Abstract
The GTPase Rab5a regulates the homotypic and heterotypic fusion of membranous organelles during the early stages of endocytosis. Many of the molecules which regulate the Rab5a cycle of association with membranes, activation, deactivation and dissociation are known. However, the extent to which these molecular scale activities are coordinated on membranes to affect the behavior of individual organelles has not been determined. This study used novel Förster Resonance Energy Transfer (FRET) microscopic methods to analyze the Rab5a cycle on macropinosomes, which are large endocytic vesicles that form in ruffled regions of cell membranes. In Cos-7 cells and mouse macrophages stimulated with growth factors, Rab5a activation followed immediately after its recruitment to newly formed macropinosomes. Rab5a activity increased continuously and uniformly over macropinosome membranes then decreased continuously, with Rab5a deactivation preceding dissociation by 1–12 min. Although the maximal levels of Rab5a activity were independent of organelle size, Rab5a cycles were longer on larger macropinosomes, consistent with an integrative activity governing Rab5a dynamics on individual organelles. The Rab5a cycle was destabilized by microtubule depolymerization and by bafilomycin A1. Overexpression of activating and inhibitory proteins indicated that active Rab5a stabilized macropinosomes. Thus, overall Rab5a activity on macropinosomes is coordinated by macropinosome structure and physiology.
Keywords: Macropinocytosis, Rab5a, FRET, fluorescence, Rabex-5, Rin-1, RabGAP-5
Introduction
Rab5 regulates the fusion, trafficking and recycling of early endosomes (1, 2). The Rab5 isoforms Rab5a, b and c are regulated by activating guanine nucleotide exchange factors (GEFs), inhibitory GTPase-activating proteins (GAPs) and proteins which facilitate Rab5 delivery to and removal from membranes (3). Rab5 functions in a multi-step cycle in which it associates with endosomal membranes in an inactive form, is activated by a GEF, and binds to effector proteins such as Rabaptin-5 (4), EEA1 (5), Rabankyrin-5 (6), Rabenosyn-5 (7) or the type III phosphoinositol 3-kinase Vps34 (8). Rab5 is deactivated at the membrane by a GAP and then dissociates from the membrane as other Rab proteins increase their association. Membrane association is regulated by GDP-dissociation inhibitors (GDI) (9) and GDI-displacement factors (GDF) (10).
Despite consensus about the Rab5 cycle of membrane association and activation, the mechanisms which coordinate Rab5 dynamics on endocytic membranes remain largely unexplained. Rab5 GEFs and GAPs are regulated by proteins which are themselves regulated by other enzymes or by phosphatidylinositol 3-phosphate (PI3P) on vesicle membranes. Rab5 activation may involve positive feedback amplification through the Rab5 GEF Rabex-5 and the Rab5 effector Vps34, a type III PI 3-kinase which synthesizes PI3P (11). Rab5 deactivation could result from GAP activities of other signaling proteins on endocytic vesicles; for example, p85α, the regulatory subunit of phosphatidylinositol 3-kinase IA, is a Rab5 GAP (12). Rab5 deactivation occurs coordinately with the arrival of the late endosomal protein Rab7, through Rab7-dependent interference with Rabex-5 function (1, 13). However, it remains unknown how much individual Rab5 molecular cycles are coordinated with each other on any individual organelle and the extent to which organelle physiology or structure modulate Rab5 activity cycles.
Macropinosomes are large endocytic organelles which form in response to various stimuli. Some cells exhibit macropinocytosis spontaneously, while in other cells macropinocytosis is initiated by growth factors or membrane-permeabilizing peptides (14). Rab5 regulates macropinosome formation and maturation (6, 15). As macropinosomes mature, they either recycle to the cell surface or lose Rab5 and acquire Rab7 before fusing with lysosomes (16, 17). The relationship between Rab5 activity and macropinosome dynamics is not known.
To analyze the regulation of the Rab5a cycle on macropinosomes, we developed a FRET microscopy system to visualize and quantify Rab5a cycle dynamics. Using fluorescent protein chimeras of Rab5a (CFP-Rab5a), the Rab5-binding domain from the amino terminus of EEA1 (YFP-RBD) and IRES vectors expressing monomeric Cherry (mCherry) and regulatory proteins, we measured the Rab5a cycle on individual macropinosomes. In Cos-7 cells stimulated with epidermal growth factor (EGF) and in murine bone-marrow-derived macrophages (BMM) stimulated with macrophage colony-stimulating factor (M-CSF), quantitative fluorescence microscopy indicated that Rab5a association, activation, deactivation and dissociation occurred coordinately on individual macropinosomes. This coordination was disrupted by the microtubule depolymerizing drug nocodazole and by the proton ATPase inhibitor bafilomycin A1. Modulation of Rab5a cycling by coexpression of Rabex-5, Rin1, the GTPase-defective mutant CFP-Rab5a(S34N) or the GAP RabGAP-5 (18) indicated that active Rab5a stabilizes macropinosomes.
Results
The Rab5a Cycle on Macropinosomes
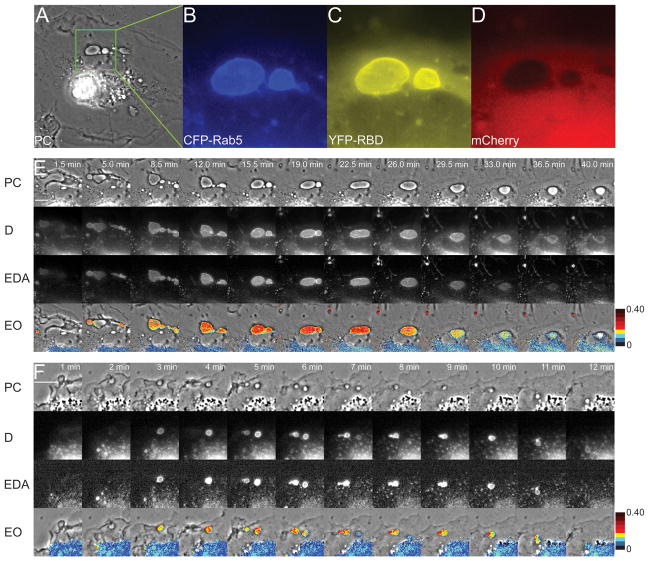
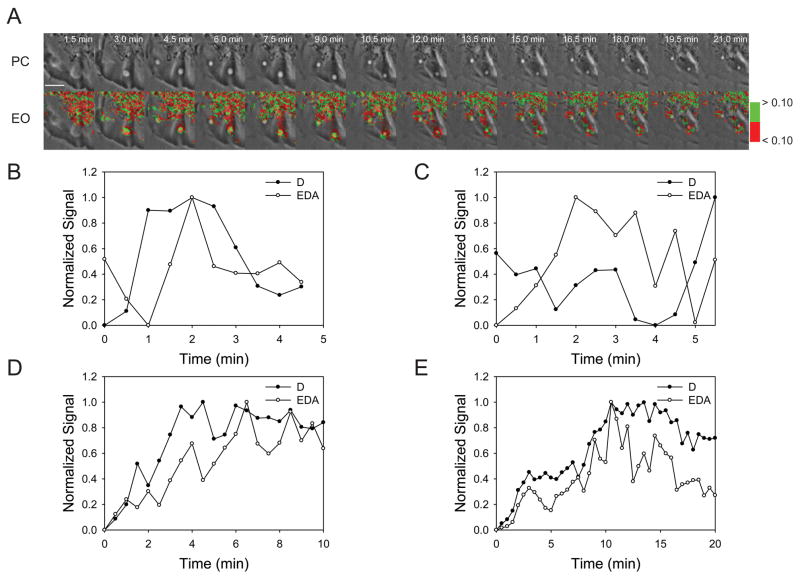
