 was anti-Armenian hamster (Jackson Immunoresearch).
Estrogen receptor
Recombinant human ERα and ERβ1 (long form) were prepared as described (23).
Cell Culture
The 5 HBEC cell lines, their maintenance and characterization were described (23, 24) and HBECs were used at passages < 8. MCF-7 cells were purchased from ATCC and used at passages < 10 from ATCC. MCF-7 were maintained as described (3). Prior to treatment, cells were placed in phenol red-free media supplemented with 5% dextran-coated charcoal stripped FBS (DCC-FBS) for 24–48 h. Ethanol (EtOH) was the vehicle control.
MUC1 genotyping
PCR primers to detect the MUC1 splice variants MUC1/A and MUC1/B were P1 and P2 (25). Products were analyzed on a DNA 500 chip of the Agilent 2100 Bioanalyzer.
Immunofluorescence imaging
H1793 cells were incubated with 10 μM of PMIP-FITC for 1, 4 and 24 h, or 10 μM of PMIP-FITC for 24 h plus 10 nM E2 for the last 4 h. Cells on coverslips were fixed with 4% paraformaldehyde for 15 min. After washing and permeabilization with 0.2 % Triton X-100 in PBS and blocking with 10% BSA in PBS, primary antibody MUC1 (CT2); ERα (HC-20); or ERβ (H150) was added at a 1:1500, 1:1000 and 1:500 dilution, respectively, overnight at 4°C. Cells were stained with secondary antibodies at a 1:200 dilution. The secondary AffiniPure Goat anti-armenian hamster antibody was labeled with R- Phycoerythyin (R-PE) 566 (red color, Jackson ImmunoResearch) or Fluoresein (FITC) and secondary anti-rabbit antibody was labeled with Zenon™ Alexa Fluor 633 (red color, Molecular Probes). Cells were incubated with Hoechst (2,5′-Bi-1H-benzimidazole, Invitrogen). Immunofluorescence imaging used a Zeiss Axiovert 200 inverted microscope with a 40x objective lens and AxioVision Release 4.3 software. Image were taken at the same exposure.
Protein Isolation
Whole cell extracts (WCE) were prepared in modified RIPA buffer (3). Protein concentrations were determined using the Bio-Rad DC Protein Assay (Bio-Rad Laboratories).
Western blotting
Western analysis was performed as described (3). The membranes were stripped and reprobed for α-tubulin. Immunoblots were scanned using a Microtek ScanMaker VII scanner. Un-Scan-It ver. 6.1 (Silk Scientific) quantitated the integrated optical densities (IOD) for each band which was divided by concordant α-tubulin IOD in the same blot. For comparison between experiments, the MUC1 CD/α-tubulin normalized pixel ratios for MCF-7 cells was set to 1.
Coimmunoprecipitation
Nuclear lysates were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) according to the manufacturer’s protocol. Nuclear lysates (400 μg) were incubated with the indicated antibodies in RIPA buffer (20 mM Tris pH 8, 100 mM NaCl, 1 mM DTT, 0.2% NP40, 0.2% DOC and 0.2% Triton X100) supplemented with protease and phosphatase inhibitors for 1 h at 4°C. Protein G-Sepharose 4B (Zymed) was added and incubated overnight with rotation at 4°C. The beads were sedimented at 10,000 × g, washed 3X with RIPA buffer, resuspended in 2X Tris-Glycine buffer (Invitrogen), and incubated at 85 °C for 2 min. Proteins in the resulting supernatant were separated on 14% Tris-Glycine gels (Invitrogen) and transferred onto PVDF membranes (Millipore). The membranes were blocked and immunoblotted. HyGlo Chemiluminescent HRP antibody detection reagent (Denville Scientific) was used to detect protein bands. The membranes were visualized on a Carestream Imager using Carestream Molecular Imaging software.
MTT assays
MDA-MB-231, H1793, H23, HBEC2-KT, and HBEC4-KT were plated as described (3). Cell lines were treated with 0.1 – 20μM CP or PMIP, +/− E2, 4-OHT, or ICI 182,780, for 5 days. Treatments were changed every 24 h. Within individual experiments, each treatment was in quadruplicate and experiments were performed at least 3 times. MTT assays used Cell-Titer96 from Promega.
Transient transfection assays
Transient transfection of H1793 cells used FuGene6 (Roche). Each well received 5 ng of a Renilla luciferase reporter (pRL-tk, Promega) and 250 ng of pGL3-2EREc38-luciferase reporter (26). The cells were harvested 30 h post-treatment using Promega’s Passive Lysis buffer. Luciferase and Renilla luciferase activities were determined using Promega’s Dual Luciferase assay and calculations were performed (3).
RNA Isolation, RT-PCR and Quantitative Real-Time-PCR (QRT-PCR)
RNA was extracted from cells using Trizol (Invitrogen) or RNeasy (Qiagen). The High Capacity cDNA archive kit (PE Applied Biosystems) was used to reverse transcribe total RNA using random hexamers. QRT-PCR for MUC1 used ABI Taqman primers (27) and was normalized by 18S rRNA. Analysis and fold differences were determined using the comparative CT method. Data are presented as relative to expression in EtOH-treated and control transfected cells.
miRNA Isolation and QRT-PCR
miRNA-enriched total RNA was extracted from MCF-7, H1793, and H23 cells using the miRNA isolation kit (Exiqon). The quality and quantity of the isolated RNA was analyzed using a NanoDrop spectrophotometer and Agilent Bioanalyzer. cDNA was synthesized using the miRCURY LNA™ first strand cDNA synthesis kit and QPCR performed using the miRCURY LNA™ SYBR Green master mix using the miRNA primer sets for miR-125b and -145 (Exiqon). 5S rRNA was used for normalization of miRNA expression. Analysis and fold change was determined using the comparative CT method. The change in miRNA expression was calculated as fold-change, i.e., relative to MCF-7 cells.
