Abstract
Obesity results from interactions between environmental and genetic factors. Despite a relatively high heritability of common, non-syndromic obesity (40–70%), the search for genetic variants contributing to susceptibility has been a challenging task. Genome wide association (GWA) studies have dramatically changed the pace of detection of common genetic susceptibility variants. To date, more than 40 genetic variants have been associated with obesity and fat distribution. However, since these variants do not fully explain the heritability of obesity, other forms of variation, such as epigenetics marks, must be considered.
Epigenetic marks, or “imprinting”, affect gene expression without actually changing the DNA sequence. Failures in imprinting are known to cause extreme forms of obesity (e.g. Prader–Willi syndrome), but have also been convincingly associated with susceptibility to obesity. Furthermore, environmental exposures during critical developmental periods can affect the profile of epigenetic marks and result in obesity.
We review the most recent evidence for genetic and epigenetic mechanisms involved in the susceptibility and development of obesity. Only a comprehensive understanding of the underlying genetic and epigenetic mechanisms, and the metabolic processes they govern, will allow us to manage, and eventually prevent, obesity.
Abbreviations: BMI, body mass index; WC, waist circumference; WHR, waist:hip ratio; T2D, type-2-diabetes; LD, linkage disequilibrium; CNV, copy number variants; PWS, Prader–Willi syndrome; QTL, quantitative trait loci; SNP, single nucleotide polymorphism
Keywords: Genetic, Epigenetic, Fat distribution, Obesity, Methylation, Imprinting
1. Introduction
Overweight and obesity are becoming more widespread with global projections of more than 2.16 billion overweight and 1.12 billion obese individuals by 2030 [1]. This clearly presents a worldwide clinical and public health burden, associated with social and personal criticism. It is also correlated with an increased risk of type-2-diabetes (T2D), cardiovascular disease, cancer and mortality [2,3]. Despite intensive research, current efforts to reduce obesity by diet, exercise, education, surgery and drug therapies are failing to provide effective long-term solutions to this epidemic.
At an individual level, obesity occurs when abnormal amounts of triglycerides are stored in adipose tissue and released from adipose tissue as free fatty acids (FFA) with detrimental effects [4]. Excess fat accumulation occurs when energy intake exceeds energy expenditure, although individuals respond differently to this imbalance owing to genetic predisposition. Twin studies estimate heritability of body mass index (BMI) to be 40–70% in children and adults [5–7], and other anthropometric measures of obesity and regional fat distribution [skinfold thickness, waist circumference (WC) and waist:hip ratio (WHR)] show similar heritability [5–12]. Furthermore, there are ethnic differences in obesity; admixture mapping studies demonstrate that obesity correlates closely with the percentage of ancestry derived from ethnic groups with elevated prevalence [13,14]. The goal of obesity research is to elucidate pathways and mechanisms that control obesity and to improve prevention, management and therapy.
Here we review recent advances in identifying factors contributing to obesity susceptibility. We focus on:
-
(a)
recent successes in identification of genetic variation affecting obesity trait susceptibility;
-
(b)
emerging evidence connecting epigenetic (heritable changes which affect gene function but do not modify DNA sequence) events with obesity.
We discuss the impact of recent findings in these two areas and their joint potential to enhance understanding of obesity susceptibility mechanisms and aetiology.
2. The identification of susceptibility loci for obesity
Until 2006, the main approaches used to track down common variants influencing obesity, involved either hypothesis-free genome-wide linkage mapping in families with multiple obese subjects or association studies within ‘candidate’ genes using case–control samples or parent–offspring trios. The former suffered from being underpowered for any sensible susceptibility models, as linkage is best placed to detect variants with high penetrance. As far as we can tell, common variants with high penetrance do not contribute substantially to risk of common forms of obesity and few, if any, robust signals have emerged from such efforts [15,16]. The latter candidate-gene association approach has historically been compromised by difficulties in selecting credible candidates. Selection was typically based on hypotheses about biological mechanisms putatively involved in obesity pathogenesis but, as the function of much of the genome is poorly characterized, it remains almost impossible to make fully informed decisions. In addition, all too often these candidate-gene studies were conducted in sample sets far too small to offer confident detection of variants with the range of effect sizes that are now known to be realistic. With hindsight, it is easy to appreciate why these approaches yielded few examples of genuine obesity–susceptibility variants.
Consequently, over the last two decades, efforts in identifying and replicating genetic variants predisposing individuals to common forms of obesity were largely characterized by slow progress and limited success, in sharp contrast to the successful gene identification in monogenic and syndromic forms of obesity [15]. The most recent edition of the “Human Obesity Gene Map” gives an excellent overview of this; it lists 11 single gene mutations, 50 loci related to Mendelian syndromes relevant to human obesity, 244 knockout or transgenic animal models and 127 candidate genes, of which slightly less than 20% are replicated by 5 or more studies [15]. A total of 253 quantitative trait loci (QTL), for different obesity-related phenotypes, have been reported from 61 genome-wide linkage scans and of these, only ∼20% are supported by more than one study [15].
Over the past three years it has become possible, from technical and economic perspectives, to undertake hypothesis-free GWA testing in samples of sufficient size to generate convincing association results. The advent of the GWA approach was the result of three components. The first was the human genome sequence which subsequently enabled cataloguing genome-sequence variation. Secondly, the International HapMap Consortium (http://www.hapmap.org) [17] taught us that, in non-African-descent populations, extensive correlations (linkage disequilibrium, LD) between neighbouring single nucleotide polymorphisms (SNPs) constrain the number of independent genetic tests required to survey the genome, such that ∼80% of all common variation can be sampled using ∼500 000 carefully selected SNPs [18,19]. Lastly, novel genotyping methods address the challenges of massively parallel SNP-typing at high accuracy and low cost [20]. The GWA approach has been hugely successful in identifying loci harbouring common forms of obesity (as defined by anthropometric measures: BMI, WC and/or WHR) susceptibility genes and hereto results from a total of 15 ‘high-density’ GWAs (i.e. ≥300 000 SNPs, offering genome-wide coverage >65%) have been published (Table 1). These studies combined, have yielded over 50 loci associated with obesity (p-values <5 × 10−8 in genotyped and imputed data sets, or <5 × 10−7 in directly genotyped data only) (Table 2).