To analyze the Rab5a activation cycle on individual organelles, a widefield fluorescence microscopy system was designed for ratiometric and FRET microscopic analysis. Fluorescent chimeras of Rab5a and the amino terminal Rab5a-binding domain of EEA1 (RBD), which binds to GTP-Rab5a (19, 20), were developed and characterized. To study the activation cycle of Rab5a on macropinosomes, Cos-7 cells and murine bone marrow-derived macrophages (BMM) were transfected with plasmids encoding CFP-Rab5a, YFP-RBD and a pIRES2-mCherry vector. The mCherry reported cytoplasmic volume distribution and marked the cells expressing non-fluorescent proteins from pIRES2 transcripts. Macropinocytosis was stimulated by addition of EGF to serum-starved Cos-7 cells expressing fluorescent chimeras (21). The formation and intracellular movements of macropinosomes were observed by widefield phase-contrast and fluorescence microscopy (Fig. 1A–D). FRET stoichiometry (22) was used to measure the essential parameters of Rab5a dynamics on macropinosomes. Informative parameters included D - the total fluorescence of CFP-Rab5a corrected for fluorescence loss due to FRET, EDA - the total fluorescence attributable to FRET and EAVG -the average of the apparent donor and acceptor FRET efficiencies (Fig. 1E, F). By correcting for variations in the average ratios of CFP-Rab5a and YFP-RBD, EAVG measures the level of Rab5a activation, with EAVG of 0.40 approximating full activation. EAVG images were thresholded using binary masks made from the D images (to identify CFP-Rab5a-positive organelles), then a pseudocolor scale was applied to the masked EAVG images, assigning different colors at 5% EAVG steps (0 to 40% EAVG). Overlaying the masked, pseudocolored EAVG images onto the phase-contrast images afforded study of the movements and activation patterns of CFP-Rab5a on individual macropinosomes (EO panels Fig. 1E). Cos-7 cells formed macropinosomes of various sizes, sometimes as large as 12 μm diameter (Fig. 1A–E, Movie S1). The levels of Rab5a activation varied between macropinosomes in a single cell, but the FRET signal on each individual macropinosome was relatively uniform over the entire organelle, indicating signal integration at the organelle level. After reaching maximal EDA values, the FRET signals declined before the CFP-Rab5a dissociated from the macropinosome. Similar patterns of Rab5a dynamics were observed on macropinosomes in BMM stimulated with M-CSF (Fig. 1F, Movie S2).
Figure 1. Visualization of the Rab5a activation cycle on macropinosomes.
(A) A Cos-7 cell transfected with CFP-Rab5a, YFP-RBD and mCherry, stimulated with EGF and viewed by phase-contrast microscopy (PC). The enlarged region shows a macropinosome labeled with CFP-Rab5a (B) and YFP-RBD (C), and the distribution of cytoplasm indicated by mCherry (D). (E) A time series showing a region of the cell shown in panels A–D, processed for ratiometric imaging of CFP-Rab5a (D = ID + EDA*ξ) and for total fluorescence due to FRET between CFP-Rab5a and YFP-RBD (EDA = IF -α*IA -β*ID). In the bottom row, EAVG values are presented as pseudocolor overlaid on the phase-contrast images (EAVG = (EA + ED)/2). Color scale indicates EAVG values. EGF (100 ng/mL) was added after acquisition of the first frame. (F) BMM expressing CFP-Rab5a, YFP-RBD and mCherry, stimulated with M-CSF. Image processing for FRET stoichiometry obtained the CFP-Rab5a distribution (D), total FRET signal (EDA) and EAVG. For both cells, CFP-Rab5a recruitment to macropinosomes coincided with the increase in FRET, but FRET decreased before CFP-Rab5a dissociated from the macropinosome. Scale bars = 10 μm.
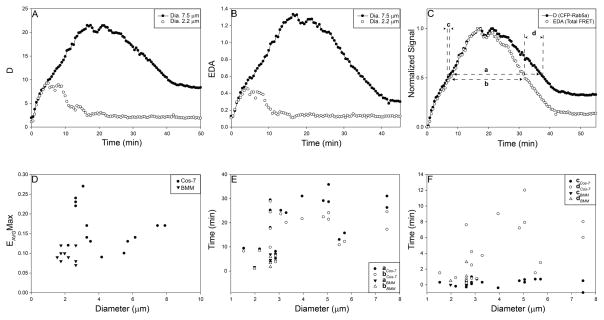
The Rab5a activation cycle was quantified and analyzed using algorithms developed in MetaMorph. CFP-Rab5a recruitment to macropinosomes increased gradually, reaching higher total levels on larger organelles (D, Fig. 2A). Similar patterns were observed for total levels of FRET (EDA, Fig. 2B). The cycle of Rab5a on each macropinosome was studied by normalizing curves to the maximal levels attained for D and EDA (Fig. 2C). The ascending curves for Rab5a association (D; CFP-Rab5a recruitment) and activation (EDA; YFP-RBD/CFP-Rab5a FRET) were nearly coincident, indicating that CFP-Rab5a was activated immediately as it associated with the macropinosome membrane. However, the EDA curve decreased sooner than the D curve, indicating that Rab5a was deactivated several minutes before it dissociated from the macropinosome.
Figure 2. The Rab5a cycle is slower on larger macropinosomes.
(A–B) Quantification of the CFP-Rab5a activation cycle on individual macropinosomes of Cos-7 (closed circles) and BMM (open circles). (A) CFP-Rab5a association and dissociation from macropinosomes. The larger macropinosome in the Cos-7 cell was labeled longer with CFP-Rab5a. (B) FRET from macropinosome-associated CFP-Rab5a and YFP-RBD (EDA). (C) Comparison of CFP-Rab5a association (closed symbols, from panel A) and activation (open symbols, from panel B) profiles on a single macropinosome from Cos-7 cells (circles) and BMM (triangles), normalized to the maximum and minimum values and plotted as Fraction of the Maximal Signal versus Time. Measured parameters were: a, for CFP-Rab5a association (D), the interval between the time the macropinosome reached half-maximal value ascending to the time it reached half-maximal value descending; b, for CFP-YFP FRET signals (EDA), the interval between the time the macropinosome reached half-maximal value ascending to the time it reached half-maximal value descending; c, for the ascending phase of D and EDA curves, the interval between the time of half-maximal D and half-maximal EDA; d, for the descending phases of the D and EDA curves, the interval between the time of half-maximal D and half-maximal EDA. (D) Maximal FRET signals (EAVGMax) on individual macropinosomes, plotted vs. macropinosome diameter. Although maximal FRET signals were lower in BMM (triangles) than in Cos-7 cells (circles) for macropinosomes of comparable sizes, maximal FRET signals did not correlate with macropinosome size. (E) Half-maximal widths for Rab5a cycles were measured in macropinosomes of Cos-7 cells (circles) and BMM (triangles). CFP-Rab5a FRET cycles (a, closed symbols) were shorter than CFP-Rab5a association cycles (b, open symbols). Cycles were generally longer on larger macropinosomes. (F) For both Cos-7 cells (circles) and BMM (triangles), CFP-Rab5a activation coincided with association for all sizes of macropinosomes (c, closed symbols), whereas deactivation preceded dissociation by up to 12 min, with larger macropinosomes exhibiting longer lag times (d, open symbols).