Statistical Analysis
Statistical analyses were performed using Student’s t-test or one-way ANOVA followed by Student-Newman-Keuls or Dunnett’s post-hoc tests using GraphPad Prism.
Results
MUC1 in lung adenocarcinoma cell lines and human bronchial epithelial cell lines
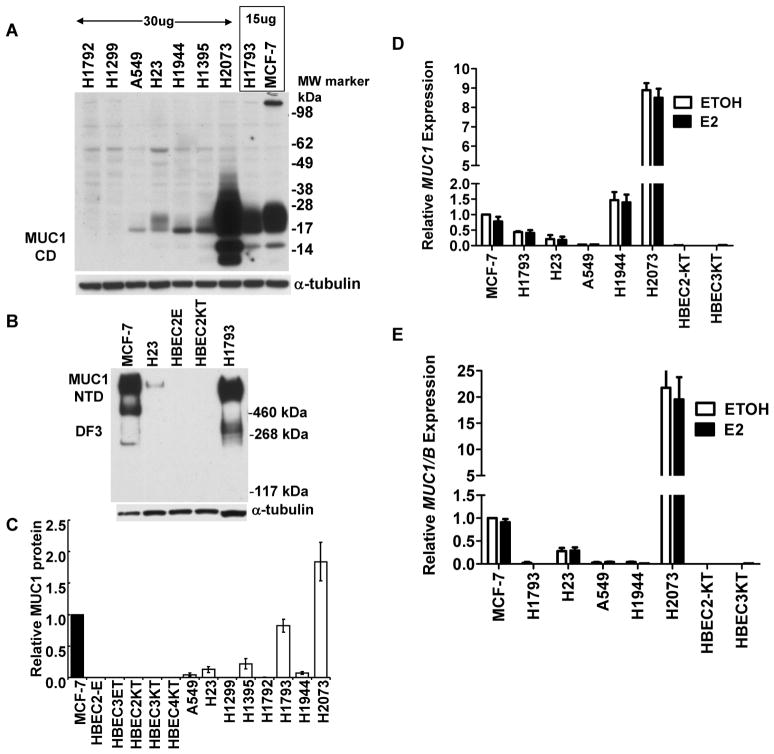
MUC1 expression was examined in WCE prepared from eight lung adenocarcinoma cell lines, five immortalized HBEC cell lines with similar phenotypic and genotypic properties as primary HBECs (24) and MCF-7 breast (28) cancer cells (Fig. 1). In agreement with other reports (29), MCF-7 cells express high MUC1. Interestingly, H2073 and H1793 cells, derived from female patients, also express high MUC1 protein. All four lung adenocarcinoma cell lines from males (H1792, H1299, A549, and H23) have lower MUC1 than MCF-7, as do H1944 and H1395 cell lines from females. MUC1 was not detected in HBECs (Fig. 1B and Supplemental Fig. 1).
Figure 1. MUC1 expression in lung adenocarcinoma cells and HBECs.
A) 15 μg WCE protein for H1793 and MCF-7 cells and 30 μg protein for all other cell lines was probed for MUC1 CD. B) All lanes contained 35μg protein for the MUC1 NTD blot using DF3 antibody. The membranes were stripped and reprobed for α-tubulin for normalization. C) The sum of all MUC1 CD bands were quantitated as the ratio of MUC1/α-tubulin and normalized to MCF-7 within each gel (set to one). Values are the average of 6 separate gels ± SEM. D and E) Q-PCR of MUC1 and MUC1/B mRNA expression in the indicated cell lines that were treated with EtOH or 10 nM E2 for 3 h. Values are the average of 3 separate experiments ± SEM and are normalized to MCF-7 EtOH.
MUC1 splice variants in lung adenocarcinoma cells and HBECs
Although MUC1 is used as a clinical marker for lung adenocarcinoma (13, 30), to our knowledge, no one has examined MUC1 splice variants in lung adenocarcinomas or cell lines derived from lung tumors. RT-PCR was performed to detect MUC1/A and MUC1/B splice variants (31). The results indicate no apparent gender-dependent difference in MUC1/A or MUC1/B splice variant expression (Supplemental Table 1), although a larger population would be needed to confirm these data.
Total MUC1 mRNA expression was evaluated using a primer set that recognizes all variants of MUC1 (Fig. 1D) and a primer set that recognizes only MUC1B (Fig. 1E). Previous investigators reported that E2 increased MUC1 in MCF-7 cells (28); however, these results were based on semi-quantitative PCR that detected two MUC1 bands, one of which the authors called a precursor form, and no quantitation of the single point experiment was performed. Here E2 had no significant effect on MUC1/B or MUC1 mRNA expression in MCF-7, H1793, H23, H1944, H2073, HBEC2-KT or HBEC3-KT cells. The expression of total MUC1 and MUC1/B correspond to the MUC1 genotyping in Supplemental Table 1.
