Abstract
The natural product quercetin is a flavonoid found in many fruits and vegetables. Previous research has shown that quercetin has antitumor, anti-inflammatory, antiallergic, and antiviral activities. In the present investigation we studied the effect of quercetin on the ability of prostate cancer cell lines with various degrees of aggressive potential to form colonies in vitro. Specifically, we examined the molecular mechanisms underlying this effect, including the expression of cell cycle and tumor suppressor genes as well as oncogenes. We observed that quercetin at concentrations of 25 and 50 μM significantly inhibited the growth of the highly aggressive PC-3 prostate cancer cell line and the moderately aggressive DU-145 prostate cancer cell line, whereas it did not affect colony formation by the poorly aggressive LNCaP prostate cancer cell line or the normal fibroblast cell line BG-9. Using the gene array methodology, we found that quercetin significantly inhibited the expression of specific oncogenes and genes controlling G1, S, G2, and M phases of the cell cycle. Moreover, quercetin reciprocally up-regulated the expression of several tumor suppressor genes. In conclusion, our results demonstrate that the antitumor effects of quercetin directly correlate with the aggressive potential of prostate cancer cells and that the mechanism(s) of quercetin-mediated antitumor effects may involve up-regulation of tumor suppressor genes and reciprocal down-regulation of oncogenes and cell cycle genes. The results of these studies provide a scientific basis for the potential use of flavonoids as nutraceuticals in the chemoprevention of cancer.
The flavonoids comprise a large class of low-molecular-weight, natural products of plant origin ubiquitously distributed in foods. These dietary antioxidants exert significant antitumor, antiallergic, and anti-inflammatory effects and have been extensively reviewed (14, 25, 38). Although various flavonoids, including quercetin, have been shown to have significant antitumor activities, the molecular mechanisms underlying these effects are generally unknown. We hypothesize that the antitumor effects of quercetin, as manifested by its ability to selectively suppress colony formation by prostate cancer cells in vitro, are mediated by its ability to regulate the expression of various genes controlling the cell cycle, tumor suppression, and oncogenesis. The present study was undertaken to investigate the effect of quercetin on the colony-forming abilities of three prostate cancer cell lines with different malignant potentials. Our results show that quercetin selectively inhibited the growth of the highly malignant PC-3 prostate cancer cell line and the moderately malignant DU-145 prostate cancer cell line but had no effect on poorly malignant LNCaP cells and normal fibroblast control cultures.
MATERIALS AND METHODS
Cell culture.
The human prostate cancer cell lines PC-3, DU-145, and LNCaP were obtained from the American Type Culture Collection (Manassas, Va.). DU-145 and PC-3 cells were isolated from metastases in brain and lumbar vertebra, respectively (2, 8). DU-145 and PC-3 are androgen independent (11, 38). DU-145 cells are less invasive in in vitro assays and exhibit a relatively low potential for metastasis in vivo compared to the metastatic potential of the more malignant PC-3 cells. The LNCaP cell line was established from a metastatic lesion of a human prostatic adenocarcinoma and is poorly aggressive (11). While LNCaP cells are androgen responsive, their growth is not androgen dependent. Cell cultures were maintained in RPMI 1640 medium supplemented with nonessential amino acids, l-glutamine, a twofold vitamin solution (Life Technologies, Grand Island, N.Y.), sodium pyruvate, Earl's balanced salt solution, 10% fetal bovine serum, and penicillin and streptomycin (Flow Laboratories, Rockville, Md.). Approximately 400 cells/60-mm plastic dish (Falcon; Becton Dickinson, Lincoln Park, N.J.) were cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air. At the end of 24 h of incubation, quercetin (Sigma Co, St. Louis, Mo.) was added at different concentrations (0 to 50 μM) and the cultures were divided into two sets. After incubation for another 24 h, one set was used for RNA extraction. The second set was harvested for colony formation after 8 days of culture. Colonies were fixed with methanol, treated with Giemsa stain, and counted by using Gene Snap Automatic Colony Counter software (Syngene, Frederick, Md.). Colonies were defined as consisting of ≥16 cells, as described previously (17, 18, 28).
Gene arrays.
Total RNA was isolated from quercetin-treated and untreated control PC-3 cells by the TRIzol reagent-phenol chloroform procedure (Gibco BRL, Rockville, Md.). To prepare cDNA, we used 4 μg of total RNA in a 10-μl volume containing 50 mmol of Tris-HCl (pH 8.3) per liter, 75 mmol of KCl per liter, 3 mmol of MgCl2 per liter, 0.5 mmol of a mixture of deoxynucleoside triphosphates except dATP per liter, 5 mmol of dithiothreitol per liter, gene-specific primer mix (Super Array Inc., Bethesda, Md.), and Maloney murine leukemia virus reverse transcriptase (Promega, Madison, Wis.) in the presence of 35 μCi of [α-32P]dCTP (3,000 Ci/mmol; Amersham, Piscataway, N.J.). After incubation at 50°C for 25 min, the reaction was stopped by addition of 0.01 mol of EDTA (pH 8.0) per liter. The radiolabeled cDNA thus obtained was used to probe the gene array membranes, as follows. Gene expression was analyzed by using GEArray kits (Super Array Inc., Bethesda, Md.). Two arrays, one for quercetin-treated samples and the other for untreated control samples, were processed simultaneously under identical conditions according to the instructions of the manufacturer to obtain gene expression patterns. The genes on the membranes were hybridized with the cDNA probes (5 × 106 to 6 × 106 cpm/membrane, equivalent to 4 μg of total RNA) described above, washed, and exposed to Kodak Biomac MS film overnight at −20°C. The resultant autoradiograph was analyzed with Gene Tool software (version 2.11a; Syngene, Frederick, Md.) and normalized by using the internal control β-actin. The values for each gene are expressed as the percent change in optical density for quercetin-treated cultures compared with the optical density for untreated control cultures after normalization with the values for the corresponding housekeeping gene.