Table 1.
Overview of GWA scans or meta-analysis thereof for obesity phenotypes.
| Reference | Study name (if any) | Number of samples in discovery cohort | Ancestry of discovery cohort | Phenotype |
|---|---|---|---|---|
| Frayling et al. [20] | WTCCC | 1924 | Europeans | BMI – quantitative analysis |
| Scuteri et al. [26] | Sardinia | 4741 | Europeans | BMI – waist circumference (WC) quantitative analysis |
| Chambers et al. [28] | LOLIPOP | 2684 | Indian Asians | Insulin resistance and related quantitative phenotypes |
| Loos et al. [27] | – | 16 876 | Northern European | BMI – quantitative analysis |
| Heard-Costa et al. [36] | The CHARGE consortium | 31 373 | Europeans | WC – quantitative analysis |
| Lindgren et al. [35] | The GIANT consortium | 38 580 | Europeans | WC and waist:hip-ratio (WHR) – quantitative analysis |
| Cotsapas et al. [33] | 775 cases and 3197 unascertained controls | Europeans | Extreme obesity/BMI | |
| Meyre et al. [32] | 1380 and 1416 age-matched normal-weight control | Europeans | Early onset and morbid adult obesity | |
| Thorleifsson et al. [31] | DeCODE | 37 347 | Europeans + African Americans | BMI – quantitative analysis |
| Willer et al. [30] | The GIANT consortium | 32 387 | Europeans | BMI – quantitative analysis |
| Hinney et al. [79] | 487 extremely obese young cases and 442 healthy lean controls | Europeans | Extreme obesity/BMI | |
| Scherag et al. [34] | 453 extremely obese young cases and 435 healthy lean controls | Europeans | Extreme obesity/BMI | |
| Cho et al. [41] | KARE | 8842 | Asian | BMI, WHR – quantitative analysis |
| Heid et al. [43] | MAGIC | 77 167 | European | WHR – quantitative analysis |
| Speliotes et al. [37] | 123 865 | European | BMI – quantitative analysis |
Table 2.
The 54 loci associated to anthropometric obesity phenotypes.
| Closest gene(s) | Chromosomal location | Phenotype | Associated lead SNP(s) | Proposed molecular or cellular function | Additional phenotypes | References |
|---|---|---|---|---|---|---|
| TBX15–WARS2 | 1p12 | WHR | rs984222 | Transcription factor involved in adipocyte and specific adipose depot development | Implicated in Cousin syndrome | Heid et al. [43] |
| PTBP2 | 1p21.3 | BMI | rs1555543 | – | Speliotes et al. [37] | |
| NEGR1 | 1p31 | BMI | rs2815752, rs3101336, rs2568958 | Neuronal outgrowth | Thorleifsson et al. [31], Willer et al. [30], Speliotes et al. [37] | |
| TNNI3K | 1p31.1 | BMI | rs1514175 | – | Speliotes et al. [37] | |
| DNM3–PIGC | 1q24.3 | WHR | rs1011731 | Dominant, negative mutations in DNM enzymes promote GLUT6 and GLUT8 transporters to adipocyte cell surface in rats. | Heid et al. [43] | |
| SEC16B, RASAL2 | 1q25 | BMI | rs10913469 | – | Thorleifsson et al. [31], Speliotes et al. [37] | |
| LYPLAL1; ZC3H11B | 1q41 | WHR | rs2605100 | Encodes protein thought to act as triglyceride lipase and is upregulated in subcutaneous adipose tissue in obese patients | Lindgren et al. [35], Heid et al. [43] | |
| SDCCAG8 | 1q43–q44 | BMI | rs12145833 | – | Scherag et al. [34] | |
| FANCL | 2p16.1 | BMI | rs887912 | – | Speliotes et al. [37] | |
| RBJ–ADCY3–POMC | 2p23.3 | BMI | rs713586 | – | Rare POMC mutations cause human obesity | Speliotes et al. [37] |
| TMEM18 | 2p25 | BMI | rs6548238, rs2867125, rs4854344, rs7561317, rs11127485 | Neural development | Associated with T2D | Willer et al. [30], Thorleifsson et al. [31], Scherag et al. [34], Speliotes et al. [37] |
| ZNRF3–KREMEN1 | 2q12.1 | WHR | rs4823006 | – | Kremen1 protein forms a complex with LDL receptor-related protein 6 | Heid et al. [43] |
| LRP1B | 2q22.2 | BMI | rs2890652 | – | LRP1B deletions seen in several types of human cancers | Speliotes et al. [37] |
| GRB14 | 2q24.3 | WHR | rs10195252 | – | Associated with triglyceride and insulin levels. GRB14-deficient mice exhibit increased body weight | Heid et al. [43] |
| ADAMTS9 | 3p14.1 | WHR | rs6795735 | Important for spatial distribution of cells in embryonic development | Associated with T2D | Heid et al. [43] |
| NISCH–STAB1 | 3p21.1 | WHR | rs6784615 | Interacts with insulin receptor substrate | Heid et al. [43] | |
| CADM2 | 3p21.1 | BMI | rs13078807 | – | Speliotes et al. [37] | |
| ETV5 (locus with three genes, strongest association in ETV5) | 3q27 | BMI | rs7647305 | – | Thorleifsson et al. [31], Speliotes et al. [37] | |
| Gene desert; GNPDA2 is one of three genes nearby | 4p13 | BMI | rs10938397 | – | Associated with T2D | Willer et al. [30], Speliotes et al. [37] |
| SLC39A8 | 4q24 | BMI | rs13107325 | – | Speliotes et al. [37] | |
| FLJ35779 | 5q13.3 | BMI | rs2112347 | – | Speliotes et al. [37] | |
| ZNF608 | 5q23.2 | BMI | rs4836133 | – | Speliotes et al. [37] | |
| CPEB4 | 5q35.2 | WHR | rs6861681 | Regulates polyadenylation elongation | Heid et al. [43] | |
| TFAP2B | 6p12 | WC, BMI | rs987237 | – | Lindgren et al. [35], Speliotes et al. [37] | |
| Locus containing NCR3, AIF1 and BAT2 | 6p21 | BMI | rs2844479, rs2260000, rs1077393 | – | Associated with weight, not BMI | Thorleifsson et al. [31] |