Because macropinosomes form asynchronously after addition of growth factor (23), we could not use biochemical methods to measure rates of Rab5 activation on macropinosomes. Net Rab5 activation could be estimated by the rate of FRET increase on individual macropinosomes. The maximal rate of EAVG increase was 0.033 per minute (± 0.003). However, this is not easily translated into a rate of Rab5 activation.
The relationships between macropinosome size and Rab5a cycle parameters were measured in several ways. Although macropinosome size in Cos-7 cells did not correlate with the maximal level of Rab5a activation (EAVGMax, Fig. 2D), the duration of the Rab5a cycle increased with macropinosome size. By measuring cycle duration as the widths of the association and activation curves (parameters a and b of Fig. 2C), we determined that macropinosomes smaller than 3 μm diameter showed shorter cycle times than did larger macropinosomes (Fig. 2E). Rab5a activation coincided with organelle association (parameter c of Fig. 2C), regardless of macropinosome size (solid symbols, Fig. 2F). However, on the descending curves, the EDA signal declined earlier than the D signal, indicating that Rab5a deactivation preceded Rab5a removal from the macropinosome membrane. The interval between Rab5a deactivation and dissociation increased with macropinosome size (open symbols, Fig. 2F). Macropinosomes formed by BMM were all less than 3 μm diameter. Although macropinosomes in BMM showed lower maximal EAVG than similarly sized macropinosomes from Cos-7 cells (Fig. 2D), their overall Rab5a cycle dynamics were similar to macropinosomes of the same size in Cos-7 cells (Fig. 2E, F).
We quantified the extent to which probe expression altered Rab5 dynamics on macropinosomes. To examine the effects of YFP-RBD expression on the Rab5a association with macropinosomes, we compared Cos-7 cells expressing YFP-RBD, CFP-Rab5a and mCherry with Cos-7 cells expressing YFP-Rab5a and CFP. The presence of the YFP-RBD increased the rate of Rab5a association with macropinosomes. In cells expressing the FRET pair, the rate of change was 0.263 ΔRM/RC per minute (+/−0.09, n=5), whereas for cells expressing the YFP-Rab5a and CFP ΔRM/RC the rate was 0.293 per minute (+/−0.06, n=4). Moreover, expression of YFP-RBD prolonged the total Rab5a cycle. Without YFP-RBD, a = 3.67 min, with YFP-RBD, a = 17.75 min. This indicates that YFP-RBD enhanced Rab5 association with macropinosomes and inhibited dissociation. However, it remains possible that overexpression of FP-Rab5a accelerated Rab5 cycle dynamics on macropinosomes, but this could not be measured by our methods.
Rab5a activation cycle modulation by GEFs
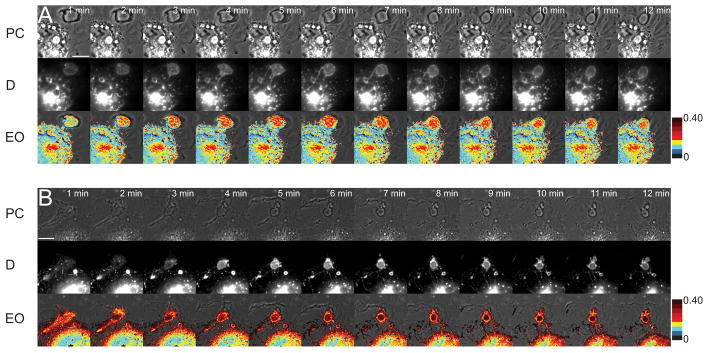
To modulate the Rab5a activation cycle, two GEFs were introduced into a pIRES2-mCherry vector and expressed in Cos-7 cells along with the Rab5a FRET reporter constructs. Use of the pIRES2 vectors expressing mCherry and non-fluorescent GEFs allowed us to use mCherry fluorescence to identify cells expressing the non-fluorescent proteins. GEF effects on the Rab5a cycle were assessed by obtaining EAVGMax for 15 macropinosomes (Fig. 3). In control macropinosomes expressing empty pIRES2-mCherry vector, EAVGMax was 0.19 ± 0.018. Cos-7 cells expressing YFP-RBD, CFP-Rab5a and pIRES2-Cherry-Rabex-5 contained enlarged macropinosomes with significantly higher FRET values (EAVGMax = 0.24 ± 0.014; p < 0.001). Cells expressing pIRES2-mCherry-Rin1 did not contain enlarged macropinosomes, but reached higher FRET levels than control macropinosomes (EAVGMax = 0.32 ± 0.01; p < 0.0001). These were similar to levels observed after expression of constitutively active Rab5a (data not shown). Expression of Rabex-5 or Rin1 also increased the abundance of FRET-positive tubular extensions from macropinosomes (Fig. 3A, Movie S3, B, Movie S4). Thus, overexpression of Rab5a GEFs increased and prolonged Rab5a activity on macropinosomes and increased tubular endosome formation.
Figure 3. Rab5a GEFs increase formation of macropinosome-associated tubules.
(A) Cos-7 cell expressing FRET probes and pIRES2-mCherry-Rabex-5. (B) Cos-7 cell expressing FRET probes and pIRES2-mCherry-Rin1. Top row, phase-contrast; middle row, D (CFP-Rab5a); bottom row, EO (EAVG Overlay). Overexpression of both GEFs increased CFP-Rab5a FRET on macropinosomes and remained higher than control. Macropinosomes extended tubules with high FRET signals. Color scale indicates EAVG FRET signals. Scale bar = 10 μm.
