Abstract
Homologous recombination is a high-fidelity DNA repair pathway. Besides a critical role in accurate chromosome segregation during meiosis, recombination functions in DNA repair and in the recovery of stalled or broken replication forks to ensure genomic stability. In contrast, inappropriate recombination contributes to genomic instability, leading to loss of heterozygosity, chromosome rearrangements, and cell death. The RecA/UvsX/RadA/Rad51 family of proteins catalyzes the signature reactions of recombination, homology search and DNA strand invasion 1,2. Eukaryotes also possess Rad51 paralogs, whose exact role in recombination remains to be defined 3. Here we show that the budding yeast Rad51 paralogs, the Rad55-Rad57 heterodimer, counteract the anti-recombination activity of the Srs2 helicase. Rad55-Rad57 associate with the Rad51-ssDNA filament, rendering it more stable than a nucleoprotein filament containing Rad51 alone. The Rad51/Rad55-Rad57 co-filament resists disruption by the Srs2 anti-recombinase by blocking Srs2 translocation involving a direct protein interaction between Rad55-Rad57 and Srs2. Our results demonstrate an unexpected role of the Rad51 paralogs in stabilizing the Rad51 filament against a biologically important antagonist, the Srs2 anti-recombination helicase. The biological significance of this mechanism is indicated by a complete suppression of the ionizing radiation sensitivity of rad55 or rad57 mutants by concomitant deletion of SRS2, as expected for biological antagonists. We propose that the Rad51 presynaptic filament is a meta-stable reversible intermediate, whose assembly and disassembly is governed by the balance between Rad55-Rad57 and Srs2, providing a key regulatory mechanism controlling the initiation of homologous recombination. These data provide a paradigm for the potential function of the human RAD51 paralogs, which are known to be involved in cancer predisposition and human disease.
Rad51 protein and its homologs RecA, UvsX, and RadA form nucleoprotein filaments with ssDNA that perform homology search and DNA strand invasion during homologous recombination. The Rad51 paralogs share the RecA core with the Rad51 protein featuring unique N- and C-terminal extensions (Supplementary Fig. 2), but themselves do not form filaments and are unable to perform homology search and DNA strand invasion 2-4. While humans contain five paralogs (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3), the budding yeast Saccharomyces cerevisiae contains two clearly identifiable paralogs, Rad55 and Rad57 (Supplementary Fig. 2). Rad55 and Rad57 in yeast as well as the five human RAD51 paralogs have unique non-redundant functions in recombination, and mutations in any one of them lead to recombination defects, chromosomal instability, sensitivity to DNA damage, and meiotic defects 1-3. Defects in the budding yeast RAD55 and RAD57 genes lead to identical and epistatic phenotypes in DNA repair and recombination, consistent with the formation of a stable Rad55-Rad57 heterodimer 4,5. Rad55-Rad57 were inferred to function as mediator proteins (ref. 6) allowing assembly of the Rad51 nucleoprotein filament on ssDNA covered by the eukaryotic ssDNA-binding protein RPA 4. This suggested that Rad55-Rad57 are involved in the nucleation of the Rad51 filament, which is otherwise inhibited on RPA-covered ssDNA. This nucleation model is akin to the role of RecFOR or BRCA2 in nucleating RecA or human RAD51 filaments 7-9. Rad51 filament formation in vivo can be monitored cytologically as Rad51 focus formation at the site of DNA damage 10. Unexpectedly, Rad51 focus formation after IR in yeast was demonstrated to be independent of Rad55-Rad57 and formation of visible Rad55-Rad57 foci required Rad51 10. These results are difficult to reconcile with the nucleation model derived from the biochemical results and suggest an alternative function of Rad55-Rad57 in vivo.