| VEGFA | 6p21.1 | WHR | rs6905288 | Involved in vascular development. Key mediator of adipogenesis | VEGFA variants nominally associated with T2D | Heid et al. [43] |
| NUDT3–HMGA1 | 6p21.31 | BMI | rs206936 | – | Speliotes et al. [37] | |
| PRL | 6p22.2–p21.3 | BMI | rs4712652 | – | Meyre et al. [32] | |
| LY86 | 6p25.1 | WHR | rs1294421 | Plays a role in recognition of lipopolysaccharide | Associated with asthma | Heid et al. [43] |
| RSPOS | 6q22.33 | WHR | rs9491696 | Promotes angiogenesis and vascular development | Oncogene in mouse mammary epithelial cells | Heid et al. [43] |
| NFE2L3 | 7p15.2 | WHR | rs1055144 | – | Heid et al. [43] | |
| MSRA | 8p23.1 | WC, BMI | rs7826222, rs17150703 | – | Lindgren et al. [35], Scherag et al. [34] | |
| LRRN6C | 9p21.3 | BMI | rs10968576 | – | Speliotes et al. [37] | |
| PTER | 10p12 | BMI | rs10508503 | – | Meyre et al. [32] | |
| MTCH2 (locus with 14 genes) | 11p11.2 | BMI | rs10838738 | Cellular apoptosis | Willer et al. [30], Speliotes et al. [37] | |
| BDNF (locus with four genes, strongest association near BDNF) | 11p14 | BMI | rs4074134, rs4923461, rs925946, rs10501087, rs6265 | BDNF expression is regulated by nutritional state and MC4R signalling | Associated with T2D. Individuals with WAGR syndrome with BDNF deletion have BMI >95th percentile. BDNF knockdown in mouse hypothalamus causes hyperphagia and obesity | Thorleifsson et al. [31], Speliotes et al. [37] |
| RPL27A | 11p15.4 | BMI | rs4929949 | – | Speliotes et al. [37] | |
| ITPR2–SSPN | 12p21.1 | WHR | rs718314 | – | Mice lacking ITPR2 and ITPR3 exhibited hypoglycaemia and lean body type | Heid et al. [43] |
| HOXC13 | 12q13.13 | WHR | rs1443512 | Transcription factor important in cell spatial distribution in embryonic development | Heid et al. [43] | |
| FAIM2 (locus also contains BCDIN3D) | 12q13 | BMI | rs7138803 | Adipocyte apoptosis | Thorleifsson et al. [31], Speliotes et al. [37] | |
| C12orf51 | 12q24 | WHR | rs2074356 | – | Cho et al. [41] | |
| MTIF3–GTF3A | 13q12.2 | BMI | rs4771122 | – | Speliotes et al. [37] | |
| PRKD1 | 14q12 | BMI | rs11847697 | – | Speliotes et al. [37] | |
| NRXN3 | 14q31 | WC, BMI | rs10146997 | – | Heard-Costa et al. [36], Speliotes et al. [37] | |
| MAP2K5 | 15q23 | BMI | rs2241423 | – | Speliotes et al. [37] | |
| SH2B1 (locus with 19–25 genes) | 16p11.2 | BMI | rs7498665, rs8049439, rs4788102, rs7498665 | Neuronal role in energy homeostasis | Sh2b1-null mice are obese and diabetic | Willer et al. [30], Thorleifsson et al. [31], Speliotes et al. [37] |
| GPRC5B | 16p12.3 | BMI | rs12444979 | – | Speliotes et al. [37] | |
| MAF | 16q22–q23 | BMI | rs1424233 | Transcription factor involved in adipogenesis and insulin–glucagon regulation | Meyre et al. [32] | |
| FTO | 16q22.2 | BMI | rs9939609, rs6499640, rs8050136, rs3751812, rs7190492, rs8044769, rs1558902 | Neuronal function associated with control of appetite | Associated with T2D | Frayling et al. [20], Scuteri et al. [26], Chambers et al. [28], Willer et al. [30], Thorleifsson et al. [31], Meyre et al. [32], Scherag et al. [34], Speliotes et al. [37] |
| NPC1 | 18q11.2 | BMI | rs1805081 | Intracellular lipid transport | NPC1-null mice show late-onset weight loss and poor food intake. NPC1 interferes with function of raft-associated insulin receptor signalling | Meyre et al. [32] |
| MC4R | 18q22 | BMI | rs17782313, rs12970134, rs17700144 | Hypothalamic signalling | Haplo-insufficiency in humans is associated with morbid obesity. MC4R-deficient mice show hyperphagia and obesity | Willer et al. [30], Thorleifsson et al. [31], Meyre et al. [32], Loos et al. [27], Chambers et al. [28], Scherag et al. [34], Speliotes et al. [37] |
| KCTD15 | 19q13.11 | BMI | rs11084753, rs29941 | – | Willer et al. [30], Thorleifsson et al. [31], Speliotes et al. [37] | |
| QPTCL-GIPR | 19q13.32 | BMI | rs2287019 | Encodes incretin receptor | Associated with fasting and 2-h glucose | Speliotes et al. [37] |
| TMEM160 | 19q13.32 | BMI | rs3810291 | – | Speliotes et al. [37] |
The first gene unequivocally associated to common, non-syndromic obesity, FTO (fat mass and obesity associated) [21], was initially identified as a result of a GWA of T2D [22]. While it was the second strongest associated locus, the association was completely abolished when adjusting for T2D. The association of the FTO region to obesity explains ∼1% of BMI heritability, such that adults homozygous for the risk allele, have a 2–3 kg higher weight compared to non-risk allele homozygous [21]. Interestingly, FTO is reported to operate on fat mass and was suggested to encode a 2-oxoglutarate-dependent nucleic acid demethylase involved in regulation of food intake [23]. In parallel, it was reported to be involved in decreased lipolytic effect in adipocytes [24]. It is unclear whether the association effect acts through FTO or the adjacent FTM gene and the precise role of the FTO locus in obesity needs further investigation.