Bafilomycin A1 increases tubule formation and destabilizes the Rab5a cycle
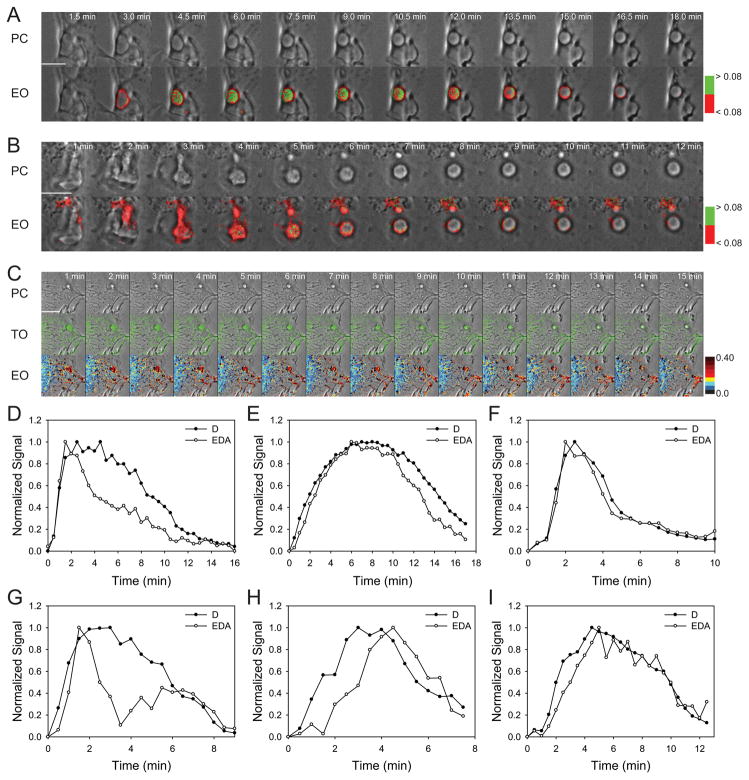
Inhibition of the proton ATPase by bafilomycin A1 inhibits acidification of endocytic compartments (24) and increases endosome tubulation (25). To examine the role of pH in the Rab5a cycle, Cos-7 cells transfected with YFP-RBD, CFP-Rab5a and mCherry were treated with bafilomycin A1 and stimulated with EGF. Bafilomycin A1 showed a range of effects on the Rab5a cycle. Macropinosomes showed increased active Rab5a on tubular endosomes protruding from macropinosomes near the center of the cell (Fig. 4C), suggesting that vacuolar pH affects Rab5-dependent tubule formation. The Rab5a cycle on macropinosomes was irregular. Compared to macropinosomes in control cells, which exhibited uniform cycles of Rab5 activation (Fig. 4A, D–F), macropinosomes in bafilomycin A1-treated cells exhibited erratic patterns of Rab5a association and activation (Fig. 4B, G–I). FRET signals on macropinosomes had a variegated appearance (Fig. 4C). CFP-Rab5a association and dissociation sometimes occurred more than once on a macropinosome and Rab5a association was not consistently accompanied by increased FRET (Fig. 4G–I).
Figure 4. Bafilomycin A1 increases tubule formation and destabilizes the Rab5a cycle on macropinosomes.
Macropinosomes in control cells (A) showed a single cycle of Rab5a activation and deactivation, whereas bafilomycin A1-treated cells (B) showed erratic patterns of CFP-Rab5a association, activation and deactivation and multiple tubular extensions with high Rab5a activity. Top row, phase-contrast; middle row, D (CFP-Rab5a); bottom row, EO [phase-contrast with overlay showing inactive CFP-Rab5a (red) and active CFP-Rab5a (green)], with the threshold for CFP-Rab5a activation set at EAVG = 8%. (C) Top row, phase-contrast; middle row, TO, phase-contrast with overlay showing active Rab5 on tubular extensions (green) of cell treated with bafilomycin A1; bottom row; EO, EAVG values presented as pseudocolor overlaid on phase-contrast. Scale bars = 10 μm. (D–I) Individual traces from macropinosomes in control (D–F) and bafilomycin A1-treated (G–I) Cos-7 cells showing CFP-Rab5 association (closed circles) and CFP-Rab5 FRET signals (open circles).
Microtubule depolymerization destabilizes the Rab5a cycle
To examine whether microtubule-based motility affects the Rab5a cycle on macropinosomes, cells expressing YFP-RBD, CFP-Rab5a and mCherry were treated with the microtubule depolymerizing agent nocodazole. The few macropinosomes which did form in these cells did not contain tubular extensions, but like the bafilomycin A1-treated cells they showed irregular patterns of Rab5 activation (Fig. 5). FRET signals were uneven and sometimes not restricted to the macropinosome itself (Fig. 5A). This was also evident from the quantitative analysis (Fig. 5B–E).
Figure 5. Nocodazole destabilizes the Rab5a cycle on macropinosomes.
(A) Rab5a activation on a macropinosome of a cell in nocodazole; top, phase-contrast; bottom, EO, phase-contrast with overlay showing inactive CFP-Rab5a (red) and active CFP-Rab5a (green), with the threshold for CFP-Rab5a activation set at EAVG = 10%. Rab5a activity fluctuates on the macropinosome. Scale bar = 5 μm. (B–E) Individual traces from macropinosomes in nocodazole-treated cells.
Inhibition of Rab5a activity destabilizes macropinosomes
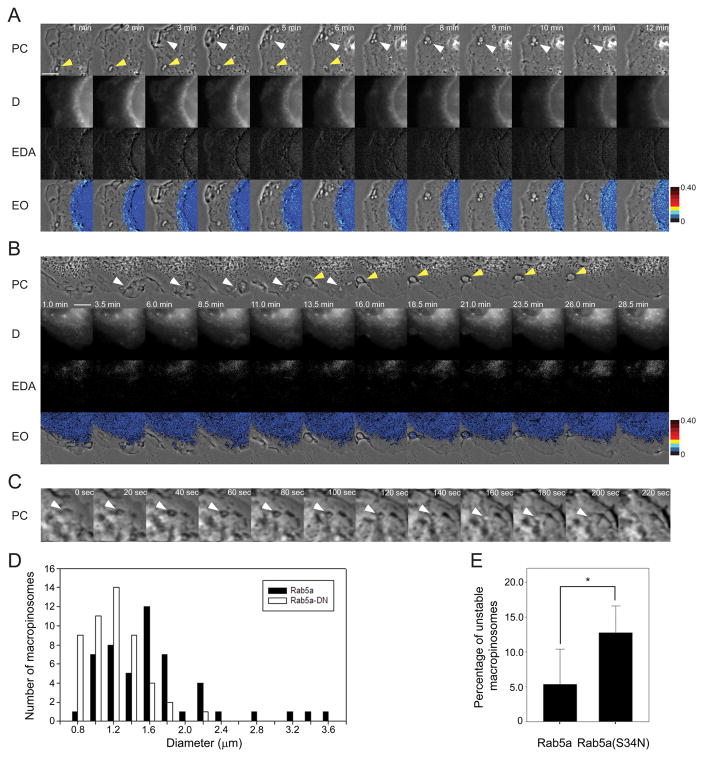
To examine the roles of small GTPases in macropinosome formation and Rab5a activation, we expressed from pIRES2-mCherry vectors GTPase-deficient forms of small GTPases which affect motility and macropinocytosis, including Arf6(T27N), Cdc42(N17), Rac1(N17) and Rab5a(S34N), and measured their effects on macropinosome formation and the Rab5a cycle. Expression of pIRES2-mCherry constructs of Arf6(T27N), Cdc42(N17) and Rac1(N17) inhibited the cells ability to ruffle and form macropinosomes (data not shown). In Cos-7 cells which were transfected with YFP-RBD, CFP-Rab5a(S34N) and pIRES2-mCherry and stimulated with EGF, no FRET was observed and the majority of macropinosomes that formed were unstable, fusing back with the membrane shortly after closure (Fig. 6A).