Real-time, quantitative reverse transcription-PCR.
The relative abundance of each mRNA species was assessed by the 5′ fluorogenic nuclease assay to perform real-time, quantitative PCR as described previously (23). The ABI Prism 5700 instrument (PE Applied Biosystems, Foster City, Calif.) detects and plots the increase in fluorescence versus the PCR cycle number to produce a continuous measure of PCR amplification. To provide a precise quantification of the initial target in each PCR mixture, the amplification plot is examined at a point during the early log phase of product accumulation. This is accomplished by assigning a fluorescence threshold above the background fluorescence and determining the time point at which each sample's amplification plot reaches the threshold (defined as the threshold cycle number [CT]). The relative expression of mRNA species was calculated by the comparative CT method described previously (23). All data were controlled for the quantity of RNA input by obtaining measurements for an endogenous reference gene, that for β-actin. For each RNA sample a difference in CT values (ΔCTs) was calculated for each mRNA species by taking the mean CT for duplicate tubes and subtracting the mean CT for the duplicate tubes for the reference RNA (β-actin) measured with an aliquot from the same reverse transcription reaction mixture; that is, relative expression is equal to 2−ΔΔCT. This calculation assumes that all PCRs are working with 100% efficiency.
RESULTS
Effect of quercetin on viabilities of prostate cancer cell lines.
To eliminate the possibility that quercetin may cause any nonspecific, toxic effects on prostate cancer cell lines, we determined the viabilities of PC-3 cells cultured with quercetin for 8 days. PC-3 cells treated with quercetin at concentrations ranging from 1.6 to 50 μM showed viabilities comparable to the 89% viability of untreated control cultures, as determined by the trypan blue dye exclusion assay. Similar viability assays were also performed with other prostate cell lines, DU-145 and LNCaP, and with the normal skin fibroblast cell line BG-9. These results were consistent with the data obtained with PC-3 cells and demonstrated that quercetin is nontoxic at the concentrations used. The lack of toxicity of quercetin was confirmed by our gene array data, which demonstrated that quercetin does not affect the constitutive expression of the β-actin housekeeping gene. These data are consistent with those from other studies, which demonstrated that similar quercetin concentrations are nontoxic to other cells in vitro (3, 15, 19).
Quercetin inhibits colony formation by prostate cancer cell lines.
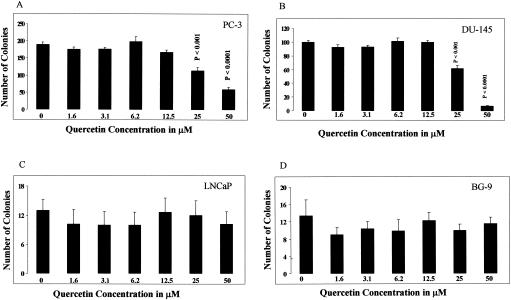
The level of colony formation by untreated prostate cancer cells was proportional to the aggressive potential of the specific cell line. Highly aggressive PC-3 cells produced the largest number of colonies, 190 ± 14 (Fig. 1A). Moderately aggressive DU-145 cells produced fewer colonies, 99 ± 5 (Fig. 1B), and minimally aggressive LNCaP cells produced the smallest number of colonies, 12 ± 6 (Fig. 1C). Quercetin at concentrations of 25 and 50 μM significantly inhibited the colony-forming abilities of both highly aggressive PC-3 cells (Fig. 1A) and moderately aggressive DU-145 cells (Fig. 1B) compared to its level of inhibition of untreated control cultures. Untreated PC-3 cells produced 190 ± 14 colonies, and the colony numbers were suppressed to 114 ± 20 (P < 0.001) and 59 ± 13 (P < 0.0001) with 25 and 50 μM quercetin, respectively. Untreated DU-145 cells produced 99 ± 5 colonies, and the colony numbers were suppressed to 64 ± 11 (P < 0.001) and 7 ± 2 (P < 0.0001) with 25 and 50 μM quercetin, respectively. Lower concentrations of quercetin (1.6 to 12.5 μM) did not have any significant effect on the colony-forming abilities of any of the cell lines. Quercetin at every concentration used, including the highest concentrations of 25 and 50 μM, did not affect the colony-forming abilities of poorly aggressive LNCaP cells (Fig. 1C) or BG-9 normal skin fibroblast control cells (Fig. 1D). The latter results demonstrate the selective antitumor activity of quercetin on prostate cancer cells with higher aggressive potential.
FIG. 1.