With reports of the first, robust dichotomous trait associations [25] and the discovery of FTO [21,26,27], came the realization that the effect sizes detected would be smaller than anticipated and that successful analysed would require larger sample sizes than previously considered. This insight catalyzed large-scale international collaboration and meta-analyses of existing data. Through the first collaboration in obesity research, a strong association was detected between SNPs located 188 kilobases (kb) downstream from the melanocortin 4 receptor gene (MC4R) and BMI (Tables 1 and 2 [28]). In parallel, it was reported to be associated with WC in individuals of Indian Asian or European ancestry (Table 1 [29]). The risk variant has subsequently been associated with higher energy and fat intake [30] and the increased BMI reported in children, is consistent with early onset obesity caused by MC4R mutations [31]. Larger GWA meta-analysis, through the Genetic Investigation of Anthropometric Trait (GIANT) Consortia and deCODE (Table 1) followed reporting 8 novel obesity loci, as well as confirming the MC4R and FTO associations (Table 2 [32]). Several of the likely causal genes in the associated regions are highly expressed or known to act in the central nervous system (CNS), suggesting, as in rare monogenic forms of obesity, the role of CNS pathways in predisposition to overall obesity [33].
A few smaller GWAs [34–36] focused on other forms of obesity (early onset, extreme obesity and/or morbid adult obesity) and replicated FTO, MC4R and TMEM18 BMI-associations. These studies also identified four novel associations (Table 2). Cotsapas et al. [35] observed nominal evidence of association for 7 of the 13 loci previously reported to influence BMI [32–34]. This suggests that variants influencing BMI might also contribute to more severe forms of obesity, which represents the extreme, of the phenotypic spectrum rather than a distinct condition, although, this needs confirmation in appropriately powered studies.
Recently, a large-scale meta-analysis of 249 769 individuals confirmed 14 of these previously identified obesity susceptibility loci and identified 18 novel loci associated with BMI and overall adiposity (Table 2) [37]. As a consequence of the increased power of this study, new signals with lower minor allele frequencies and smaller effect sizes, compared to previously identified variants, were discovered. These results suggest that genome-wide association studies are only the first step in the identification of the causal variants that play a role in common, overall obesity and weight regulation and further insights into the underlying biological mechanisms and pathways will be needed for the effective treatment and management of these traits [37].
While no consistent association has been shown between height and BMI-associated variants as a group, three of the loci (MC4R, RBJ–ADCY3–POMC and MTCH2–NDUFS3) show individual associations with height [37]. The BMI-increasing alleles of the variants near POMC and MC4R were associated with decreased and increased height respectively, and this is analogous to the effects of severe mutations in POMC and MC4R on height and weight [31,38].
2.1. Genetics of fat distribution
As described above, efforts to identify common and rare variants influencing BMI and risk of obesity have emphasized a key role for neuronal regulation of overall obesity [21–24,26–30,32–35], but until recently provided few clues to processes responsible for variation in central obesity and fat distribution [29,39,40]. Measures of central and general adiposity are highly correlated: BMI has an r2 ∼ 0.9 with WC and ∼0.6 WHR. Also, WC and WHR are correlated with measures of intra-abdominal fat, measured by magnetic resonance imaging in obese women (r2 ∼ 0.6 and 0.5, respectively). Clinically, central obesity is associated with susceptibility to T2D, cardiovascular disease and increased mortality [3]. Evidence indicates that individual variability in patterns of fat distribution involve local, depot-specific and body-shape processes, which are probably independent of the mechanisms that control overall energy balance and general obesity. First, anthropometric measures of central adiposity are highly heritable [41] and, after BMI correction, heritability estimates remain high (∼0.6 for WC and ∼0.45 for WHR) [10,11,42]. Second, substantial gender-specific differences in fat distribution, reflect specific genetic influences [43,44]. Third, inherited lipodystrophies, which are monogenic syndromes, demonstrate that DNA variants can have specific effects on the development and/or maintenance of specific regional fat-depots and body shape [45]. Hereto, five GWA studies of central obesity (WC and WHR) have been published [29,39,40,44,46] (Table 1) and have identified 17 novel common obesity loci. The associations of three of these loci (MSRA; TFAP2B and NRXN3, Table 2) to WC appear to be mediated through BMI [36,37,39,44], implying their involvement in overall adiposity. The remaining 14 loci, however, showed significant associations with WHR, after correction for BMI, suggesting their role in body fat distribution, independent of overall obesity. Interestingly, some of these loci showed strong sex-specific effects in women [39,44] and in a study by Heid et al. [44], the WHR-increasing allele was shown to be associated with an increase in WC for 14 loci in women, but only six men. This evident heterogeneity between men and women across many of these WHR loci is a reflection of the sex-specific genetic effects driving individual patterns of body fat.
3. Epigenetics and obesity
Epigenetics is loosely defined as the study of heritable changes which affect gene function without modifying the DNA sequence [47]. The maintenance of epigenetic marks through generations is poorly understood and the notion of their transmission is contentious [48]. Epigenetic marks are tissue specific and include DNA methylation and histone modifications which mediate biological processes such as imprinting. As many imprinted genes are growth factors, or regulators of gene expression controlling growth, imprinting disorders often feature obesity as one of their clinical characteristics.
Genomic imprinting determines expression of alleles according to their maternal or paternal origin [49] and establishes a balance between the expression of the parental alleles influencing growth [50], resulting in counteracting growth effects of paternal and maternal genomes [51]. In addition to growth, imprinted genes are also involved in differentiation, development, viability and metabolic functions [52]. Two main clusters of genomic imprinting are known in humans: a region at 11p15 containing several imprinted genes including IGF2, INS, KCNQ10T1 (LIT1) (paternally expressed) and H19, KCNQ1, CDKN1C, PHLDA2, KVLQT1 (maternally expressed) [53]. The second cluster at 15q11–q12 contains at least 7 imprinted genes, including; MKRN3, MAGEL2, NDN, SNURF–SNRPN (paternally expressed) and UBE3A, ATP10A (maternally expressed) [54].