Figure 6. RabGAP-5 and Rab5a(S34N) destabilize macropinosomes.
(A) Cos-7 cells expressing YFP-RBD, CFP-Rab5a(S34N) and mCherry were stimulated by addition of EGF after the first frame was acquired. Small macropinosomes formed rarely. Most macropinosomes showed no association with CFP-Rab5a(S34N), no FRET and did not persist. (B) Cos-7 cells expressing YFP-RBD, CFP-Rab5a and pIRES2-mCherry-RabGAP-5 formed macropinosomes, which showed low CFP-Rab5 association and low FRET signals. The vast majority of the macropinosomes were unstable and fused back with the membrane. Some small macropinosomes that were able to form showed low FRET signals. (C) Macrophages expressing CFP-Rab5a(S34N) (black bars) formed smaller macropinosomes in response to M-CSF than macrophages expressing CFP-Rab5a (white bars). (D) Macrophages expressing CFP-Rab5a(S34N) formed unstable macropinosomes more often than did macrophages expressing CFP-Rab5a.
Expression of Rab5a GAP, RabGAP-5, lowered FRET signals and destabilized macropinosomes. Like cells expressing CFP-Rab5a(S34N), cells expressing Rab5a FRET reporters and pIRES2-mCherry-RabGAP-5 formed small macropinosomes with little CFP-Rab5a association and very low FRET signals (Fig. 6B, Movie S5). Many macropinosomes would start to form but fused back with the membrane. A small amount of CFP-Rab5a localized to these unstable macropinosomes, but EAVGMax was very low (0.04 +/− 0.007, p value < 0.0001, compared to controls). Macropinosome size and stability were measured in BMM expressing CFP and either YFP-Rab5a or YFP-Rab5a(S34N). YFP-Rab5a(S34N) decreased macropinosome size (Fig. 6D) and increased the fraction of unstable macropinosomes (Fig. 6E). Thus, the increase in unstable macropinosomes in Cos-7 and BMM expressing dnRab5a or RabGAP-5 indicated a role for Rab5a in stabilizing the macropinosome.
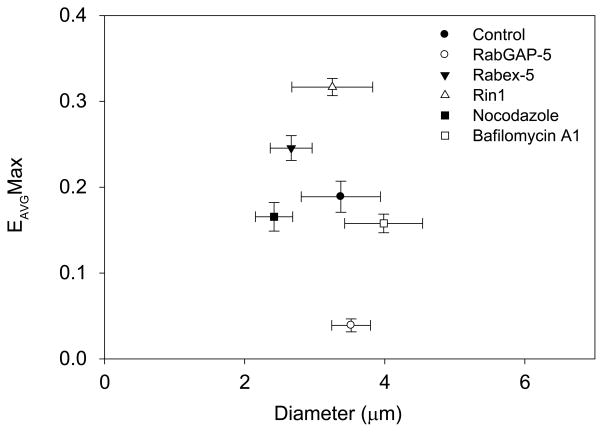
The effects of these overexpressed proteins on the Rab5a cycle in Cos-7 cells were analyzed relative to macropinosome size (Fig. 7). Macropinosomes in control cells were a wide range of sizes, all of which exhibited single cycles of Rab5a activation. Rabex-5 and Rin1 increased the overall levels of EAVGMax. RabGAP-5 and dnRab5a showed low levels of Rab5a recruitment to nascent macropinosomes and little if any Rab5a activation. Bafilomycin A1 increased macropinosome size and Rab5a activity on tubular extensions. Although both bafilomycin A1 and nocodazole destabilized the Rab5a cycle on macropinosomes, neither significantly decreased overall FRET levels.
Figure 7. The Rab5a cycle is modulated by GEFs and GAPs.
Cos-7 cells were transfected with YFP-RBD, CFP-Rab5a and pIRES2-mCherry co-expressing GEFs (Rin1 and Rabex-5), GAP (RabGAP-5) or nothing (control). Cells were stimulated and macropinosomes were imaged and analyzed as in Figures 1 and 2. For each condition, the maximum EAVG values were calculated for >9 macropinosomes and plotted as functions of macropinosome diameter. Relative to control cells expressing YFP-RBD, CFP-Rab5a and mCherry only, the GEFs, Rabex-5 and Rin1, increased EAVGMax, whereas the GAP, RabGAP-5, decreased EAVGMax. Treatment of control cells with bafilomycin A1 or nocodazole showed lower EAVGMax levels, but these were not significant.
Thus, the analysis of Rab5a cycle dynamics on macropinosomes indicated novel mechanisms that regulate the level and overall uniformity of Rab5a activity on large areas of membrane.
Discussion
Using new tools to elucidate the dynamics of Rab5 in living cells, this study revealed a novel feature of organelle biology, which is that physiological parameters affect overall molecular cycles. These tools allowed us to observe and quantify the Rab5 cycle of association with macropinosomes, activation, deactivation and dissociation. The ability to measure all of these parameters on individual organelles permitted analysis of the overall Rab5 cycle with respect to morphological transitions. Visualization of Rab5a dynamics revealed a single, continuous sequence of Rab5a localization, activation and deactivation covering the entire organelle. This indicated a large-scale coordination of Rab5 activity on macropinosomes. It is important to note that these methods measure net activities on organelles. Individual molecular cycles are likely more rapid.
The overall Rab5 cycle
Although expression of the fluorescent chimeras altered Rab5a dynamics, general patterns were evident. As the macropinosome formed, the D and EDA signals increased at the same rate, beginning just as the macropinosome closed into the cell. This indicated that the mechanisms of Rab5 association and activation are independent of the timing of growth factor addition (closure occurred at various times after EGF addition). The slope of EAVG was not affected by the size of the macropinosome. Rab5 dissociation from the membrane occurred more slowly than deactivation. This delay was partly due to expression of YFP-RBD, which significantly slowed the Rab5a cycle, but it could also be the result of phosphoinositide turnover dynamics or delayed recruitment or access of Rab5-dissociation factors.
Size-dependent activities
Different sizes of macropinosomes showed similar overall patterns of Rab5 cycle dynamics. Small macropinosomes generally cycled faster than large macropinosomes. The principal size-dependent effect was on Rab5 dissociation from the macropinosome (Fig. 2F), which indicated a role for organelle size in maturation rates or Rab conversion (13). Movement of smaller endosomes around the forming macropinosome area suggested that mechanisms such as piranhalysis (26) could play a role in reducing the cargo and diameter of the macropinosome to a size which can mature and fuse with the lysosomes.