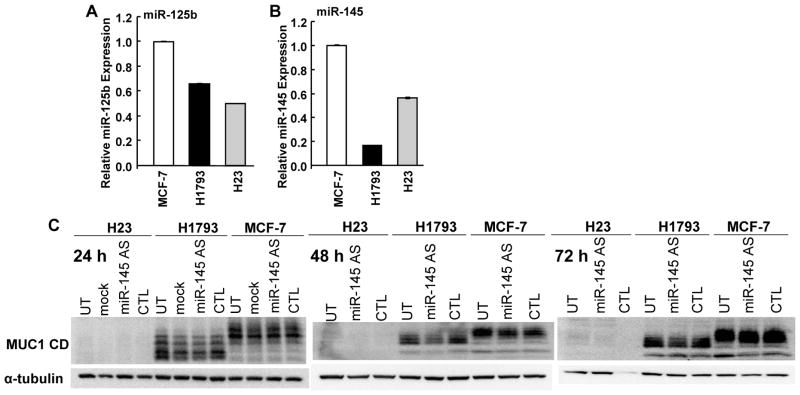
miR-125b and miR-145 expression does not correspond to differences in MUC1 protein between H23 and H1793 cells
MUC1 is downregulated by miR-125b (32) and miR-145 (33) which interact with the 3′UTR of MUC1 mRNA and reduce MUC1 protein in ZR-75-1 breast cancer cells and HEK-293T cells, respectively. To determine if the levels of miR-125b and miR-145 corresponded inversely with MUC1 protein in H23 and H1793 cells, miR-125b and miR-145 expression was evaluated using MCF-7 cells as a positive control. Given that MCF-7 and H1793 have ~ 8-fold more MUC1 protein than H23, we anticipated lower miR-125b and miR-145 in MCF-7 and H1793 cells relative to H23. MCF-7 has higher miR-125b and miR-145 than either H1793 or H23 cells (Fig. 2A and B). The lower miR-145 expression in H1793 relative to H23 suggests that miR-145 may reduce MUC1 protein in H23 cells. To test this idea, H23, H1793, and MCF-7 were transfected with control AS and AS-miR-145 and MUC1 protein was examined by western blot (Fig. 2C). Supplemental Fig. 2 shows that AS-miR-145 reduced miR-145 and that neither the AS control or mock transfection affected miR-145 in any cell line. However, AS-miR-145 had no effect on MUC1 protein, indicating that the miR-145 is not responsible for the lack of MUC1 in H23 cells and does not regulate MUC1 in H1793 or MCF-7 cells.
Figure 2. Expression of miR-125b and miR-145 does not correspond inversely with MUC1 protein expression in MCF-7, H1793, and H23 cells.
A and B) miR-125b and miR-145 expression. Values are the average of triplicate determinations and were normalized by 5S rRNA and are expressed as fold relative to MCF-7 cells. C) Western blot of MUC1 CD in the indicated cell lines either untransfected (UT) or 24, 48, or 72 h after mock transfection (mock), control AS (CTL), or AS miR-145 transfection (miR-145 AS). 15 μg of WCE were separated on 14% Tris-glycine gels and immunoblotted first for MUC1 CD and then the membranes were stripped and reprobed with α-tubulin as a loading control.
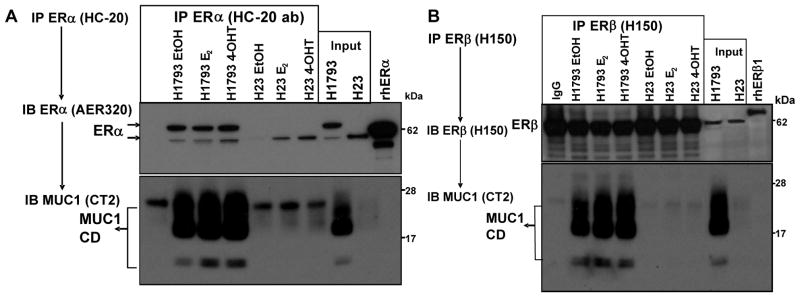
MUC1 interacts with ERα and ERβ in lung adenocarcinoma cell lines
Since MUC1 interacts with ERα in the nucleus of MCF-7 cells (17), we evaluated the interaction of ERα and ERβ with MUC1 in NE prepared from H1793 and H23 cells and used MCF-7 cells as a positive control (Fig. 3). ERα and ERβ are located in the nucleus, cytoplasm, and mitochondria of H1793, H23, and MCF-7 cells (34). MUC1 interacted with nuclear ERα and ERβ in H1793 cells in accordance with higher basal MUC1 expression. The MW estimate of the two ERα bands detected in the IP was 66 and 46 kDa. Notably, only the 46 kDa band (ERσ46) was detected in H23 cells (also see input lane), despite the detection of full length ERα using another antibody (Supplemental Fig. 3). These data reflect the expression of ERα variants in lung adenocarcinoma cells as reported by us (3, 23, 34) and others (35). Low MUC1-ERα46 and MUC1-ERβ interaction was seen in H23 cells, reflecting low MUC1 protein.
Figure 3. MUC1- ERα and ERβ interaction in lung adenocarcinoma cells.
H23 and H1793 lung adenocarcinoma cells were grown in medium with 5% DCC-FBS for 72 h prior to treatment with ethanol vehicle control (EtOH), 10 nM E2, or 100 nM 4-OHT for 1 h. NE, 400 μg protein, were immunoprecipitated with HC-20 anti- ERα (A) or H150 anti-ERβ polyclonal antibodies. The membranes were probed with the specified antibodies indicated. Baculovirus-expressed recombinant human ERα and ERβ1 were separated in parallel as a control.
PMIP localizes to the nucleus of lung adenocarcinoma cell lines
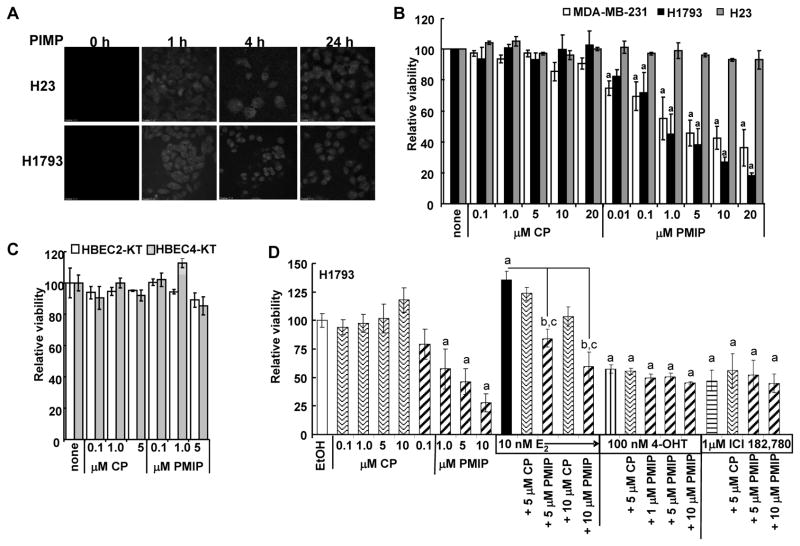
PMIP (10 μM) was taken up by BT-20 cells without any transfection reagent after 4 h incubation (21). H23 and H1793 cells were incubated with 10 μM FITC-PMIP for 1, 4, and 24 h (Fig. 4A). PMIP-FITC was distributed throughout the cell with greater nuclear uptake at 4 and 24 h compared to 1 h and no apparent difference in immunofluorescence signal between 4 and 24 h.