Failures in imprinting which result in obesity by altering expression of growth and cellular differentiation factors can arise due to numerous genetic events: translocation, inversion, duplication, paternal disomy and hyper/hypo-methylation. For instance; paternal deletion or uniparental disomy at 15q11–q13, results in Prader–Willi syndrome (PWS). A syndrome characterized by severe (sometimes life threatening), early onset obesity caused by hyperphagia (due to dysfunction in the satiety centre [55]). Moderate obesity appears in Albright hereditary osteodystrophy (AHO), due to disruption of imprinting at the GNAS gene (20q13.11 [50]).
3.1. Mediators of genomic imprinting
3.1.1. DNA methylation
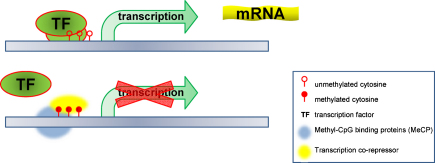
Genomic imprinting is mediated by DNA methylation as exemplified in the H19 and IGF2 loci [56]. Methylation is a widespread feature of the genome, and is obtained through the addition of a methyl group (CH3) to a cytosine positioned next to a guanine nucleotide (CpGs), usually in regions with a high presence of CpG dinucleotides (>60%). Methylation in a promoter region results in the repression (silencing) of gene expression [57], this effect may be achieved by a number of mechanisms including: obstructing access to transcription factors/activators and recruitment of co-repressors (like histone deacetylases) which alter chromatin structure [58] resulting in failure to initiate transcription (Fig. 1).
Fig. 1.
CpG methylation and regulation of gene expression. Unmethylated or hypomethylated DNA (usually in the promoter region) allows binding of the transcription factors (TF) and other regulatory mechanisms which results in transcription and mRNA production. Methylated DNA (bottom panel) obstructs binding of the TF, and in some cases might recruit methyl-CpG binding proteins and other transcription co-repressors, blocking access of the transcription enzymes and resulting in gene silencing.
3.1.2. Histone modifications
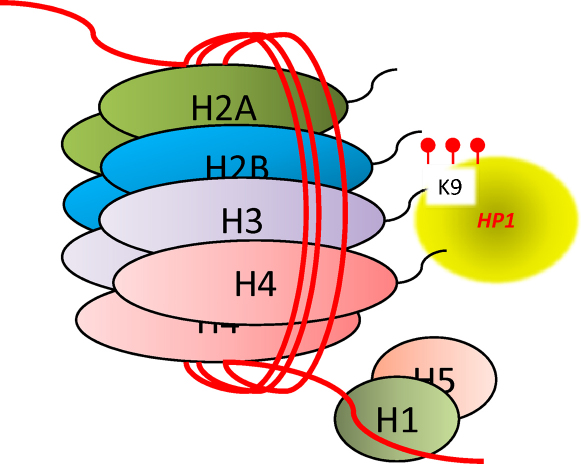
DNA in cells is packaged as chromatin in a “beads on a string” configuration. A length of 147 DNA base pairs wraps around a core histone octamer (a tetramer of histones H3 and H4, flanked by two H2A–H2B dimmers) and these nucleosome “beads” are separated by DNA “strings” between 20 and 60 base pairs. Linker histones (H1/H5) occupy the exit and entry of the DNA into the histone, these structures are in turn coiled into a compact helical “closed” configuration [59]. Packaging DNA in this manner allows its efficient storage and is paramount for regulation of gene expression, as the “closed” configuration does not allow access to transcriptional enzymes. Post translational modification of the histones by: H3 and H4 hyperacetylation in the promoter [60], methylation of the lys4 and lys36 of histone H3 open the structure and allow gene expression. In contrast, methylation of lys9, lys20 and lys27 on H3 [60,61]) and ubiquitination of H2A present at high density CpG promoters [62] result in gene silencing “closed configuration”. Further complexity is achieved by the level of methylation, so mono-, bi- or tri-methylation may also effect the control of gene expression [63].
A feature of both methylation and histone modifications is that they are both tissue specific and can vary with age (and developmental stage). Therefore, in order to place findings in an appropriate context, it is of paramount importance that evaluation of epigenetic factors be carried out on suitable tissues extracted at specified times [64,65] (Fig. 2).
Fig. 2.
Histone modification. This simplified diagram of a nucleosome shows a histone octamer “bead” surrounded by a DNA strand and try-methylation at lysine-9, this kind of modification exemplifies modifications found at promoter regions of silenced genes.
3.2. Epigenetic changes introduced during early development may increase the risk of obesity
Although birth weight (BW) is an imperfect summary index of growth, however, it has been widely used as a proxy for foetal nutrition and intrauterine growth. Although better measurements of foetal growth exist, BW measurements are used because they are non-intrusive, easily obtained and often found in birth records, which enabled its use as in retrospective studies linking BW with adult onset diseases [66]. Many studies focus on the hypothesis that early environmental influences induce epigenetic variation, thereby permanently affecting metabolism and chronic disease risk. Specifically, for obesity, it has been shown that obese mothers tend to have obese children [67], it has been shown that clinical intervention to cause maternal weight loss can have a positive effect on reducing risk of obesity in the offspring [68]. The mechanisms by which nutritional challenges affect the risk of disease in later life are poorly understood. However, evidence indicates that the establishment of the epigenome can be affected by environmental factors during critical developmental periods [69]. Possible disturbances of methylation may arise during foetal development due to lack of availability of dietary methyl donors [70–72]. Potential interactions between the environment and epigenetic mechanisms mediating the expression of genes associated with increased BMI and adiposity, may also be possible as suggested for; the FTO locus is a DNA-demethylase enzyme [23], the MC4R gene which has reduced methylation following long-term exposure to a high fat diet [73], the PPARγ protein which interacts with histone acetyltransferases [58] during adipogenesis and on the effect of diet on methylation of POMC [74] and Leptin [75].
With this in mind, it is important to consider the effect of assisted reproduction technology on epigenetics and subsequently on obesity susceptibility. Despite small numbers, increased incidence of PWS, when compared to background population prevalence, has been shown [76] and subtler adverse effects may only become apparent later on in life. We are currently gaining more insight through the use of high-throughput sequencing methods for the detection of DNA methylation patterns, and chromatin immunoprecipitation (ChIP) used to detect histone modifications and new integrated genomics-based technologies which are currently being developed by the human epigenome project (http://www.epigenome.org) initiative.