Cycle modulation by GEFs and GAPs
The effects of Rab5 activity on macropinosome dynamics were analyzed by expressing GEFs and GAPs from mCherry-pIRES vectors. Overexpression of the GEFs Rabex-5 and Rin1 increased EAVGMax values and the persistence of active Rab5a on macropinosomes. Sustained Rab5 association and activation on macropinosomes in these cells did not allow measurement of cycle times (i.e., D and EDA remained elevated). Interestingly, cells expressing Rabex-5 contained enlarged active Rab5a-positive vesicles that had formed prior to stimulation and image acquisition, a phenotype resembling the effect of overexpression of constitutively active Rab5a (data not shown). Rin1 overexpression did not result in enlarged endosomes, indicating that the two GEFs function in different aspects of macropinosome maturation. Both GEFs increased the number of active Rab5a-positive tubular endosomes, which appeared only rarely in control cells.
Conversely, cells expressing RabGAP-5 contained macropinosomes with low levels of FRET. Such cells also contained more cytosolic CFP-Rab5a, indicating a higher fraction of inactive Rab5a in those cells. Most macropinosomes were unstable. Typically, nascent macropinosomes would close into cells with an amorphous shape, but would fail to mature into circular organelles. Rather, the irregularly shaped phase-bright organelles persisted for up to 2 minutes then collapsed, presumably by finally fusing with the plasma membrane. These unstable macropinosomes recruited little or no Rab5a and the Rab5a that did accumulate was inactive. Similar inhibition was observed in cells expressing YFP-RBD and CFP-Rab5a(S34N). BMM expressing YFP-Rab5a(S34N) and CFP increased the frequency of unstable macropinosomes when compared to BMM expressing YFP-Rab5a. These results indicate that activation of Rab5a plays a critical role in macropinosome stability. The failure of unstable macropinosomes to round up may be related to their ability to form tubular extensions. A study by Kerr and Teasdale (27) showed that sorting nexins-1 and 5 (SNX1, SNX5) contribute to the formation of Rab5a-positive tubular extensions and the rounding of macropinosomes. Rab5a may facilitate the SNX-mediated tubulation necessary for macropinosome rounding.
Irregular Rab5a cycles
The uniformity of the Rab5a cycle on macropinosomes became more obvious with the appearance of irregular Rab5a cycles. Bafilomycin A1 treatment led to erratic cycles of Rab5a and irregular localization and activation of Rab5a on the macropinosome membrane. Treatment of cells with nocodazole also caused irregular and erratic patterns of Rab5a localization and activation on macropinosomes. Bafilomycin A1- and nocodazole did not affect the maximum levels of FRET on the macropinosomes (Fig. 7). These variegated patterns of Rab5a activation indicate the existence of signal-integrating activities on organelle membranes and suggest that luminal pH and microtubule integrity affect overall cycle dynamics.
Tubular endosomes
Bafilomycin A1 increased the appearance of Rab5a-positive tubular endosomes that stretched from the nuclear area toward the edges of the cell and exhibited movements typical of microtubule-based motility. Conditions which caused sustained activation of Rab5a (Rin1 or Rabex-5) also created tubules with active Rab5a. Tubular endosome morphologies were shown by earlier studies to be microtubule-dependent (25). The increase in FRET-positive tubular endosomes after overexpression of Rabex-5 and Rin1 and after bafilomycin A1-treatment indicates a relationship between macropinosome pH, Rab5a activity and microtubule-based endosome trafficking. This could be attributable to the Rab5 effector Rabenosyn-5, which mediates microtubule-based extension of endosomal membranes (28).
Thus, imaging of Rab5a cycles on macropinosomes revealed the existence of mechanisms which shape the overall Rab5a cycle. We speculate that organelle size affects the rate of formation of 3′ phosphoinositides and concentrations of 3′ phosphoinositides affect overall levels of Rab5 GEF or GAP activities. Furthermore, these studies indicate that Rab5a activity facilitates the extension of tubules and the rounding of the macropinosome, which may help to stabilize the organelle.
Materials and Methods
Tissue culture, Treatment and Transfection
The kidney fibroblast-like cell line Cos-7 (American Type Culture Collection) was maintained at 37°C with 5% CO2. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (HIFBS), 100 U/ml penicillin, 100 μg/mL and 4 mM L-glutamine (Invitrogen). Bone marrow-derived macrophages (BMM) were prepared from femurs of C57/BL6 mice and cultured 6 days, as described previously (29). For microscopy, Cos-7 cells were plated one day prior to imaging onto 25 mm, No. 1.5 circular coverslips (Fisher Scientific) at 2.0 × 105 cells per coverslip. Cells were incubated for 4 hours to allow attachment to the coverslips, transfected with plasmids using Roche FuGENE HD, following the manufacturer’s protocol (Roche Diagnostics) and serum-starved overnight prior to imaging. BMM were transfected using Mouse Macrophage Nucleofector kit (Lonza) according to the manufacturer’s protocol, plated on coverslips and incubated in RPMI-1640 with 20% HIFBS, 4 mM L-glutamine, 20 U/ml penicillin and 20 μg/ml streptomycin for 3 hours. Cells were incubated for 20 hours in DMEM without added M-CSF.
For imaging, coverslips with cells were placed in a Leiden chamber (Harvard Apparatus) with Ringer’s buffer (155 mM NaCl, 5 mM KCl, 2 mM CaCl, 1 mM MgCl, 2 mM NaH2PO4, 10 mM glucose, 10 mM HEPES at pH 7.2) at 37°C. Macropinocytosis was stimulated by addition of 100 ng/mL epidermal growth factor (EGF; R&D Systems) to Cos-7 cells, or 200 μg/mL macrophage colony-stimulating factor (M-CSF) to BMM. Bafilomycin A1 (0.5 μM, Sigma-Aldrich) and nocodazole (5 μM, Sigma-Aldrich) were added to Cos-7 cells 30 minutes prior to image acquisition.
Plasmids
Monomeric versions of the cyan fluorescent protein (CFP), citrine (YFP) and Cherry (mCherry) were used for all chimeric constructs (30). Rab5a, Rab5a(Q79L), Rab5a(S34N), Arf6(T27N), Cdc42(N17), Rac1(N17), RabGAP-5, Rin1 and Rabex-5 were cloned in YFP, CFP or mCherry vectors. RabGAP-5 plasmid was provided by F.A. Barr (Max-Planck Institute of Biochemistry, Martinsried, Germany). Constructs for Rin1 and Rabex-5 were provided by A.R. Saltiel (University of Michigan, Ann Arbor, Michigan). Polymerase chain reaction was used to amplify the Rab5-binding domain (RBD) from the amino terminus of EEA1 (residues 36–218 aa) which was subcloned into mCit-C1 and mCFP-C1 vectors (19, 20, 31). The Clontech pIRES2-EGFP vector was modified by replacing EGFP with mCherry. Rab5a, Arf6(T27N), Cdc42(N17), Rac1(N17), RabGAP-5, Rin1 and Rabex-5 were cloned into the pIRES2-mCherry using In-Fusion Advantage PCR Cloning Kit (Clontech Laboratories Inc.). All constructs originated from human cDNA and all sequences of constructs were confirmed by DNA sequencing at the University of Michigan DNA Sequencing Core.