To address the function of the Rad51 paralogs in yeast, we determined the effect of Rad55-Rad57 on the stability of Rad51-ssDNA nucleoprotein complexes. Deletion mutants of the RAD55 or RAD57 genes display a curious enhancement of some phenotypes at low temperature (in particular IR sensitivity; see Supplementary Fig. 12) 5, suggesting that these proteins are involved in the stabilization of a molecular complex, likely the Rad51 presynaptic filament. To test this hypothesis, we incubated subsaturating amounts of Rad51 protein with ssDNA (1 Rad51 per 15 nts) in the presence of substoichiometric amounts of Rad55-Rad57 heterodimer (1 Rad55-Rad57 per 4 Rad51) and challenged the filaments with buffer containing high salt (500 mM NaCl) (Supplementary Fig. 3a, b). Under these conditions, Rad51 does not maintain stable complexes with ssDNA during electrophoresis. However, the presence of Rad55-Rad57 resulted in stable, Rad51-containing ssDNA complexes that withstood the salt challenge. In a complementary approach, we examined the effect of Rad55-Rad57 on Rad51 filament formation at near physiological ionic strength (90 mM NaCl) (Fig. 1a, b). Under these conditions, only a fraction of the available Rad51 binds ssDNA, causing retarded mobility of the DNA (Fig. 1b, lane 3). Addition of substoichiometric amounts of Rad55-Rad57 (1 Rad55-Rad57 per 6 Rad51 in lane 4 of Fig. 1b) led to the formation of a novel, supershifted complex that contained both Rad51 and Rad55-Rad57, as demonstrated by immunoblotting. Rad55-Rad57 alone binds to DNA under these conditions, leading to the formation of protein-networks that are too large to enter the gel (Fig. 1b, lane 2). The results from both experiments (Fig. 1b; Supplementary Fig. 3) suggest that Rad55-Rad57 form a co-complex with Rad51 on ssDNA and stabilize Rad51-ssDNA filaments. Indeed, immunogold electron microscopy (EM) targeted towards Rad55 (GST-tag; see Fig. 1c) directly visualized Rad55 associated with the Rad51-ssDNA filaments (Fig. 1d). Control experiments demonstrated the specificity of the gold labeling (Supplementary Table 1) with over 90% of the gold particles associated with clearly identifiable Rad51 filaments. The remainder may have associated with filaments too short to be scored or with free Rad55-Rad57. Gold particles were found either at the filament terminus (n=40) or interstitially (n=43) (Supplementary Table 1). Negative controls with Rad51 filaments assembled in the absence of Rad55-Rad57 showed negligible gold labeling (Supplementary Table 1). These data show that Rad55-Rad57 are associated with the Rad51-ssDNA filament, but the exact disposition of the heterodimer with the filament remains to be determined (see Fig. 1e).
Figure 1. Rad55-Rad57 is associated with and stabilizes Rad51-ssDNA filaments.
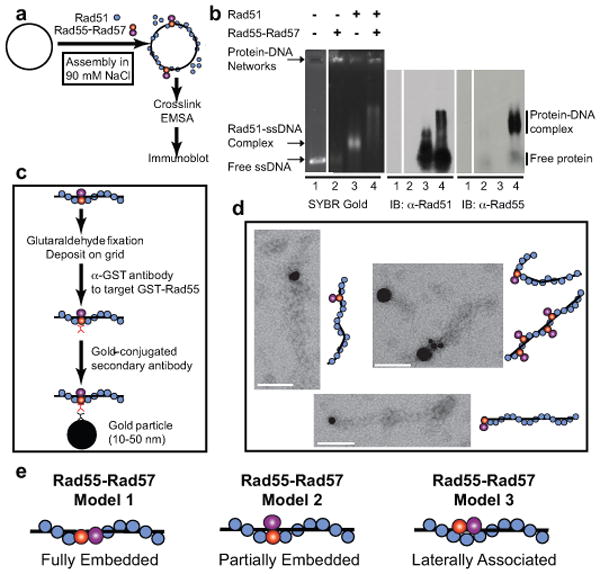
a, Rad51-ssDNA filament assembly assay. b, 0.67 μM Rad51 ± 0.11 μM Rad55-Rad57 was incubated with 4 μM φX174 ssDNA. The migration position of free protein was confirmed in controls lacking DNA (Supplementary Fig. 3c). c, Reaction scheme of immunoaffinity gold labeling of Rad55. d, EM images of gold-labeled Rad55 associated with Rad51-ssDNA filament (1:3 Rad51/nucleotide; 2.34 μM Rad51 ±0.43 μM Rad55-Rad57, 7 μM (nt) ssDNA). Scale bars: 100 nm. e, Models for the disposition of Rad55-Rad57 with the Rad51 filament. For simplicity, only model 2 is drawn in all illustrations.