Effect of quercetin on colony-forming abilities of prostate cancer cell lines with different aggressive potentials. (A) Highly aggressive PC-3 cell line; (B) moderately aggressive DU-145 cell line; (C) minimally aggressive LNCaP cell line; (D) BG-9 normal skin fibroblasts. The data represent means ± standard errors of six experiments performed in sextuplicate. The statistical significance of the difference between control and treated cultures was calculated by Student's t test.
Quercetin inhibits expression of cell cycle genes by prostate cancer cell lines.
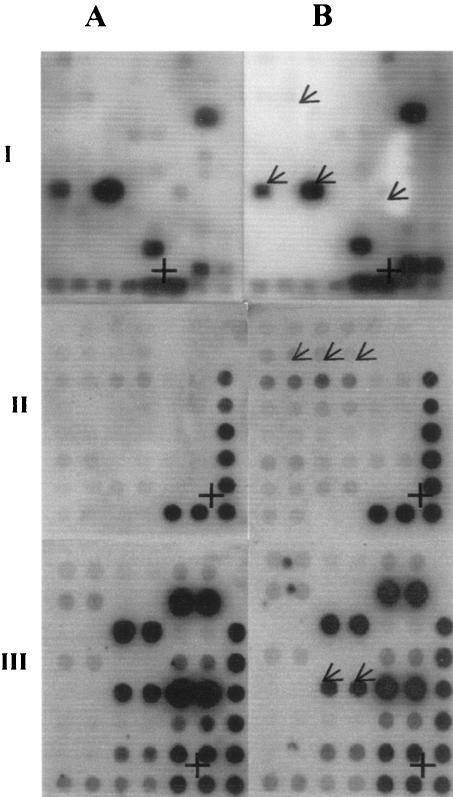
To test the hypothesis that quercetin-induced inhibition of colony formation by prostate cell lines is mediated by modulation of the expression of cell cycle genes, we treated PC-3 cells with 25 μM quercetin and determined the expression of a variety of cell cycle genes by the gene array methodology. These experiments demonstrate that quercetin treatment suppressed the expression of a wide range of genes involved in the regulation of the cell cycle (Fig. 2, panel IB, and Table 1). Quercetin inhibited the expression of several G1-specific genes, including three human D-type cyclin genes (CCND1, CCND2, and CCND3), two human E-specific cyclin genes (CCNE1 and CCNE2), two cell division cycle-dependent kinase genes (CDK2 and CDK4), and two transcription factor genes (E2F2 and E2F3). Quercetin inhibited the following S-phase-specific genes: those for cycle dependent kinase 8 (CDK8), cell division cycle 7 (CDC7L1), proliferating cell nuclear antigen (PCNA), and the G2-specific gene CCNF. The M-phase-specific genes CDC2 and CDC16 were also significantly inhibited by quercetin. Of note, CUL4B, a member of the cullin gene family that is also known to be involved in control of the cell cycle, was significantly up-regulated by quercetin. The significance of this paradoxical finding is unknown.
FIG. 2.
Autoradiographs of cDNA gene array of regulatory genes from untreated control (column A) and quercetin-treated (column B) cells. (I) Cell cycle genes; (II) tumor suppressor genes; (III) oncogenes. An arrow identifies the specific genes that are modulated, and a plus sign (+) indicates the position of the β-actin housekeeping gene. The arrow in the second row of panel IB indicates the positions of CCND1, the arrows in the sixth row indicate the positions of CDK8 and CDC16 (left and right, respectively), and the arrow in the seventh row indicates the position of E2F2. The arrows in panel IIB indicate the positions of the CREB-binding protein, and the arrows in panel IIIB indicate the positions of v-akt murine thymoma viral oncogene homolog 1. Data are from a representative experiment, and two other experiments produced similar results.
TABLE 1.
Genes from PC-3 cells that are significantly up- or down-regulated after treatment with quercetin, as analyzed by the gene array methodologya
| Gene array | Genes whose level is modulated less than or more than 50% | % Increase or decrease in gene expression |
|---|---|---|
| Cell cycleb | CCND1 | 65 |
| CCND2 | CS | |
| CCND3 | CS | |
| CCNF1 | CS | |
| CCNE2 | CS | |
| CDK2 | CS | |
| CDK4 | CS | |
| E2F2 | CS | |
| E2F3 | CS | |
| CDK8 | 50 | |
| CDC 7L1 | CS | |
| PCNA | CS | |
| CCNF | CS | |
| CDC2 | CS | |
| CDC16 | CS | |
| CUL4B | NI | |
| Tumor suppressorc | CREB-binding protein | 71 |
| Phospatase and tensin homolog | 72 | |
| MutS homolog 2 | 71 | |
| Cyclin-dependent kinase inhibitor 1A (p21, CiP1) | 57 | |
| CREB-binding protein (p300) | 72 | |
| Von Hippel-Lindau syndrome | 67 | |
| Breast Cancer 1, early onset | 82 | |
| Tuberous sclerosis 1 | 52 | |
| Transforming growth factor β2 receptor | 92 | |
| Adenomatosis polyposis coli | NI | |
| Transforming growth factor, beta receptor 11 | NI | |
| Tumor suppressor protein p53 | NI | |
| Retinoblastoma 1 | NI | |
| Cyclin-dependent kinase inhibitor 1C | NI | |
| Tuberous sclerosis 2 | NI | |
| Oncogeneb | v-akt murine thymoma viral oncogene homolog 1 | 61 |
| v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 | 89 | |
| Breakpoint cluster region | CS | |
| v-myb avian myeloblastosis viral oncogene homolog | CS |
Gene expression was normalized to the expression of the β-actin gene, provided as a housekeeping gene in the array kit. The results are expressed as the percent increase or decrease of gene expression in PC-3 cells treated with quercetin compared to that in control PC-3 cells, after normalization. See Fig. 2. CS, complete suppression; NI, newly induced.