Figure 4. PMIP uptake by H23 and H1793 cells and inhibition of H1793 proliferation.
A) H23 and H1793 cells were incubated with 10 μM PMIP-FITC for 1, 4 and 24 h. Cells were washed, fixed and images were captured at the same exposure setting. B-D) MTT assays were performed and values are an average of 2–6 ± SEM. B) MDA-MB-231 breast cancer cells and H1793 and H23 lung adenocarcinoma cells, and C) HBEC2-KT and HBEC4-KT were incubated with the indicated concentrations of control peptide (CP) or PMIP for 5 d with fresh PMIP added every 24 h. D) H1793 cells were treated with the indicated concentrations of CP or PMIP alone +/− E2, 4-OHT, or ICI 182,780 for 5 d. Significantly different (p < 0.05) from: a control; b the same concentration of PMIP alone; c 10 nM E2 alone.
PMIP inhibits H1793 cell proliferation
To determine if PMIP inhibits cell proliferation, H23, H1793, HBEC2-KT, and HBEC4-KT were treated with control peptide (CP) or PMIP (Fig. 4B). Since PMIP inhibited MDA-MB-231 breast tumor xenograft growth in mice (21), MDA-MB-231 cell viability was determined in parallel and was inhibited in a concentration-dependent manner (IC50 = 1.5 μM). Notably, H1793, but not H23, HBEC2-KT, or HBEC3-KT, cells were selectively inhibited by PMIP (Fig. 4B and C, IC50 = ~ 0.1 μM). These data correspond to the relative MUC1 protein in each cell line (Fig. 1). E2 reduced the PMIP inhibition of cell viability (Fig. 4D), but not completely. PMIP did not synergize with inhibition of cell viability by ER antagonists 4-OHT or ICI 182,780 (Fig. 4D).
PMIP inhibits ER-activated gene transcription in H1793 cells
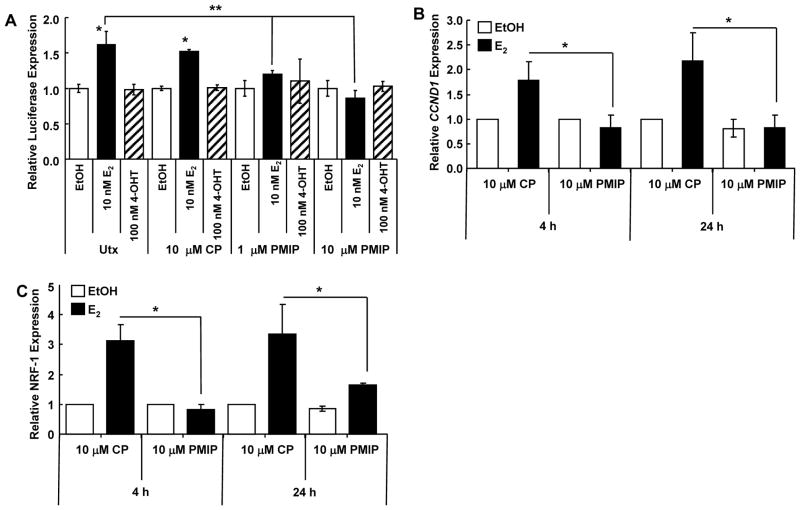
Because MUC1 was reported to be an ERα coactivator in MCF-7 cells (17), we examined if PMIP inhibited ER-mediated transcription in H1793 cells (Fig. 5A). PMIP did not affect basal luciferase activity, but significantly inhibited E2-induced ERE-driven luciferase activity indicating that endogenous MUC1 plays a role in ER-mediated transcription in H1793 cells.
Figure 5. PMIP inhibits E2-induced transcription in H1793 cells.
A) H1793 cells were transfected with an ERE-luciferase reporter, a Renilla luciferase reporter, and treated with 10 μM CP, 1 μM PMIP, or 10 μM PMIP and with EtOH, 10 nM E2, or 100 nM 4-OHT for 24 h. Values are firefly luciferase/Renilla luciferase normalized to untreated/EtOH. * significantly different from * EtOH or ** 10 nM E2/Utx, p < 0.05. B and C) H1793 cells were treated with 10 μM CP or 10 μM PMIP and with EtOH or 10 nM E2 for 4 or 24 h. Q-PCR for CCND1 (B) and NRF1 mRNA expression. * significantly different from CP +E2, p < 0.05. In all panels, values are the average of three separate experiments ± SEM.
E2 increases cyclin D1 (CCND1) (23, 34) and NRF-1 (NRF1) (36) transcription in H1793 cells. Here we found that 10 μM PMIP inhibited E2-induced CCND1 and NRF-1 transcription (Fig. 5B and C), a result in agreement with PMIP inhibition of cell viability (Fig. 4B).