Ratiometric and fluorescence resonance energy transfer (FRET) microscopy
Cells were co-transfected with plasmids encoding YFP-RBD, CFP-Rab5a, mCherry or pIRES2-mCherry. The pIRES2 vector contains an internal ribosomal entry site between its multiple cloning site and the fluorophore site which allows the translation of both in a single bicistronic mRNA. The mCherry allowed for detection of cells transfected with non-fluorescent proteins. mCherry served as a cytosolic volume marker which corrected for optical pathlength due to cell shape. mCherry also allowed identification of cells expressing non-fluorescent Rab5-modifying proteins (GAPs and GEFs). Five images were acquired: Phase-contrast, IA, which contains the FRET-independent fluorescence from the acceptor, ID, which contains the fluorescence from the donor, IF, which contains a mixture of donor, acceptor, and FRET fluorescence and IR, which contains the fluorescence from mCherry. These images were acquired on a Nikon TE300 widefield inverted fluorescence microscope with a 60× 1.4 Planapo objective, excitation and emission filters in filter wheels, a temperature-controlled stage, shutters for both phase-contrast and epifluorescence and a cooled digital charge-coupled camera (Quantix; Photometrics, Tucson, AZ). The source of epifluorescent light was a mercury arc lamp (OSRAM GmbH, Augusburg, Germany). Images were corrected for exposure times and shade and bias corrections were applied before processing. All images were acquired using MetaMorph version 7.7r1 (Molecular Devices, Sunnyvale, CA).
FRET and ratiometric measurements were calculated for each cell using MATLAB R2009a (The Mathworks, Natick, MA), the DIPImage toolbox for MATLAB (http://www.diplib.org, Quantitative Imaging Group, Delft University of Technology, Netherlands) and FRET Calculator (available from the Center for Live Cell Imaging at the University of Michigan). Ratiometric images of YFP-and CFP-chimeras were calculated from three images, the YFP image IA, the CFP image ID, and mCherry image IR. The ratio of YFP- or CFP-chimera to mCherry reported the localization of each chimera normalized to cell volume. FRET stoichiometry (22) was used to measure the proportions of donor, acceptor and donor-acceptor complex in each pixel. FRET calibration parameters were determined using Cos-7 cells expressing YFP only (α), CFP only (β) or a linked YFP-CFP molecule of known FRET efficiency (γ and ξ). FRET Calculator was used to obtain the total concentrations of acceptor (A) and donor (D = ID + EDA*ξ), the FRET efficiency times the concentration of donor-acceptor complex (EDA = IF -α*IA -β*ID), the fraction of the acceptor in complex times the FRET efficiency (EA), the fraction of the donor in complex times the FRET efficiency (ED), the average FRET efficiency (EAVG = (E A+ED)/2), the ratio of acceptors to cytoplasmic marker (RYR), the ratio of donors to cytoplasmic marker (RCR) and molar ratio of acceptors to donors (RM). The EO image was created by thresholding the EAVG image with different binary masks made from D images. The masks were thresholded to identify only the CFP-Rab5a-positive organelles and a pseudocolor scale was applied to the masked EAVG images, assigning different colors at 5% EAVG steps (0 to 40% EAVG), the new scaled images were overlayed onto the phase-contrast images (EO in Figs. 1, 3, 4C and 6). In Figures 4A, B and 5 the EAVG images were thresholded with two different binaries from D images; the threshold was set to red for values from 0–8% and green from 8–40%. In figure 4C, the TO images were created by averaging the EDA images and subtracting the average noise from the original EDA images, the new EDA images were eroded and dilated in Metamorph to improve the visualization of the active Rab5a-positive tubules. The images were then overlaid as green on phase-contrast images.
Particle Tracking
The centroid-tracking algorithm TRACKOBJ of Metamorph was used to measure signals associated with macropinosomes, as previously described (22). Using the phase-contrast image, the center of the macropinosome was identified; a region encompassing the macropinosome was drawn in the phase-contrast, EA, ED, EDA, EAVG, A, D and Ratio images. To compare multiple image series, the macropinosome data traces were aligned by designating the first phase-contrast image containing a fully rounded macropinosome as the 1-min time point.
Data Analysis
To quantify the Rab5a activation cycle, the particle tracking plots for D (total Rab5a signal) and EDA (total FRET signal) images were selected and all values were normalized to the maximum and minimum values. D and EDA signals for each macropinosome were analyzed for the interval between half-maximal values ascending and descending (i.e., the width of the association or activation profiles). D and EDA signals were also analyzed to determine the intervals between D and EDA at half-maximal ascending and half-maximal descending parts of the curve (Fig. 2C). The mean EAVGMax for each modulator was used to compare the effect on Rab5a FRET during macropinocytosis. To measure EAVGMax and diameter on the unstable macropinosomes in RabGAP-5-expressing cells, the diameter of the circular ruffle was measured prior to collapse and fusion with the membrane.
Statistical analysis
A paired two-tailed Student’s t-test (Two-Sample Assuming Equal Variances) was used to compare the effect on the EAVGMax average of the macropinosomes formed in control cells, cells expressing pIRES2-mCherry-RabGAP-5, pIRES2-mCherry-Rin1 and cells expressing pIRES2-mCherry-Rabex-5.
Supplementary Material
A Cos-7 cell transfected with CFP-Rab5a, YFP-RBD and mCherry. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase-contrast images. Color scale indicates EAVG values below 12% (red) and above 12% (green). EGF (100 ng/mL) was added after acquisition of the first frame and frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 10 μm)
Primary murine bone marrow-derived macrophage transfected with CFP-Rab5a, YFP-RBD and mCherry, stimulated with M-CSF. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase-contrast images. Color scale indicates EAVG values below 8% (red) and above 8% (green). Frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 5μm)
Cos-7 cell transfected with CFP-Rab5a, YFP-RBD and pIRES2-mCherry-Rabex- 5. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase- contrast images. Color scale indicates EAVG values below 12% (red) and above 12% (green). EGF (100 ng/mL) was added after acquisition of the first frame and frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 10 μm)
Cos-7 cell transfected with CFP-Rab5a, YFP-RBD and pIRES2-mCherry-Rin1. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase-contrast images. Color scale indicates EAVG values below 12% (red) and above 12% (green). EGF (100 ng/mL) was added after acquisition of the first frame and frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 10 μm)
Cos-7 cell transfected with CFP-Rab5a, YFP-RBD and pIRES2-mCherry-RabGAP-5. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase-contrast images. Color scale indicates EAVG values below 6% (red) and above 6% (green). EGF (100 ng/mL) was added after acquisition of the first frame and frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 10 μm)
Acknowledgments
The authors gratefully acknowledge technical support and advice from Adam Hoppe, Lad Dombrowski, Michael Davis, Tim Welliver and Youxin Zhang. Supported by NIH grant AI-79414 to J.S.