For this array, genes are listed in order of decreasing expression.
For this array, genes are listed in order of increasing expression.
Quercetin up-regulates the expression of tumor suppressor genes by prostate cancer cell lines.
Since quercetin showed significant inhibition of colony formation by the PC-3 and DU-145 cell lines and tumor suppressor genes are known to be involved in inhibition of colony formation, we investigated the effect of quercetin on the expression of several tumor suppressor genes using gene arrays. Our results (Fig. 2, panel IIB, and Table 1) show that the expression of nine different tumor suppressor genes in PC-3 cells was up-regulated by quercetin by more than 50% (range, 52 to 92%) compared to the levels of expression in untreated control cultures. These included the genes for the human cyclic AMP response element binding protein (CBP); phosphatase and tensin homolog (PTEN); mutS homolog 2 (MSH2); cyclin-dependent kinase inhibitor 1A (p21, ciP1); CREB-binding protein (p300); Von Hippel-Lindau syndrome (VHS); breast cancer 1, early onset (BRCA1); neurofibromin 2 (NF2); tuberous sclerosis 1 (TSC-1); and transforming growth factor β receptor 1 (TGFβR1, ALK-5) genes. Furthermore, the six tumor suppressor genes adenomatosis polyposis coli (APC), transforming growth factor β receptor 11 (TGFβR2), tumor suppressor protein p53 (p53), retinoblastoma 1 (Rb), cyclin-dependent kinase inhbitor 1C (p57Kip2), and tuberous sclerosis 2 (TSC-2) were induced by quercetin, whereas undetectable levels of these genes were expressed by control, untreated PC-3 cells. These studies suggest that quercetin can induce certain tumor suppressor genes as well as up-regulate the expression of existing or endogenous tumor suppressor genes in PC-3 cells.
Quercetin down-regulates expression of oncogenes by prostate cancer cell lines.
Oncogenes are involved in the regulation of cell growth, and increased expression of these genes may contribute to the development of cancer. Since quercetin up-regulated a number of tumor suppressor genes in the gene array analysis, we also examined the effect of quercetin on the expression of several oncogenes. The data presented (Fig. 2, panel IIIB, and Table 1) show that, compared to the levels of expression in untreated control cultures, quercetin significantly down-regulated (61 to 100%) the expression of the following oncogenes: v-kt murine thymoma viral oncogene homolog 1 (akt-1), v-erb-b2 avian erythoblastic leukemia viral oncogene homolog 2 (erb-2), breakpoint cluster region (bcr), and v-myb avian myeloblastosis viral oncogene homolog (c-myc) (Fig. 2, panel IIIA). It is known that oncogenes such as akt-1 promote cancer cell survival through multiple mechanisms both in vitro and in vivo (1, 22). Thus, our studies demonstrate that quercetin down-regulates the expression of several oncogenes and reciprocally up-regulates the expression of various tumor suppressor genes in prostate cancer cells with increased metastatic potential, thereby decreasing their survival abilities.
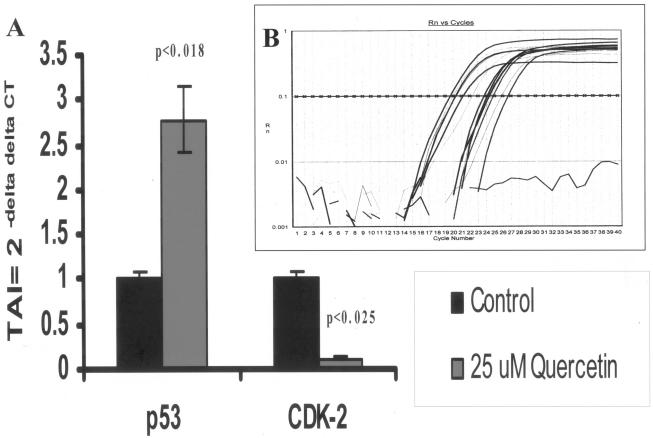
To confirm the results obtained by gene array analysis, we performed quantitative, real-time PCR using PCR primers specific for two randomly selected genes, the gene for p53 and CDK2. Our results, presented in Fig. 3, show that PC-3 cells treated with 25 μM quercetin showed reciprocal up-regulation of the p53 tumor suppressor gene (178% up-regulation; P < 0.008) and reciprocal suppression of the CDK2 cell cycle gene (89% down-regulation; P < 0.025) (Fig. 3) compared to the levels of expression in untreated control cultures, thus confirming the results of gene array analysis presented in Table 1 and Fig. 2.
FIG. 3.