4. Remaining challenges
4.1. Identifying novel loci
Despite successes in susceptibility loci identification for obesity through the first wave of GWAs, the combined effect of the loci explains only 2–3% of the inherited contribution to obesity risk.
4.2. Collaborative studies to for larger GWA meta-analysis
The GIANT consortium is finalising the next wave of GWA meta-analysis for obesity related traits incorporating >100 000 samples. This increase in sample size should attain sufficient power to allow identification of common variants with even smaller effect sizes than hereto. Also, conditional analysis using the previously identified variants will reveal whether some of the already identified loci contain more than one independent association with obesity, which would be an additional source of unexplored variation contributing to the missing heritability.
4.3. Obesity susceptibility associated to rare-low-frequency variants
The obesity risk alleles that have been identified so far by GWAs are quite common (27–91%) in European populations but the current approaches are limited in the range of minor allele frequencies (MAF) that are detected due to the range of allele frequencies (>5% MAF) present on current genotype arrays. The 1000 genomes project (http://www.1000genomes.org/) is designed to catalogue low and rare frequency variants and this will lead to the design of new genotype arrays including wider spectra of allele frequencies.
4.4. Copy number variations
Copy number variants (CNVs) are hard to detect due to technical constrains but recently [74], two complementary approaches have delivered the first examples of CNVs associated to obesity. The SNPs associated with obesity at the NEGR1 locus are in strong LD with a nearby CNV [32], tagging a 45-kb deletion polymorphism which is a candidate causal-variant at this locus. Recently by analysing SNP genotype arrays that are enriched in CNV information and applying novel CNV detecting algorithms on these arrays deletions on chromosome 16p11.2 have been reported to be associated with extreme, highly penetrant obesity [77,78]. Further applications of these approaches in larger sample sets should allow a better understanding of total CNVs contribution to overall variation in obesity susceptibility.
4.5. Characterization of associated loci and causal variants
It is not clear how much of the remaining variation will be uncovered by the aforementioned steps. Beyond identifying more susceptibility loci, finding the causal gene(s) and variant(s) in each locus will be necessary to allow the detailed molecular and physiological investigations that are necessary to determine the mechanisms and pathways through which these contribute to obesity susceptibility and ultimately allow this knowledge to be translated into better prediction and treatment options.
4.6. Integration of genetic and epigenetic information
Genetic and epigenetic factors are intimately intertwined, as epigenetic marks and DNA modifications are the direct consequence of sequence-specific interactions between proteins and DNA [79]. The completion of the human epigenome project will increase our understanding of the genetic and epigenetics underlying cellular homeostasis and the integration of this knowledge with the known environmental risks to obesity, must be applied so that it may eventually be exploited and manipulated to appease the obesity epidemic.
5. Conclusions
The current obesity epidemic is clearly not of genetic origin per se, but due to unfavourable changes in lifestyle and environment (the ‘obesogenic’ environment). The obesogenic environment has different effects on different individuals in the same environment, highlighting an underlying, inherited susceptibility to obesity and fat-distribution. For more than a decade, the genetics underlying common forms of obesity have remained elusive although the advent of the GWA approach has started to deliver robust associations to obesity. The epigenetic contribution to common forms of obesity are still largely unknown but, from rare syndromes and animal models we conclude that it is likely that both genetic and environmental effects on epigenetics will in turn be associated with obesity. We have started to identify an emerging pattern of effects acting through CNS, suggesting that a component of an individual's response to the obesogenic environment is partly neurobehaviourally driven. There is also some evidence of effects acting more peripherally in the adipose tissue. Despite the success of GWAs in obesity loci identification, we still only explain a low fraction of the inter-individual variation of obesity. Extensive work including identification of more obesity susceptibility loci, a better understanding of the gene(s) through which the effect is executed, as well as further molecular and physiological characterization of the associated genes, is now necessary before any of these findings will lead to any useful therapeutic interventions.
Contributors and their role
All the authors were involved in the drafting, editing and approval of this manuscript.
Blanca M. Herrera and Sarah L. Keildson were involved in the construction of the tables and figures.
Competing interests
None.
Provenance and peer review
Commissioned and externally peer reviewed.
Acknowledgement
C.M.L. is a Wellcome Trust Research Career Development Fellow (086596/Z/08/Z).