PMIP affects MUC1, ERα, and ERβ intracellular location in a time-dependent manner
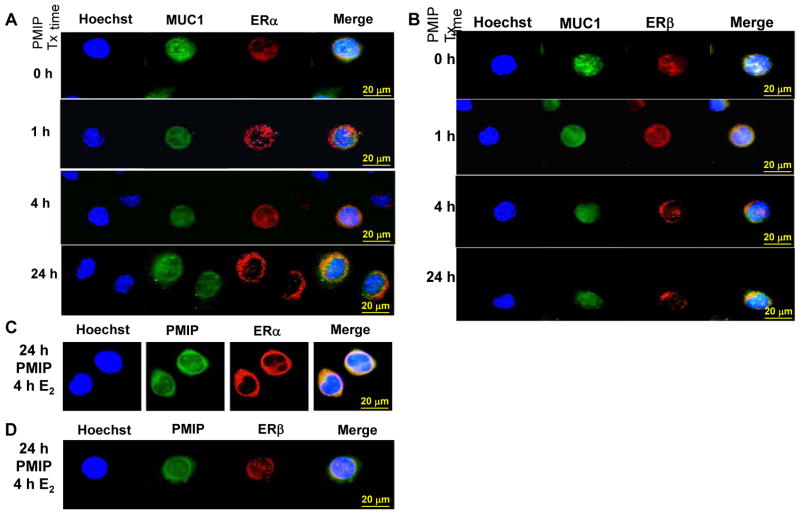
To examine how PMIP affected the intracellular localization of MUC1 in H1793 cells, cells were treated with 10 μM PMIP for 1, 4, or 24 h (Fig. 6A). Treatment with PMIP increased MUC1 nuclear localization with 1 and 4 h treatment. After 24 h of PMIP, MUC1 was detected in the nucleus and cytoplasm. Notably, colocalization of ERα and ERβ with MUC1 is seen in the merged images (Fig. 6A and B). We also examined MUC1 and PMIP intracellular localization (Supplemental Fig. 4). MUC1 and PMIP were colocalized at all time points.
Figure 6. PMIP affects MUC1, ERα, and ERβ subcellular location in H1793 cells.
Immunofluorescent microscopic imaging of H1793 cells. A and B) H1793 cells were treated with 10 μM PMIP for the indicated time and incubated with MUC1 antibody (green), ERα (HC-20, red in A), and ERβ (H150, red in B). C and D) H1793 cells treated with 10 μM FITC-PMIP (green) for 20 h and then 10 nM E2 was added for the last 4 h. MUC1 antibody (green), ERα (HC-20, red in C) and ERβ (H150, red in D). Nuclei are stained with Hoechst (blue color). Bars (gold) are 20 μm. The merged image is indicated.
We examined how PMIP affects ERα and ERβ cellular distribution in H1793 cells (Fig. 6A and B). ERα was predominantly cytoplasmic in the absence of any treatment and there was an increase in nuclear ERα with 4 h PMIP treatment followed by a return to the cytoplasmic localization at 24 h PMIP treatment (Fig. 6A). In contrast, ERβ was primarily nuclear in the absence of PMIP treatment and 1 h of PMIP increased ERβ nuclear intensity (Fig. 6B). However, 4 and 24 h of PMIP resulted in ERβ translocation from the nucleus to the cytoplasm (Fig. 6B). Treatment of H1793 cells with 10 nM E2 for 4 h increased nuclear MUC1 (Supplemental Fig. 4B). E2 did not alter the reduction in nuclear ERα seen in H1793 cells treated with PMIP (Fig. 6C). These data are in agreement with PMIP’s inhibition of E2-activated CCND1 and NRF1 transcription (Fig. 5B and C). Interestingly, 75 of 107 H1793 cells analyzed after PMIP + E2 treatment showed ERβ retention in the nucleus (Fig. 6C). These data indicate that E2 reduces the PMIP-mediated distribution of ERβ from the nucleus to the cytoplasm in ~ 70 % of cells. ERα and ERβ colocalize with PMIP (Fig. 6C and D), reflecting the colocalization of ERα and ERβ with MUC1 (Fig. 6A and B).
Discussion
Since MUC1 is an oncogene and its overexpression is a marker for reduced survival of lung adenocarcinoma patients (37), inhibiting MUC1 function has become an important target for therapeutic intervention (18). At the same time, MUC1, when appropriately expressed on the apical surface, is important for normal lung function and protective mucus production (38). Inhibition of MUC1 reduced MDA-MB-231 xenograft tumor growth in (16, 21). Here we used the MUC1 inhibitory peptide PMIP created by Schroder’s group to be taken up by cells without any sort of transfection (21). Similar to that study, we found that H1793 and H23 cells took up PMIP without transfection within 1 h of treatment. We observed that PMIP only inhibited the proliferation of H1793 cells that expressed significantly higher levels of MUC1 protein compared to H23 cells and not HBECs that are MUC1 null. While this report was in preparation, Kufe’s group reported that MUC1 inhibitors called GO- 201, 202, 203 that bind the MUC1-CD, inhibited the proliferation of lung adenocarcinoma cell lines including A549 and H1795 without affecting normal human lung epithelial cells (22). Thus, this is the second report that inhibition of MUC1 function reduces lung adenocarcinoma cell proliferation. At present, there is not a clear way to compare the efficacy of GO- 201, 202, 203 to PMIP since these peptides were not tested in the same cell lines, but the IC50 for PMIP for H1793 reported here is 0.1 μM whereas 5 μM GRO-202 and GRO-201 inhibited H1975 lung cancer cell proliferation by 50% (no IC50 values were provided)(22). Since MUC1 interacts with many proteins and has multiple roles in cells, the precise mechanisms by which inhibiting MUC1 with PMIP reduces H1793 proliferation, remains to be defined. We demonstrated that PMIP inhibited E2-induced transcriptional responses, indicating that ER is one target of PMIP’s blockade of MUC1 action. Indeed, PMIP treatment resulted in retention of ERα, but not ERβ in the cytoplasm. Additional MUC1 targets may also be involved in PMIP activity in H1793 cells. One such MUC1 interacting partner is EGFR as indicated in recent MUC1 inhibitor studies in NSCLC with EGFR and Ras mutation (22). EGFR is overexpressed and mutated in lung adenocarcinoma. Both H1793 and H23 cell lines have wild type EGFR and mutant p53 (39). A focus of future studies will be to identify the interacting protein partners of intracellular MUC1 in lung adenocarcinoma cell lines and how PMIP affects those interactions.