(A) CDK2 and p53 gene expression levels, as quantitated by real-time quantitative PCR with SYBR Green master mix (Stratagene, La Jolla, Calif.). The relative expression of the mRNA species was calculated by the comparative CT method, as described in Material and Methods. (B) Specific gene amplicon plots in PC-3 control and quercetin-treated cells for β-actin, p53, and CDK2 genes. Rn, normalized fluorescence; TAI, transcript accumulation index.
DISCUSSION
Food-derived flavonoids, in particular, quercetin, can modulate a variety of immune functions. In addition to their immunologic effects, previous studies (37) showed that flavonoids also act as enhancers of the effect of radiation. Fruits and vegetables, particularly citrus fruits, apples, onions, parsley, tea, and red wine, are the primary dietary sources of quercetin. Olive oil, grapes, dark cherries, and dark berries, such as blueberries, blackberries, and bilberries, are also high in flavonoids, including quercetin (43). Mahmoud et al. (24) reported that several plant-derived phenolics, including quercetin, prevented the formation of intestinal tumors in murine models. Others have shown a biphasic inhibitory effect of quercetin on the growth and proliferation of the human oral carcinoma cell line SCC-25 (3).
Propolis, a resinous, alternative medicine derived from poplar trees and collected from bees, has been proposed to have antimicrobial and anti-inflammatory activities. In a clinical study, Suzuki and coworkers (33) showed that when crude, water-soluble, propolis-containing quercetin was used together with chemotherapeutic agents, quercetin increased the antitumor effects of the agents and decreased the frequency of postchemotherapeutic complications. Kaneuchi et al. (15) reported that quercetin suppressed the proliferation of human endometrial adenocarcinoma (Ishikawa) cells by down-regulating the expression of epidermal growth factor receptor and cyclin DI genes. Recent studies also showed that quercetin can inhibit the proliferation of other cancers by interfering with tubulin binding and subsequent microtubule assembly (9). The cytocidal activities of flavonoids such as quercetin were observed to correlate reciprocally with their lipophilicities (16). In a phase I clinical trial, quercetin demonstrated significant antitumor effects, and it was proposed that these effects were mediated by inhibition of lymphocyte protein tyrosine phosphorylation (5). We recently reported that quercetin inhibited epidermal growth factor receptor tyrosine kinase activity while stimulating apoptosis in a pancreatic tumor model (19). Although quercetin and other polyphenols have been shown to inhibit carcinogenesis and tumor growth in various organs, this is the first report of their antitumor potentials in cancer of the prostate.
The cyclin D gene encodes a regulatory subunit of the CDK4 and CDK6 holoenzyme complex, which phosphorylates and inactivates the tumor suppressor protein pRB as well as the pRB-related proteins p107 and p130 (33). Phosphorylation of pRB in middle to late G1 results in its inactivation and the release of the E2F transcription factor and other transcription factors that are sequestered by the unphosphorylated, active form of pRB. Once liberated by pRB inactivation, E2Fs then proceed to activate genes essential for progression into late G1 and S phases (12, 41). It is interesting that in the present study the expression of all these genes was significantly inhibited by quercetin. In addition to its inhibitory effect on cell cycle gene expression, quercetin significantly down-regulated the expression of the antiapoptosis gene bcl-2 (data not presented). This effect of quercetin on bcl-2 gene expression with the reciprocal maintenance of the viability of tumor cells is inconsistent with available reports of bcl-2 function. Previous studies suggest that the bcl-2 family is composed of both proapoptotic and antiapoptotic genes (44). Among the factors determining whether apoptosis occurs are the balance between pro- and antiapoptotic genes in the bcl-2 family of genes and the levels of various cytokines and growth factors produced under the given culture conditions (44). It is possible that under our culture conditions quercetin did not affect the viability of the culture yet down-regulated bcl-2 gene expression due to the imbalance between pro- and antiapoptotic gene expression or due to the influence of cytokines and/or growth factors that are known to be induced by quercetin. Furthermore, quercetin significantly suppressed the colony-forming abilities of prostate cancer cells in vitro. This inhibition is associated with down-regulation of the expression of several cell cycle genes and oncogenes and reciprocal up-regulation of tumor suppressor genes. The quercetin-induced modulation of gene expression was not due to toxic effects, as cells treated with quercetin demonstrated viabilities comparable to those of control cells. While quercetin did not affect the expression of housekeeping genes, it selectively up-regulated the expression of tumor suppressor genes. The concentrations of quercetin used in the present investigation are consistent with the concentrations used by other investigators in vitro (3, 15, 19).
As predicted, we found that colony formation by untreated prostate cancer cells was proportional to the aggressive potential of each cell line. We also noted a similar correlation of the effects of quercetin on colony formation with the aggressive potential of the cell lines used. While quercetin significantly inhibited the colony-forming abilities of moderately (DU-145) and highly (PC-3) aggressive cells, quercetin did not have any effect on poorly aggressive LNCaP cells. Previous studies suggest that metastasis is dependent, in part, on the expression of pro- and antiangiogenic factors (6, 7, 10, 21; R. Aalinkeel, M. P. N. Nair, G. S. Sufrin, S. D. Mahajan, K. C. Chadha, and S. A. Schwartz, submitted for publication). Recently, we found that angiogenic regulatory factors are also differentially expressed by and correlate with the metastatic potential of different prostate cancer cell lines (Aalinkeel et al., submitted). We are examining whether quercetin has any effect on angiogenesis by prostate cancer cells.