References
- 1.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 2.Perskvist N, Roberg K, Kulyte A, Stendahl O. Rab5a GTPase regulates fusion between pathogen-containing phagosomes and cytoplasmic organelles in human neutrophils. J Cell Sci. 2002;115:1321–1330. doi: 10.1242/jcs.115.6.1321. [DOI] [PubMed] [Google Scholar]
- 3.Bucci C, Lutcke A, Steele-Mortimer O, Olkkonen VM, Dupree P, Chiariello M, Bruni CB, Simons K, Zerial M. Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett. 1995;366:65–71. doi: 10.1016/0014-5793(95)00477-q. [DOI] [PubMed] [Google Scholar]
- 4.Stenmark H, Vitale G, Ullrich O, Zerial M. Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell. 1995;83:423–432. doi: 10.1016/0092-8674(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 5.Mu FT, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EP, Toh BH. EEA1, an early endosome-associated protein. EEA1 is a conserved alpha-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 6.Schnatwinkel C, Christoforidis S, Lindsay MR, Uttenweiler-Joseph S, Wilm M, Parton RG, Zerial M. The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2004;2:E261. doi: 10.1371/journal.pbio.0020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson DH, Chamberlain MD. Assay and stimulation of the Rab5 GTPase by the p85 alpha subunit of phosphatidylinositol 3-kinase. Methods Enzymol. 2005;403:552–561. doi: 10.1016/S0076-6879(05)03048-X. [DOI] [PubMed] [Google Scholar]
- 9.Ullrich O, Stenmark H, Alexandrov K, Huber LA, Kaibuchi K, Sasaki T, Takai Y, Zerial M. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem. 1993;268:18143–18150. [PubMed] [Google Scholar]
- 10.Dirac-Svejstrup AB, Sumizawa T, Pfeffer SR. Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. Embo J. 1997;16:465–472. doi: 10.1093/emboj/16.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 12.Chamberlain MD, Berry TR, Pastor MC, Anderson DH. The p85alpha subunit of phosphatidylinositol 3′-kinase binds to and stimulates the GTPase activity of Rab proteins. J Biol Chem. 2004;279:48607–48614. doi: 10.1074/jbc.M409769200. [DOI] [PubMed] [Google Scholar]
- 13.Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Swanson JA, Watts C. Macropinocytosis. Trends Cell Biol. 1995;5:424–428. doi: 10.1016/s0962-8924(00)89101-1. [DOI] [PubMed] [Google Scholar]
- 15.Lanzetti L, Palamidessi A, Areces L, Scita G, Di Fiore PP. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 2004;429:309–314. doi: 10.1038/nature02542. [DOI] [PubMed] [Google Scholar]
- 16.Racoosin EL, Swanson JA. Macropinosome maturation and fusion with tubular lysosomes in macrophages. J Cell Biol. 1993;121:1011–1020. doi: 10.1083/jcb.121.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hewlett LJ, Prescott AR, Watts C. The coated pit and macropinocytic pathways serve distinct endosome populations. J Cell Biol. 1994;124:689–703. doi: 10.1083/jcb.124.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas AK, Fuchs E, Kopajtich R, Barr FA. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- 19.Galperin E, Sorkin A. Visualization of Rab5 activity in living cells by FRET microscopy and influence of plasma-membrane-targeted Rab5 on clathrin-dependent endocytosis. J Cell Sci. 2003;116:4799–4810. doi: 10.1242/jcs.00801. [DOI] [PubMed] [Google Scholar]
- 20.Stenmark H, Aasland R, Toh BH, D’Arrigo A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J Biol Chem. 1996;271:24048–24054. doi: 10.1074/jbc.271.39.24048. [DOI] [PubMed] [Google Scholar]
- 21.Collins VP. The fine structure of growing and non-growing whole glia cell preparations. Cytobiologie. 1978;18(2):327–338. [PubMed] [Google Scholar]
- 22.Hoppe A, Christensen K, Swanson JA. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys J. 2002;83:3652–3664. doi: 10.1016/S0006-3495(02)75365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida S, Hoppe AD, Araki N, Swanson JA. Sequential signaling in plasma-membrane domains during macropinosome formation in macrophages. J Cell Sci. 2009;122:3250–3261. doi: 10.1242/jcs.053207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 25.D’Arrigo A, Bucci C, Toh BH, Stenmark H. Microtubules are involved in bafilomycin A1-induced tubulation and Rab5-dependent vacuolation of early endosomes. Eur J Cell Biol. 1997;72:95–103. [PubMed] [Google Scholar]
- 26.Willingham MC, Yamada SS. A mechanism for the destruction of pinosomes in cultured fibroblasts. Piranhalysis J Cell Biol. 1978;78:480–487. doi: 10.1083/jcb.78.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerr MC, Lindsay MR, Luetterforst R, Hamilton N, Simpson F, Parton RG, Gleeson PA, Teasdale RD. Visualisation of macropinosome maturation by the recruitment of sorting nexins. J Cell Sci. 2006;119:3967–3980. doi: 10.1242/jcs.03167. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- 29.Swanson JA. Phorbol esters stimulate macropinocytosis and solute flow through macrophages. J Cell Sci. 1989;94:135–142. doi: 10.1242/jcs.94.1.135. [DOI] [PubMed] [Google Scholar]
- 30.Henry RM, Hoppe AD, Joshi N, Swanson JA. The uniformity of phagosome maturation in macrophages. J Cell Biol. 2004;164:185–194. doi: 10.1083/jcb.200307080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453:241–245. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A Cos-7 cell transfected with CFP-Rab5a, YFP-RBD and mCherry. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase-contrast images. Color scale indicates EAVG values below 12% (red) and above 12% (green). EGF (100 ng/mL) was added after acquisition of the first frame and frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 10 μm)
Primary murine bone marrow-derived macrophage transfected with CFP-Rab5a, YFP-RBD and mCherry, stimulated with M-CSF. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase-contrast images. Color scale indicates EAVG values below 8% (red) and above 8% (green). Frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 5μm)
Cos-7 cell transfected with CFP-Rab5a, YFP-RBD and pIRES2-mCherry-Rabex- 5. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase- contrast images. Color scale indicates EAVG values below 12% (red) and above 12% (green). EGF (100 ng/mL) was added after acquisition of the first frame and frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 10 μm)
Cos-7 cell transfected with CFP-Rab5a, YFP-RBD and pIRES2-mCherry-Rin1. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase-contrast images. Color scale indicates EAVG values below 12% (red) and above 12% (green). EGF (100 ng/mL) was added after acquisition of the first frame and frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 10 μm)
Cos-7 cell transfected with CFP-Rab5a, YFP-RBD and pIRES2-mCherry-RabGAP-5. Right panel, Phase-contrast; central panel, D; left panel, EO, EAVG values are presented as pseudocolor overlaid on the phase-contrast images. Color scale indicates EAVG values below 6% (red) and above 6% (green). EGF (100 ng/mL) was added after acquisition of the first frame and frames were acquired every 30 sec. Video was speeded up 60X (2 frames per second) over real time. (Scale bars = 10 μm)