Salt stability of protein-DNA complexes is a valuable biochemical criterion. To establish biological significance, we tested whether Rad55-Rad57 stabilize Rad51-ssDNA filaments against a biologically relevant destabilizer. The Srs2 helicase was identified as a negative regulator of homologous recombination, and genetic experiments suggested that Srs2 targets Rad51 protein 11-13. Consistent with the genetic data, Srs2 translocates on ssDNA and disrupts Rad51 presynaptic filaments in vitro, providing a compelling mechanism for its function as an anti-recombinase 14-16. In the presence of 0.1 or 0.33 μM Srs2 approximately 70% of the Rad51 is dissociated as assessed by measuring Rad51 associated with ssDNA coupled to magnetic beads (Fig. 2a- c). The presence of substochiometric amounts of Rad55-Rad57 (0.1 μM) enhanced the recovery of ssDNA-bound Rad51 by ∼2-fold (from 31% to 60% in the presence of 333 nM Srs2). Rad55-Rad57 and Srs2 bound to Rad51-covered ssDNA in a quantitative and concomitant manner (Fig. 2d). Together the data show that Rad55-Rad57 inhibit Srs2 when bound to DNA and not in solution. Concentration-dependent inhibition of Srs2-mediated dissociation of Rad51 from ssDNA by Rad55-Rad57 was also observed in a topology-based assay (Supplementary Fig. 4, 5).
Figure 2. Rad55-Rad57 stabilizes Rad51-ssDNA filaments to resist disruption by Srs2.
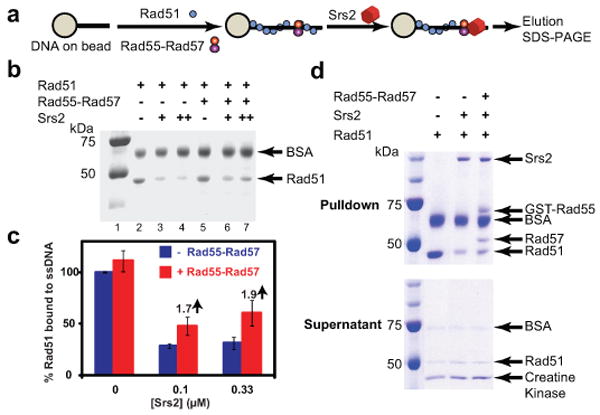
a, Pulldown assay measuring stability of Rad51-ssDNA complexes (1 Rad51: 3 nt, 1 μM Rad51 ± 0.1 μM Rad55-Rad57) against disruption by Srs2 (0.1 or 0.33 μM). b, Rad51 remaining bound to ssDNA. c, Quantitation of results in (b) and additional experiments. Shown are means ± 1 sd, n=3. d, Concomitant binding of Rad55-Rad57 and Srs2 to Rad51-covered ssDNA. Pulldown assay measuring stability of Rad51-ssDNA complexes (1 Rad51: 3 nt, 1 μM Rad51 ± 0.2 μM Rad55-Rad57) against disruption by 0.33 μM Srs2. Top, pulldowns; bottom, supernatants.