In vivo evidence indicates that the malignant state emerges from complex interactions in the tumor-host microenvironment affecting the induction, selection, and expression of neoplastic cells (20, 27). Malignant cells proliferate indefinitely and are therefore considered immortal (31, 40). Normal cells must circumvent at least two proliferative barriers, designated mortality stage 1 (M1) and mortality stage 2 (M2), to become immortal. The M1 blockade is regulated by several tumor suppressor genes (31, 32). In this investigation quercetin induced the expression of several tumor suppressor genes. Previous studies demonstrated that negative control mechanisms, mediated by various tumor suppressor genes that normally act to constrain tumor cell proliferation and colony formation, play a major role in suppression of carcinogenesis. The tumor suppressor gene phosphate and tensin homolog (PTEN), later identified as phospholipid phosphatase, suppresses tumor growth by inhibiting the phosphatidylinositol 3-kinase-Akt signaling pathway and causes G1 cell cycle arrest and cell death (36, 42). The transcriptional activators p300 and CREB-binding protein have properties of tumor suppressor proteins and can enhance the activity of the p53 tumor suppressor protein (30). It has previously been demonstrated that the tuberous sclerosis complex (TSC) gene also functions as a tumor suppressor gene and that expression of the TSC-1 and TSC-2 genes can be up-regulated by quercetin (13). While quercetin significantly up-regulated the expression of several tumor suppressor genes that were constitutively expressed at low levels, it is noteworthy that several previously unexpressed tumor suppressor genes were induced by quercetin. These studies suggest that the colony-inhibiting activity of quercetin may be due in part to its ability to induce or enhance the expression of a number of tumor suppressor genes. Although we could not confirm the modulation of all the genes studied using gene arrays by quantitative, real-time PCR, we randomly selected two genes, the gene for p53 and CDK-2, for quantitative PCR, and the gene array results correlated with those of the quantitative PCR.
Increased expression of oncogenes, which also regulate cell growth and induce oncogenesis, has been described for many different malignancies (for reviews, see references 4 and 29). In this investigation we observed an interesting, reciprocal relationship between the effects of quercetin on the expression of oncogenes and tumor suppressor genes. Thus, while quercetin significantly down-regulated oncogene expression, it concomitantly up-regulated the expression of various tumor suppressor genes. We studied a total of 136 genes, and the results for only 35 genes that were up-regulated by quercetin at levels of ≥50% are included in Table 1 and Fig. 1. However, RNA measurements do not necessarily reflect specific protein synthesis. Studies of the corresponding protein expression by all of these genes were beyond the scope of this investigation. Such studies may be the subject of further investigation.
Although quercetin, in general, has been reported to be anti-inflammatory (34), previous studies (26, 39) have shown that quercetin exerts significant modulatory effects on gene expression as well as protein synthesis of both pro- and anti-inflammatory molecules. A previous study (26) showed that quercetin up-regulated the Th-1-derived proinflammatory cytokine gamma interferon, while it reciprocally inhibited the Th-2-derived anti-inflammatory cytokine interleukin-4. Since inflammatory molecules are known to play a significant role in the migration and activation of immunocompetent cells to the tumor site to mediate antitumor effects, it is possible that suppression of anti-inflammatory molecules may lead to reciprocal up-regulation of proinflammatory gamma interferon, which is known to mediate antitumor effects. Therefore, the effect of quercetin on inhibition of the colony-forming abilities of prostate cancer cell lines, as reported in the present investigation, may be mediated in part by an interplay between pro- and anti-inflammatory molecules.
In summary, we found that the dietary bioflavonoid quercetin, present in many foods of plant origin, manifested significant, selective inhibition of the growth of prostate cancer cells in vitro. These effects were greatest on cells with the highest aggressive potentials. We also showed that the anticancer effects of quercetin are mediated by inhibition of the expression of various cell cycle genes and oncogenes and enhancement of the expression of several tumor suppressor genes. Our results correlate with those of nutritional studies that support the roles of dietary bioflavonoids as cancer chemopreventive agents.
Acknowledgments
This work was supported in part by NIH grants R01 DA10632, R01 DA14218, R01 DA12366, and R01 DA 15628 and the Margaret Duffy and Robert Cameron Troup Memorial Fund for Cancer Research of Kaleida Health.
We express our appreciation to Carol Sperry and Tracy Roth for excellent secretarial assistance.