References
- 1.Kelly T., Yang W., Chen C.S., Reynolds K., He J. Global burden of obesity in 2005 and projections to 2030. Int J Obes (Lond) 2008;32(September (9)):1431–1437. doi: 10.1038/ijo.2008.102. [DOI] [PubMed] [Google Scholar]
- 2.Flegal K.M., Graubard B.I., Williamson D.F., Gail M.H. Cause-specific excess deaths associated with underweight, overweight, and obesity. JAMA. 2007;298(November (17)):2028–2037. doi: 10.1001/jama.298.17.2028. [DOI] [PubMed] [Google Scholar]
- 3.Pischon T., Boeing H., Hoffmann K. General and abdominal adiposity and risk of death in Europe. N Engl J Med. 2008;359(November (20)):2105–2120. doi: 10.1056/NEJMoa0801891. [DOI] [PubMed] [Google Scholar]
- 4.O’Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462(November (7271)):307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 5.Stunkard A.J., Foch T.T., Hrubec Z. A twin study of human obesity. JAMA. 1986;256(July (1)):51–54. [PubMed] [Google Scholar]
- 6.Turula M., Kaprio J., Rissanen A., Koskenvuo M. Body weight in the finnish twin cohort. Diabetes Res Clin Pract. 1990;10 (Suppl. 1):S33–S36. doi: 10.1016/0168-8227(90)90137-i. [DOI] [PubMed] [Google Scholar]
- 7.Wardle J., Carnell S., Haworth C.M., Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr. 2008;87(February (2)):398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- 8.Malis C., Rasmussen E.L., Poulsen P. Total and regional fat distribution is strongly influenced by genetic factors in young and elderly twins. Obes Res. 2005;13(December (12)):2139–2145. doi: 10.1038/oby.2005.265. [DOI] [PubMed] [Google Scholar]
- 9.Moll P.P., Burns T.L., Lauer R.M. The genetic and environmental sources of body mass index variability: the muscatine ponderosity family study. Am J Hum Genet. 1991;49(December (6)):1243–1255. [PMC free article] [PubMed] [Google Scholar]
- 10.Rose K.M., Newman B., Mayer-Davis E.J., Selby J.V. Genetic and behavioral determinants of waist–hip ratio and waist circumference in women twins. Obes Res. 1998;6(November (6)):383–392. doi: 10.1002/j.1550-8528.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 11.Selby J.V., Newman B., Quesenberry C.P., Jr., Fabsitz R.R., King M.C., Meaney F.J. Evidence of genetic influence on central body fat in middle-aged twins. Hum Biol. 1989;61(April (2)):179–194. [PubMed] [Google Scholar]
- 12.Stunkard A.J., Sorensen T.I., Hanis C. An adoption study of human obesity. N Engl J Med. 1986;314(January (4)):193–198. doi: 10.1056/NEJM198601233140401. [DOI] [PubMed] [Google Scholar]
- 13.Redden D.T., Divers J., Vaughan L.K. Regional admixture mapping and structured association testing: conceptual unification and an extensible general linear model. PLoS Genet. 2006;2(August (8)):e137. doi: 10.1371/journal.pgen.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams R.C., Long J.C., Hanson R.L., Sievers M.L., Knowler W.C. Individual estimates of European genetic admixture associated with lower body-mass index, plasma glucose, and prevalence of type 2 diabetes in Pima Indians. Am J Hum Genet. 2000;66(February (2)):527–538. doi: 10.1086/302773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rankinen T., Zuberi A., Chagnon Y.C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14(April (4)):529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 16.Saunders C.L., Chiodini B.D., Sham P. Meta-analysis of genome-wide linkage studies in BMI and obesity. Obesity (Silver Spring) 2007;15(September (9)):2263–2275. doi: 10.1038/oby.2007.269. [DOI] [PubMed] [Google Scholar]
- 17.A haplotype map of the human genome. Nature. 2005;437(October (7063)):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett J.C., Cardon L.R. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38(June (6)):659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- 19.Pe’er I., de Bakker P.I., Maller J., Yelensky R., Altshuler D., Daly M.J. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38(June (6)):663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 20.Fan J.B., Chee M.S., Gunderson K.L. Highly parallel genomic assays. Nat Rev Genet. 2006;7(August (8)):632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 21.Frayling T.M., Timpson N.J., Weedon M.N. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(May (5826)):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeggini E., Weedon M.N., Lindgren C.M. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(June (5829)):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerken T., Girard C.A., Tung Y.C. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(November (5855)):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahlen K., Sjolin E., Hoffstedt J. The common rs9939609 gene variant of the fat mass- and obesity-associated gene FTO is related to fat cell lipolysis. J Lipid Res. 2008;49(March (3)):607–611. doi: 10.1194/jlr.M700448-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(June (7145)):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dina C., Meyre D., Gallina S. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(June (6)):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 27.Scuteri A., Sanna S., Chen W.M. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(July (7)):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loos R.J., Lindgren C.M., Li S. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40(June (6)):768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chambers J.C., Elliott P., Zabaneh D. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40(June (6)):716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 30.Qi L., Kraft P., Hunter D.J., Hu F.B. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum Mol Genet. 2008;17(November (22)):3502–3508. doi: 10.1093/hmg/ddn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farooqi I.S., Keogh J.M., Yeo G.S., Lank E.J., Cheetham T., O’Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348(March (12)):1085–1095. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 32.Willer C.J., Speliotes E.K., Loos R.J. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(January (1)):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorleifsson G., Walters G.B., Gudbjartsson D.F. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(January (1)):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 34.Meyre D., Delplanque J., Chevre J.C. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41(February (2)):157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 35.Cotsapas C., Speliotes E.K., Hatoum I.J. Common body mass index-associated variants confer risk of extreme obesity. Hum Mol Genet. 2009;18(September (18)):3502–3507. doi: 10.1093/hmg/ddp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scherag A., Dina C., Hinney A. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and German study groups. PLoS Genet. 2010;6(4):e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speliotes E.K., Willer C.J., Berndt S.I. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(November (11)):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farooqi I.S., Drop S., Clements A. Heterozygosity for a POMC-null mutation and increased obesity risk in humans. Diabetes. 2006;55(September (9)):2549–2553. doi: 10.2337/db06-0214. [DOI] [PubMed] [Google Scholar]
- 39.Lindgren C.M., Heid I.M., Randall J.C. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(June (6)):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heard-Costa N.L., Zillikens M.C., Monda K.L. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE consortium. PLoS Genet. 2009;5(June (6)):e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamel E.G., McNeill G., Van Wijk M.C. Usefulness of anthropometry and DXA in predicting intra-abdominal fat in obese men and women. Obes Res. 2000;8(January (1)):36–42. doi: 10.1038/oby.2000.6. [DOI] [PubMed] [Google Scholar]