To further investigate the role of Rad55-Rad57 in antagonizing disruption of Rad51 presynaptic filaments by Srs2, we utilized EM to examine nucleoprotein filaments directly (Fig. 3; Supplementary Fig. 6). Rad51 filaments were assembled on a 600 nt fragment of ssDNA and RPA was added to visualize free ssDNA. Consistent with previous observations 14,15, in the absence of Rad55-Rad57 Srs2 disrupts the Rad51-ssDNA filament efficiently, leading to binding of RPA to the newly exposed ssDNA (Fig. 3). Importantly, when sub-stoichiometric amounts of Rad55-Rad57 were co-incubated with Rad51 and ssDNA, the filaments were stabilized against disruption by Srs2, as indicated by the significantly increased mean filament length.
Figure 3. Rad55-Rad57 inhibit disruption of Rad51 presynaptic filaments by Srs2.
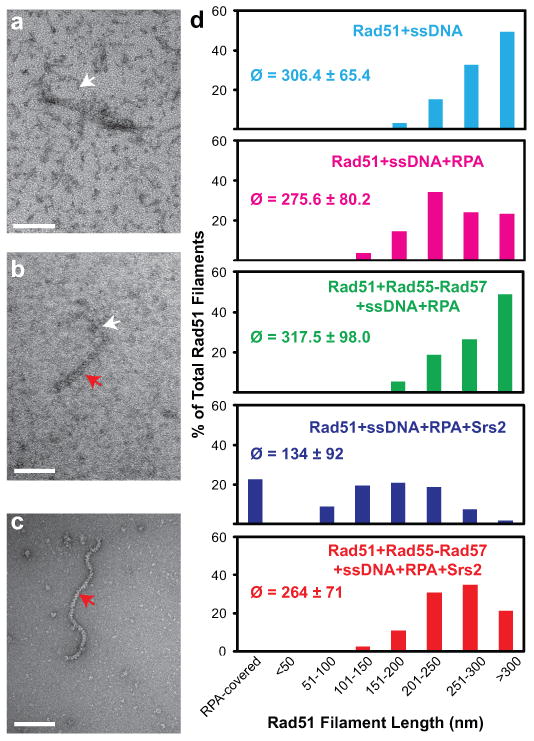
a, RPA-ssDNA complex. b, Short (145 nm) Rad51-ssDNA filament. c, Long (350 nm) Rad51-ssDNA filaments d, Quantitation of electron microscopic analysis. 300-400 filaments were analyzed for each reaction condition (2.34 μM Rad51, 7 μM (nt) 600 nt ssDNA, ± 0.43 μM Rad55-Rad57, ± 0.21 μM RPA, ± 0.4 μM Srs2), and the means (ø) ± 1 sd and distributions of filament length classes are shown. Scale bars: 100 nm. White arrows indicate RPA-ssDNA complexes and red arrows Rad51 filaments.
How do Rad55-Rad57 block Srs2 from dissociating Rad51 from ssDNA? Srs2 is known to interact with Rad51 and trigger the Rad51 ATPase leading to dissociation of Rad51 from ssDNA 16. We found that Rad55-Rad57 form a 1:1 complex with Srs2 (Fig. 4a) and have higher affinity to Srs2 than to Rad51 (Fig. 4b, c). Excess Rad51 does not compete with Srs2 binding to Rad55-Rad57 (Supplementary Fig. 7). Moreover, Rad55-Rad57 are able to simultaneously bind Rad51 and Srs2 in a 1:1:1 stoichiometry (Fig. 4d; Supplementary Figs. 7-9). We entertained the possibility that Rad55-Rad57 inhibit the Srs2 ATPase activity and by that Srs2 translocation, but Srs2 ATPase activity is barely altered by the presence of Rad55-Rad57 (data not shown). Srs2 translocase/helicase activity is stimulated by Rad51 binding to DNA 17 (Fig. 4e-g). Importantly, Rad55-Rad57 completely suppress this stimulatory effect of Rad51, leading to inhibition of the Srs2 helicase activity even at a 5-fold molar excess of Srs2 over Rad55-Rad57 (Fig. 4f, g; Supplementary Fig. 10). This substoichiometric action of Rad55-Rad57 eliminates the possibility that Rad55-Rad57 inhibition functions by binding Srs2 in solution. Rad55-Rad57 only slightly inhibit Srs2 helicase in the absence of Rad51 (Fig. 4g; Supplementary Fig. 10c). Control experiments show that this effect depends on Srs2 translocating in the expected 3′ to 5′ direction (Supplementary Fig. 10d), showing that Rad55-Rad57 inhibit Srs2 translocation on DNA to increase filament stability (Figs. 1, 2) and function (Supplementary Fig. 11). Direct visualization of human RAD51 filaments revealed that RAD51 is only able to form discontinuous short clusters on dsDNA, as a result of frequent nucleation but limited extension 18,19. If this property holds true for ssDNA, the formation of a co-filament with Rad51 by Rad55-Rad57 might provide a mechanism to form extended Rad51 filaments. This could also explain the increase in Rad55-Rad57 focus intensity over time in following IR exposure and is consistent with the dependence of Rad55-Rad57 foci on Rad51 10.