REFERENCES
- 1.Albini, A., Y. Iwamoto, H. K. Kleinman, G. R. Martin, S. A. Aaronson, J. M. Kozlowski, and R. N. McEwan. 1987. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 47:3239-3245. [PubMed] [Google Scholar]
- 2.Burgering, B. M., and P. J. Coffer. 1995. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376:599-602. [DOI] [PubMed] [Google Scholar]
- 3.Elattar, T. M., and A. S. Virji. 2000. The inhibitory effect of curcumin, genistein, quercetin and cisplatin on the growth of oral cancer cells in vitro. Anticancer Res. 20:1733-1738. [PubMed] [Google Scholar]
- 4.Ember, I., Z. Pusztai, Z. Gyongyi, and I. Kiss. 2000. 1-Nitropyrene induces elevated expression of oncogenes and tumor suppressor genes 24 hours after treatment in CBA/Ca mice. Anticancer Res. 20:1563-1566. [PubMed] [Google Scholar]
- 5.Ferry, D. R., A. Smith, J. Malkhandi, D. W. Fyfe, P. G. deTakats, D. Anderson, J. Baker, and D. J. Kerr. 1996. Phase I clinical trial of the flavonoid quercetin: pharmacokinetics and evidence for in vivo tyrosine kinase inhibition. Clin. Cancer Res. 2:659-668. [PubMed] [Google Scholar]
- 6.Fidler, I. J., and L. M. Ellis. 1994. The implications of angiogenesis for the biology and therapy of cancer metastasis. Cell 79:185-188. [DOI] [PubMed] [Google Scholar]
- 7.Folkman, J. 1995. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1:27-31. [DOI] [PubMed] [Google Scholar]
- 8.Foster, C. S., P. Cornford, L. Forsyth, M. B. Djamgoz, and Y. Ke. 1999. The cellular and molecular basis of prostate cancer. Br. J. Urol. Int. 83:171-194. [DOI] [PubMed] [Google Scholar]
- 9.Gupta, K., and D. Panda. 2002. Perturbation of microtubule polymerization by quercetin through tubulin binding: a novel mechanism of its antiproliferative activity. Biochemistry 41:13029-13038. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D., and J. Folkman. 1996. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 86:353-364. [DOI] [PubMed] [Google Scholar]
- 11.Horoszewicz, J. S., S. S. Leong, E. Kawinski, J. P. Karr, H. Rosenthal, T. M. Chu, E. A. Mirand, and G. P. Murphy. 1983. LNCaP model of human prostatic carcinoma. Cancer Res. 43:1809-1818. [PubMed] [Google Scholar]
- 12.Hunter, T., and J. Pines. 1991. Cyclins and cancer. Cell 66:1071-1074. [DOI] [PubMed] [Google Scholar]
- 13.Jaeschke, A., J. Hartkamp, M. Saitoh, W. Roworth, T. Nobukuni, A. Hodges, J. Sampson, G. Thomas, and R. Lamb. 2002. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J. Cell Biol. 159:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandaswami, C., and E. Middleton, Jr. 1994. Free radical scavenging and antioxidant activity of plant flavonoids. Adv. Exp. Med. Biol. 366:351-376. [DOI] [PubMed] [Google Scholar]
- 15.Kaneuchi, M., M. Sasaki, Y. Tanaka, N. Sakuragi, S. Fujimoto, and R. Dahiya. 2003. Quercetin regulates growth of Ishikawa cells through the suppression of EGF and cyclin D1. Int. J. Oncol. 22:159-164. [PubMed] [Google Scholar]
- 16.Krol, W., S. Dworniczak, G. Pietsz, Z. P. Czuba, M. Kunicka, M. Kopacz, and D. Nowak. 2002. Synthesis and tumoricidal activity evaluation of new morin and quercetin sulfonic derivatives. Acta Pol. Pharm. 59:77-79. [PubMed] [Google Scholar]
- 17.Landolph, J. R., R. S. Bhatt, N. Telfer, and C. Heidelberger. 1980. Comparison of adriamycin- and ouabain-induced cytotoxicity and inhibition of 86rubidium transport in wild-type and ouabain-resistant C3H/10T1/2 mouse fibroblasts. Cancer Res. 40:4581-4588. [PubMed] [Google Scholar]
- 18.Landolph, J. R., and C. Heidelberger. 1979. Chemical carcinogens produce mutations to ouabain resistance in transformable C3H/10T1/2 Cl 8 mouse fibroblasts. Proc. Natl. Acad. Sci. USA 76:930-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, L. T., Y. T. Huang, J. J. Hwang, P. P. Lee, F. C. Ke, M. P. Nair, C. Kanadaswam, and M. T. Lee. 2002. Blockade of the epidermal growth factor receptor tyrosine kinase activity by quercetin and luteolin leads to growth inhibition and apoptosis of pancreatic tumor cells. Anticancer Res. 22:1615-1627. [PubMed] [Google Scholar]
- 20.Liotta, L. A., and E. C. Kohn. 2001. The microenvironment of the tumour-host interface. Nature 411:375-379. [DOI] [PubMed] [Google Scholar]
- 21.Liotta, L. A., P. S. Steeg, and W. G. Stetler-Stevenson. 1991. Cancer metastasis and angiogenesis: an imbalance of positive and negative regulation. Cell 64:327-336. [DOI] [PubMed] [Google Scholar]
- 22.Liu, A. X., J. R. Testa, T. C. Hamilton, R. Jove, S. V. Nicosia, and J. Q. Cheng. 1998. AKT2, a member of the protein kinase B family, is activated by growth factors, v-Ha-ras, and v-src through phosphatidylinositol 3-kinase in human ovarian epithelial cancer cells. Cancer Res. 58:2973-2977. [PubMed] [Google Scholar]
- 23.Mahajan, S. D., S. A. Schwartz, and M. P. Nair. 2003. Immunological assays for chemokine detection in In-vitro culture of CNS cells. Biol. Proc. Online 5:90-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahmoud, N. N., A. M. Carothers, D. Grunberger, R. T. Bilinski, M. R. Churchill, C. Martucci, H. L. Newmark, and M. M. Bertagnolli. 2000. Plant phenolics decrease intestinal tumors in an animal model of familial adenomatous polyposis. Carcinogenesis 21:921-927. [DOI] [PubMed] [Google Scholar]
- 25.Middleton, E., Jr., and C. Kandaswami. 1993. The impact of plant flavonoids on mammalian biology: implication for immunity, inflammation and cancer, p. 619-645. In J. B. Harborne (ed.), The flavonoids: advances in research since 1986. Chapman & Hall, London, United Kingdom.