Estrogens induce differentiation and maturation of the lung, but the role of estrogens in NSCLC is controversial. Some studies, reviewed in (40), indicate a role for estrogen in lung cancer risk, but recent epidemiological data indicate a minor role for estrogens in lung cancer (41). We and others reported ERα and ERβ expression in lung adenocarcinoma cells and tumors(reviewed in (3)). Because ER was transcriptionally active in some lung adenocarcinoma cell lines (3) and MUC1 is an ERα coactivator in MCF-7 cells (17), here we examined ER-MUC1 interaction and inhibition by PMIP in lung adenocarcinoma cells.
We report that MUC1 protein expression is higher in 4 lung adenocarcinoma cell lines from females than 4 cell lines from males. MUC1 in the top 10% of overexpressed genes in lung cancer (www.oncomine.org). The factors regulating MUC1 expression appear to be cell-type specific. E2 and tamoxifen increased the secreted MUC1 isoform transcription via ERα activation in breast cancer cells (29), but E2 did not increase MUC1 in human endometrial cancer cells (42). We did not detect E2 regulation of MUC1 mRNA in any lung adenocarcinoma cells, HBECs, or MCF-7 cells and conclude that E2 does not regulate MUC1 in these cells. The difference between our results and those reported earlier in MCF-7 cells (29) is that we used Q-PCR rather than a MUC1-promoter-reporter and non-quantitative PCR. While the samples size is small, we observed no significant difference in MUC1/A and MUC1/B splice variant expression between lung adenocarcinoma cell lines derived from female versus male patients. Exploring gender- or disease- specific associations with MUC1 splice variant expression requires a large sample size and is a separate project.
As reported for MCF-7 cells (17), ERα interacts with MUC1 in H1793 cells. Although H23 has lower MUC1, MUC1 interacted with the ERα46 band detected in H23 cells. The role for ERα46 in lung cancer is unknown and not the focus of this study, but in breast cancer cells it is a dominant-negative effector of ERα66 (43). We report for the first time that nuclear ERβ interacts with MUC1 in H1793 cells. The role of ERβ is not fully understood (44). ERβ is considered antiproliferative in breast cancer but its role in lung adenocarcinoma remains undefined. Because the gene targets of ERβ are only now being identified (45), and are still unknown in lung, future studies to identify ERβ-MUC1-regulated genes in lung cancer will be informative.
Given the higher MUC1 protein in H1793 compared to H23, we were surprised to find comparable MUC1 mRNA. Although H23 expresses both MUC1/A and MUC1/B variants whereas H1793 express only MUC1/A, the variant is in the NTD which is in the extracellular space (31). There are no reports regarding differential mRNA stability of MUC1 and only 3 reports on miRNA regulation of MUC1 (32, 33, 46). A miR-1226 site at position 90–96 with an 8-mer seed match was found in TargetScan (47); Miranda (48) identified a miR-183 site in the 3′UTR of MUC1; and MUC1 was not in the PicTar (49) data base. Overexpression of a miR-1226 mimic in MCF-7 and MCF-10A cells reduced MUC1 protein (46). However, this paper failed to correlate miR-1226 expression with either mRNA or protein levels of MUC1 in 3 of 6 breast cancer cell lines, indicating that other cell-specific factors regulate MUC1 protein. miR-183 is upregulated in squamous cell lung cancer compared to normal lung (50). Despite not being identified as a high-scoring candidate, miR-145 reduced MUC1 in MDA-MB-231 cells (33). Likewise, miR-125b regulated MUC1 in BT-549 breast cancer cells (32). Although miR-145 expression was lower in H1793 than H23 cells, transfection of miR-145 AS did not increase MUC1 expression in H23 cells. Further studies are needed to determine the mechanisms regulating MUC1 protein expression in H23 cells.
Supplementary Material
Acknowledgments
Financial Support: NIH DK53220, NIH CA138410, the Kentucky Lung Cancer Research Program, and an intramural research incentive grant from the Office of the Executive Vice President for Research at the University of Louisville to C.M.K. Y.I.F. was supported by NIH F31 EY017275. S.M.A. was supported by NIH T35 DK072923.
We thank Farzan Pouranfour for performing western blots on MUC1 expression. We thank Dr. John Minna for the gift of the HBEC cell lines. We thank Dr. Barbara J. Clark for her review of this manuscript.