- 42.Selby J.V., Newman B., Quesenberry C.P., Jr. Genetic and behavioral influences on body fat distribution. Int J Obes. 1990;14(July (7)):593–602. [PubMed] [Google Scholar]
- 43.Zillikens M.C., Yazdanpanah M., Pardo L.M. Sex-specific genetic effects influence variation in body composition. Diabetologia. 2008;51(December (12)):2233–2241. doi: 10.1007/s00125-008-1163-0. [DOI] [PubMed] [Google Scholar]
- 44.Heid I.M., Jackson A.U., Randall J.C. Meta-analysis identifies 13 new loci associated with waist–hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(November (11)):949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garg A. Acquired and inherited lipodystrophies. N Engl J Med. 2004;350(March (12)):1220–1234. doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]
- 46.Cho Y.S., Go M.J., Kim Y.J. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(May (5)):527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 47.Bird A. Perceptions of epigenetics. Nature. 2007;447(May (7143)):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 48.Chong S., Youngson N.A., Whitelaw E. Heritable germline epimutation is not the same as transgenerational epigenetic inheritance. Nat Genet. 2007;39(May (5)):574–575. doi: 10.1038/ng0507-574. author reply 5–6. [DOI] [PubMed] [Google Scholar]
- 49.Reik W., Walter J. Imprinting mechanisms in mammals. Curr Opin Genet Dev. 1998;8(April (2)):154–164. doi: 10.1016/s0959-437x(98)80136-6. [DOI] [PubMed] [Google Scholar]
- 50.Butler M.G. Genomic imprinting disorders in humans: a mini-review. J Assist Reprod Genet. 2009;26(September–October (9–10)):477–486. doi: 10.1007/s10815-009-9353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haig D., Graham C. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell. 1991;64(March (6)):1045–1046. doi: 10.1016/0092-8674(91)90256-x. [DOI] [PubMed] [Google Scholar]
- 52.Smith F.M., Garfield A.S., Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res. 2006;113(1–4):279–291. doi: 10.1159/000090843. [DOI] [PubMed] [Google Scholar]
- 53.Lee M.P., Brandenburg S., Landes G.M., Adams M., Miller G., Feinberg A.P. Two novel genes in the center of the 11p15 imprinted domain escape genomic imprinting. Hum Mol Genet. 1999;8(April (4)):683–690. doi: 10.1093/hmg/8.4.683. [DOI] [PubMed] [Google Scholar]
- 54.Schweizer J., Zynger D., Francke U. In vivo nuclease hypersensitivity studies reveal multiple sites of parental origin-dependent differential chromatin conformation in the 150 kb SNRPN transcription unit. Hum Mol Genet. 1999;8(April (4)):555–566. doi: 10.1093/hmg/8.4.555. [DOI] [PubMed] [Google Scholar]
- 55.Shapira N.A., Lessig M.C., He A.G., James G.A., Driscoll D.J., Liu Y. Satiety dysfunction in Prader–Willi syndrome demonstrated by fMRI. J Neurol Neurosurg Psychiatry. 2005;76(February (2)):260–262. doi: 10.1136/jnnp.2004.039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferguson-Smith A.C., Sasaki H., Cattanach B.M., Surani M.A. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362(April (6422)):751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 57.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(January (1)):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 58.Choy J.S., Wei S., Lee J.Y., Tan S., Chu S., Lee T.H. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132(February (6)):1782–1783. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Q., Wani A.A. Histone modifications: crucial elements for damage response and chromatin restoration. J Cell Physiol. 2010;223(May (2)):283–288. doi: 10.1002/jcp.22060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Litt M.D., Simpson M., Gaszner M., Allis C.D., Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293(September (5539)):2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 61.Musri M.M., Gomis R., Parrizas M. Chromatin and chromatin-modifying proteins in adipogenesis. Biochem Cell Biol. 2007;85(August (4)):397–410. doi: 10.1139/O07-068. [DOI] [PubMed] [Google Scholar]
- 62.Kallin E.M., Cao R., Jothi R. Genome-wide uH2A localization analysis highlights Bmi1-dependent deposition of the mark at repressed genes. PLoS Genet. 2009;5(June (6)):e1000506. doi: 10.1371/journal.pgen.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lachner M., O'Sullivan R.J., Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116(June (Pt 11)):2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 64.Feinberg A.P. Epigenetics at the epicenter of modern medicine. JAMA. 2008;299(March (11)):1345–1350. doi: 10.1001/jama.299.11.1345. [DOI] [PubMed] [Google Scholar]
- 65.Feinberg A.P. Genome-scale approaches to the epigenetics of common human disease. Virchows Arch. 2010;456(January (1)):13–21. doi: 10.1007/s00428-009-0847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stein A.D., Zybert P.A., van de Bor M., Lumey L.H. Intrauterine famine exposure and body proportions at birth: the Dutch hunger winter. Int J Epidemiol. 2004;33(August (4)):831–836. doi: 10.1093/ije/dyh083. [DOI] [PubMed] [Google Scholar]
- 67.Dabelea D., Mayer-Davis E.J., Lamichhane A.P. Association of intrauterine exposure to maternal diabetes and obesity with type 2 diabetes in youth: the SEARCH case–control study. Diabetes Care. 2008;31(July (7)):1422–1426. doi: 10.2337/dc07-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith J., Cianflone K., Biron S. Effects of maternal surgical weight loss in mothers on intergenerational transmission of obesity. J Clin Endocrinol Metab. 2009;94(November (11)):4275–4283. doi: 10.1210/jc.2009-0709. [DOI] [PubMed] [Google Scholar]
- 69.Waterland R.A., Jirtle R.L. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20(January (1)):63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 70.Waterland R.A., Michels K.B. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 71.Poirier L.A. The effects of diet, genetics and chemicals on toxicity and aberrant DNA methylation: an introduction. J Nutr. 2002;132(August (8 Suppl.)):S2336–S2339. doi: 10.1093/jn/132.8.2336S. [DOI] [PubMed] [Google Scholar]
- 72.Biniszkiewicz D., Gribnau J., Ramsahoye B. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol. 2002;22(April (7)):2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Widiker S., Karst S., Wagener A., Brockmann G.A. High-fat diet leads to a decreased methylation of the Mc4r gene in the obese BFMI and the lean B6 mouse lines. J Appl Genet. 2010;51(2):193–197. doi: 10.1007/BF03195727. [DOI] [PubMed] [Google Scholar]
- 74.Plagemann A., Harder T., Brunn M. Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. J Physiol. 2009;587(October (Pt 20)):4963–4976. doi: 10.1113/jphysiol.2009.176156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Milagro F.I., Campion J., Garcia-Diaz D.F., Goyenechea E., Paternain L., Martinez J.A. High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J Physiol Biochem. 2009;65(March (1)):1–9. doi: 10.1007/BF03165964. [DOI] [PubMed] [Google Scholar]
- 76.Manipalviratn S., DeCherney A., Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91(February (2)):305–315. doi: 10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walters R.G., Jacquemont S., Valsesia A. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463(February (7281)):671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bochukova E.G., Huang N., Keogh J. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463(February (7281)):666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ptashne M., Hobert O., Davidson E. Questions over the scientific basis of epigenome project. Nature. 2010;464(March (7288)):487. doi: 10.1038/464487c. [DOI] [PubMed] [Google Scholar]