Figure 4. Rad55-Rad57 interact with Srs2 and inhibit Srs2 helicase.
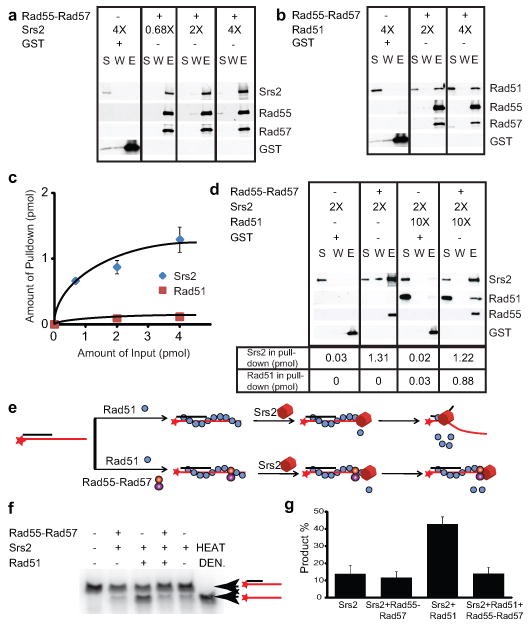
a, Pull-down with 4 nM (1 pmol) Rad55-Rad57 and 2.7, 8, or 16 nM Srs2. b, Pull-down with 4 nM Rad55-Rad57 and 8 or 16 nM Rad51. c, Quantitation of results in (a) and (b) and additional experiments. d, Pull-down with 4 nM Rad55-Rad57 and 8 nM Srs2 ± 40 nM Rad51. GST was used as control. S: supernatant, W: wash, E: eluate. e, Helicase assay. f, 28 nM Rad51 ± 25 nM Rad55-Rad57 were incubated with 1.5 nM 3′-tailed substrate before addition of 120 nM Srs2 protein. Product yields at 20 min. were quantified as shown in g. HEAT DEN.: heat denatured substrate, shown are means ± 1sd, n=3.
Our biochemical data are consistent with a model (Supplementary Fig. 1) whereby Rad51 presynaptic filament formation is modulated by a balance between the stabilizing function of Rad55-Rad57 and the destabilizing function of Srs2 anti-recombinase. This model predicts that a deletion of SRS2 should suppress the phenotypes caused by defects in Rad55-Rad57. In fact, srs2Δ completely suppresses the IR sensitivity of rad57 and rad55 mutations in quantitative survival assays (Supplementary Fig. 12), consistent with semi-quantitative results using rad57 20. However, srs2Δ only mildly suppresses the MMS sensitivity (Supplementary Fig. 13) and recombination defect (Supplementary Fig. 14) of a rad55 mutation, consistent with previous rad57 data20. The difference in suppression is likely related to that IR-induced DNA damage requires primarily DSB repair, whereas MMS-induced DNA damage and sister chromatid recombination require gap repair (Supplementary Fig. 1). We propose that the Rad51 presynaptic filament is a meta-stable reversible intermediate, whose dynamics in yeast are partially controlled by the balance of the filament-stabilizing activity of Rad55-Rad57 and the filament-destabilizing activity of the Srs2 helicase (Supplementary Fig. 1). This balance is likely influenced by the multiple post-translational modifications that have been identified to regulate Rad55-Rad57 21 and Srs2 22 functions (Supplementary Fig. 1) (ref. 1). Together with the local availability of SUMO-PCNA, which specifically recruits Srs2 23-25, post-translational modifications may determine the balance between recombination and anti-recombination in wild type cells and explain the various degrees of suppression observed in the srs2 rad55 (rad57) double mutants that depend on the type of DNA damage or genetic endpoint (DSB versus replication fork associated gap in Supplementary Fig. 1).