Abbreviation list
- 4-OHT
4-hydroxtyamoxifen
- CD
MUC1 C-terminal intracellular domain
- CP
control peptide
- DCC-FBS
5% dextran-coated charcoal stripped FBS
- EGFR
epidermal growth factor receptor
- E2
estradiol
- ERα
estrogen receptor α
- ERβ
estrogen receptor β
- ERE
estrogen response element
- EtOH
ethanol (vehicle control)
- FBS
fetal bovine serum
- HBECs
normal human bronchial epithelial cells
- ICI
ICI 182,780 (Fulvestrant)
- MUC1
mucin
- NSCLC
non-small cell lung cancer
- NRF-1
nuclear respiratory factor-1
- NTD
N-terminal domain of MUC1
- PM
plasma membrane
- PMIP
MUC1 inhibitory peptide (MIP) cloned adjacent to the protein transduction domain (PTD4)
- WCE
whole cell extracts
Footnotes
Conflicts of interest: None of the authors has potential conflicts of interest
References
- 1.Paris C, Clement-Duchene C, Vignaud JM, et al. Relationships between lung adenocarcinoma and gender, age, smoking and occupational risk factors: A case-case study. Lung Cancer. 2010;68:146–53. doi: 10.1016/j.lungcan.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Bouchardy C, Benhamou S, Schaffar R, et al. Lung cancer mortality risk among breast cancer patients treated with anti-estrogens. Cancer. 2011;117:1288–95. doi: 10.1002/cncr.25638. [DOI] [PubMed] [Google Scholar]
- 3.Dougherty SM, Mazhawidza W, Bohn AR, et al. Gender difference in the activity but not expression of estrogen receptors alpha and beta in human lung adenocarcinoma cells. Endocr-Relat Cancer. 2006;13:113–34. doi: 10.1677/erc.1.01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pietras RJ, Marquez-Garban DC. Membrane-Associated Estrogen Receptor Signaling Pathways in Human Cancers. Clin Cancer Res. 2007;13:4672–6. doi: 10.1158/1078-0432.CCR-07-1373. [DOI] [PubMed] [Google Scholar]
- 5.Siegfried JM, Hershberger PA, Stabile LP. Estrogen Receptor Signaling in Lung Cancer. Seminars in Oncology. 2009;36:524–31. doi: 10.1053/j.seminoncol.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gendler SJ. MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia. 2001;6:339–53. doi: 10.1023/a:1011379725811. [DOI] [PubMed] [Google Scholar]
- 7.Carson DD. The cytoplasmic tail of MUC1: a very busy place. Sci Signal. 2008;1:pe35. doi: 10.1126/scisignal.127pe35. [DOI] [PubMed] [Google Scholar]
- 8.Chang TH, Szabo E. Induction of differentiation and apoptosis by ligands of peroxisome proliferator-activated receptor gamma in non-small cell lung cancer. Cancer Res. 2000;60:1129–38. [PubMed] [Google Scholar]
- 9.Kim KC, Lillehoj EP. MUC1 Mucin: A Peacemaker in the Lung. Am J Respir Cell Mol Biol. 2008;39:644–7. doi: 10.1165/rcmb.2008-0169TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitsuta K, Yokoyama A, Kondo K, Nakajima M, Arita K, Kohno N. Polymorphism of the MUC1 mucin gene is associated with susceptibility to lung adenocarcinoma and poor prognosis. Oncol Rep. 2005;14:185–9. [PubMed] [Google Scholar]
- 11.Li Y, Liu D, Chen D, Kharbanda S, Kufe D. Human DF3//MUC1 carcinoma-associated protein functions as an oncogene. Oncogene. 2003;22:6107–10. doi: 10.1038/sj.onc.1206732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powell CA, Spira A, Derti A, et al. Gene Expression in Lung Adenocarcinomas of Smokers and Nonsmokers. Am J Respir Cell Mol Biol. 2003;29:157–62. doi: 10.1165/rcmb.2002-0183RC. [DOI] [PubMed] [Google Scholar]
- 13.Amelung JT, Buhrens R, Beshay M, Reymond MA. Key genes in lung cancer translational research: a meta-analysis. Pathobiology. 2010;77:53–63. doi: 10.1159/000278292. [DOI] [PubMed] [Google Scholar]
- 14.Awaya H, Takeshima Y, Yamasaki M, Inai K. Expression of MUC1, MUC2, MUC5AC, and MUC6 in Atypical Adenomatous Hyperplasia, Bronchioloalveolar Carcinoma, Adenocarcinoma With Mixed Subtypes, and Mucinous Bronchioloalveolar Carcinoma of the Lung. American Journal of Clinical Pathology. 2004;121:644–53. doi: 10.1309/U4WG-E9EB-FJN6-CM8R. [DOI] [PubMed] [Google Scholar]
- 15.Kesimer M, Kirkham S, Pickles RJ, et al. Tracheobronchial air-liquid interface cell culture: a model for innate mucosal defense of the upper airways? Am J Physiol Lung Cell Mol Physiol. 2009;296:L92–100. doi: 10.1152/ajplung.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raina D, Ahmad R, Joshi MD, et al. Direct Targeting of the Mucin 1 Oncoprotein Blocks Survival and Tumorigenicity of Human Breast Carcinoma Cells. Cancer Res. 2009;69:5133–41. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X, Xu H, Kufe D. MUC1 Oncoprotein Stabilizes and Activates Estrogen Receptor [alpha] Molecular Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Rajabi H, Kufe D. Mucin 1 C-Terminal Subunit Oncoprotein Is a Target for Small-Molecule Inhibitors. Molecular Pharmacology. 2011;79:886–93. doi: 10.1124/mol.110.070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren J, Agata N, Chen D, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163–75. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, McConnell MJ, Yu B, et al. MUC1 is a downstream target of STAT3 and regulates lung cancer cell survival and invasion. Int J Oncol. 2009;35:337–45. [PMC free article] [PubMed] [Google Scholar]
- 21.Bitler BG, Menzl I, Huerta CL, et al. Intracellular MUC1 Peptides Inhibit Cancer Progression. Clin Cancer Res. 2009;15:100–9. doi: 10.1158/1078-0432.CCR-08-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raina D, Kosugi M, Ahmad R, et al. Dependence on the MUC1-C oncoprotein in non-small cell lung cancer cells with RGFR and K-RAS mutations. Molecular Cancer Therapeutics. 2011 doi: 10.1158/1535-7163.MCT-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanova MM, Mazhawidza W, Dougherty SM, Minna JD, Klinge CM. Activity and intracellular location of estrogen receptors [alpha] and [beta] in human bronchial epithelial cells. Mol Cell Endocrinol. 2009;205:12–21. doi: 10.1016/j.mce.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez RD, Sheridan S, Girard L, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–34. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 25.Obermair A, Schmid BC, Packer LM, et al. Expression of MUC1 splice variants in benign and malignant ovarian tumours. International Journal of Cancer. 2002;100:166–71. doi: 10.1002/ijc.10456. [DOI] [PubMed] [Google Scholar]