- 26.Nair, M. P., C. Kandaswami, S. Mahajan, K. C. Chadha, R. Chawda, H. Nair, N. Kumar, R. E. Nair, and S. A. Schwartz. 2002. The flavonoid, quercetin, differentially regulates Th-1 (IFNgamma) and Th-2 (IL4) cytokine gene expression by normal peripheral blood mononuclear cells. Biochim. Biophys. Acta 1593:29-36. [DOI] [PubMed] [Google Scholar]
- 27.Reznikoff, C. A., J. S. Bertram, D. W. Brankow, and C. Heidelberger. 1973. Quantitative and qualitative studies of chemical transformation of cloned C3H mouse embryo cells sensitive to postconfluence inhibition of cell division. Cancer Res. 33:3239-3249. [PubMed] [Google Scholar]
- 28.Robbins, D. H., and S. H. Itzkowitz. 2002. The molecular and genetic basis of colon cancer. Med. Clin. N. Am. 86:1467-1495. [DOI] [PubMed] [Google Scholar]
- 29.Scolnick, D. M., N. H. Chehab, E. S. Stavridi, M. C. Lien, L. Caruso, E. Moran, S. L. Berger, and T. D. Halazonetis. 1997. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 57:3693-3696. [PubMed] [Google Scholar]
- 30.Shay, J. W. 1997. Telomerase in human development and cancer. J. Cell. Physiol. 173:266-270. [DOI] [PubMed] [Google Scholar]
- 31.Shay, J. W., O. M. Pereira-Smith, and W. E. Wright. 1991. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 196:33-39. [DOI] [PubMed] [Google Scholar]
- 32.Sherr, C. J. 1994. G1 phase progression: cycling on cue. Cell 79:551-555. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, I., I. Hayashi, T. Takaki, D. S. Groveman, and Y. Fujimiya. 2002. Antitumor and anticytopenic effects of aqueous extracts of propolis in combination with chemotherapeutic agents. Cancer Biother. Radiopharm. 17:553-562. [DOI] [PubMed] [Google Scholar]
- 34.Theoharides, T. C., M. Alexandrakis, D. Kempuraj, and M. Lytinas. 2001. Anti-inflammatory actions of flavonoids and structural requirements for new design. Int. J. Immunopathol. Pharmacol. 14:119-127. [PubMed] [Google Scholar]
- 35.Tsugawa, K., M. K. Jones, K. Sugimachi, I. J. Sarfeh, and A. S. Tarnawski. 2002. Biological role of phosphatase PTEN in cancer and tissue injury healing. Front. Biosci. 7:245-251. [DOI] [PubMed] [Google Scholar]
- 36.Van Rijn, J., and J. van den Berg. 1997. Flavonoids as enhancers of X-ray-induced cell damage in hepatoma cells. Clin. Cancer Res. 3:1775-1779. [PubMed] [Google Scholar]
- 37.Wang, H. K. 2000. The therapeutic potential of flavonoids. Expert Opin. Investig. Drugs 9:2103-2119. [DOI] [PubMed] [Google Scholar]
- 38.Webber, M. M., D. Bello, and S. Quader. 1997. Immortalized and tumorigenic adult human prostatic epithelial cell lines: characteristics and applications. Part 2. Tumorigenic cell lines. Prostate 30:58-64. [DOI] [PubMed] [Google Scholar]
- 39.Wei, B. L., C. M. Lu, L. T. Tsao, J. P. Wang, and C. N. Lin. 2001. In vitro anti-inflammatory effects of quercetin 3-O-methyl ether and other constituents from Rhamnus species. Planta Med. 67:745-747. [DOI] [PubMed] [Google Scholar]
- 40.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 41.Weng, L. P., W. M. Smith, P. L. Dahia, U. Ziebold, E. Gil, J. A. Lees, and C. Eng. 1999. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res. 59:5808-5814. [PubMed] [Google Scholar]
- 42.Wright, W. E., O. M. Pereira-Smith, and J. W. Shay. 1989. Reversible cellular senescence: implications for immortalization of normal human diploid fibroblasts. Mol. Cell. Biol. 9:3088-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young, J. F., S. E. Nielsen, J. Haraldsdottir, B. Daneshvar, S. T. Lauridsen, P. Knuthsen, A. Crozier, B. Sandstrom, and L. O. Dragsted. 1999. Effect of fruit juice intake on urinary quercetin excretion and biomarkers of antioxidative status. Am. J. Clin. Nutr. 69:87-94. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, X. D., X. Y. Zhang, C. P. Gray, T. Nguyen, and P. Hersey. 2001. Tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis of human melanoma is regulated by smac/DIABLO release from mitochondria. Cancer Res. 61:7339-7348. [PubMed] [Google Scholar]