- 26.Tyulmenkov VT, Jernigan SC, Klinge CM. Comparison of transcriptional synergy of estrogen receptors alpha and beta from multiple tandem estrogen response elements. Mol Cell Endocrinol. 2000;165:151–61. doi: 10.1016/s0303-7207(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 27.Walsh MD, Luckie SM, Cummings MC, Antalis TM, McGuckin MA. Heterogeneity of MUC1 expression by human breast carcinoma cell lines in vivo and in vitro. Breast Cancer Res Treat. 1999;58:255–66. doi: 10.1023/a:1006345301364. [DOI] [PubMed] [Google Scholar]
- 28.Ren L, Marquardt MA, Lech JJ. Estrogenic effects of nonylphenol on pS2, ER and MUC1 gene expression in human breast cancer cells-MCF-7. Chem Biol Interact. 1997;104:55–64. doi: 10.1016/s0009-2797(97)03767-8. [DOI] [PubMed] [Google Scholar]
- 29.Zaretsky JZ, Barnea I, Aylon Y, Gorivodsky M, Wreschner DH, Keydar I. MUC1 gene overexpressed in breast cancer: structure and transcriptional activity of the MUC1 promoter and role of estrogen receptor alpha (ERalpha) in regulation of the MUC1 gene expression. Mol Cancer. 2006;5:57. doi: 10.1186/1476-4598-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ring BZ, Seitz RS, Beck RA, et al. A novel five-antibody immunohistochemical test for subclassification of lung carcinoma. Mod Pathol. 2009 doi: 10.1038/modpathol.2009.60. [DOI] [PubMed] [Google Scholar]
- 31.Imbert Y, Darling DS, Jumblatt MM, et al. MUC1 splice variants in human ocular surface tissues: Possible differences between dry eye patients and normal controls. Experimental Eye Research. 2006;83:493–501. doi: 10.1016/j.exer.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 32.Rajabi H, Jin C, Ahmad R, McClary C, Joshi MD, Kufe D. MUCIN 1 oncoprotein expression is suppressed by the miR-125b oncomir. Genes Cancer. 2010;1:62–8. doi: 10.1177/1947601909357933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sachdeva M, Mo Y-Y. MicroRNA-145 Suppresses Cell Invasion and Metastasis by Directly Targeting Mucin 1. Cancer Res. 2010;70:378–87. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanova MM, Mazhawidza W, Dougherty SM, Klinge CM. Sex Differences in Estrogen Receptor Subcellular Location and Activity in Lung Adenocarcinoma Cells. Am J Respir Cell Mol Biol. 2010;42:320–30. doi: 10.1165/rcmb.2009-0059OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stabile LP, Lyker JS, Gubish CT, Zhang W, Grandis JR, Siegfried JM. Combined targeting of the estrogen receptor and the epidermal growth factor receptor in non-small cell lung cancer shows enhanced antiproliferative effects. Cancer Res. 2005;65:1459–70. doi: 10.1158/0008-5472.CAN-04-1872. [DOI] [PubMed] [Google Scholar]
- 36.Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of Nuclear Respiratory Factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. 2008;22:609–22. doi: 10.1210/me.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacDermed DM, Khodarev NN, Pitroda SP, et al. MUC1-associated proliferation signature predicts outcomes in lung adenocarcinoma patients. BMC Med Genomics. 2010;3:16. doi: 10.1186/1755-8794-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Copin MC, Buisine MP, Devisme L, et al. Normal respiratory mucosa, precursor lesions and lung carcinomas: differential expression of human mucin genes. Front Biosci. 2001;6:D1264–75. doi: 10.2741/copin. [DOI] [PubMed] [Google Scholar]
- 39.http://www.sanger.ac.uk/perl/genetics/CGP/. [cited; Available from:
- 40.Stabile LP, Siegfried JM. Sex and gender differences in lung cancer. J Gend Specif Med. 2003;6:37–48. [PubMed] [Google Scholar]
- 41.Baik CS, Strauss GM, Speizer FE, Feskanich D. Reproductive factors, hormone use, and risk for lung cancer in postmenopausal women, the Nurses’ Health Study. Cancer Epidemiol Biomarkers Prev. 2010;19:2525–33. doi: 10.1158/1055-9965.EPI-10-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horne AW, Lalani E-N, Margara RA, White JO. The effects of sex steroid hormones and interleukin-1-beta on MUC1 expression in endometrial epithelial cell lines. Reproduction. 2006;131:733–42. doi: 10.1530/rep.1.00883. [DOI] [PubMed] [Google Scholar]
- 43.Klinge CM, Riggs KA, Wickramasinghe NS, et al. Estrogen receptor alpha 46 is reduced in tamoxifen resistant breast cancer cells and re-expression inhibits cell proliferation and estrogen receptor alpha 66-regulated target gene transcription. Molecular and Cellular Endocrinology. 2010;323:268–76. doi: 10.1016/j.mce.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deroo BJ, Buensuceso AV. Minireview: Estrogen Receptor-{beta}: Mechanistic Insights from Recent Studies. Mol Endocrinol. 2010;24:1703–14. doi: 10.1210/me.2009-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao C, Dahlman-Wright K, Gustafsson J-Å. Estrogen Signaling via Estrogen Receptor β. Journal of Biological Chemistry. 2010;285:39575–9. doi: 10.1074/jbc.R110.180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin C, Rajabi H, Kufe D. miR-1226 targets expression of the mucin 1 oncoprotein and induces cell death. Int J Oncol. 2010;37:61–9. doi: 10.3892/ijo_00000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.http://targetscan.org/. [cited; Available from:
- 48.http://www.microrna.org/microrna/getGeneForm.do. [cited; Available from:
- 49.http://pictar.mdc-berlin.de
- 50.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA Classifiers for Predicting Prognosis of Squamous Cell Lung Cancer. Cancer Res. 2009;69:5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.