The human RAD51 paralogs play important roles in tumor suppression and human disease 3,26. Our studies established an unprecedented mechanism of anti-antirecombination that may serve as a paradigm for the mechanism of action of the five human RAD51 paralogs. The diversification of the human RAD51 paralogs may reflect the multiplicity of human motor proteins that may disrupt RAD51 presynaptic filaments, including the RecQ-like helicases BLM and RECQL5 as well as FBH1 and FANCJ 27-30 or indicate additional functions during recombinational repair.
Methods summary
Purification of yeast Rad51, Rad55-Rad57, RPA, and Srs2, the biochemical assays and the EM analysis are detailed in Methods.
Supplementary Material
Acknowledgments
We thank M. Alexeeva for the cell culture support. We thank P. Sung, R. Kolodner, and L. Symington for plasmids and yeast strains. We are grateful to S. Kowalczykowski, N. Hunter, D. Castaño-Diez, P. Ringler and all members of the Heyer laboratory for discussions and comments on the manuscript. This work was supported by a Postdoctoral fellowship 17FT-0046 from the Tobacco-Related Disease Research Program (J.L.), by European Community (LSHG- CT-2003-503303) and the Centre National de la Recherche Scientifique, the Commissariat à l'Energie Atomique (X.V., F.F.), by SystemsX.ch (H.S.), the National Institutes of Health (NIH) U54GM74929 (H.S.) and CA92267 and GM58015 (W.D.H.).
Footnotes
Full methods and any associated references are available in the online version of the paper at www.nature.com/nature
Author contribution: J.L. designed, performed, analyzed all experiments, except the IR survival assay, and helped write the manuscript. L.R. helped with the EM image collection and data analysis. X.V. purified Srs2 protein. F.F. performed the IR experiment. H.S. advised on the EM analysis. W.D.H. conceived the project, designed experiments, coordinated collaborations, contributed to data analysis and wrote the manuscript. All authors discussed results and edited the manuscript.
The authors declare no competing financial interests.
References
- 1.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 4.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 5.Lovett ST, Mortimer RK. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: Effects of temperature, osmotic strength and mating type. Genetics. 1987;116:547–553. doi: 10.1093/genetics/116.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beernink HTH, Morrical SW. RMPs: Recombination/replication mediator proteins. Trends Biochem Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 7.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Schiestl RH, Prakash S, Prakash L. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics. 1990;124:817–831. doi: 10.1093/genetics/124.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aboussekhra A, et al. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguilera A, Klein HL. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krejci L, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 15.Veaute X, et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 16.Antony E, et al. Srs2 Disassembles Rad51 Filaments by a Protein-Protein Interaction Triggering ATP Turnover and Dissociation of Rad51 from DNA. Mol Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupaigne P, et al. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 18.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Nat Acad Sci USA. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Modesti M, et al. Fluorescent human RAD51 reveals multiple nucleation sites and filament segments tightly associated along a single DNA molecule. Structure. 2007;15:599–609. doi: 10.1016/j.str.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Fung CW, Mozlin AM, Symington LS. Suppression of the Double-Strand-Break-Repair defect of the Saccharomyces cerevisiae rad57 mutant. Genetics. 2009;181:1195–1206. doi: 10.1534/genetics.109.100842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herzberg K, et al. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol Cell Biol. 2006;26:8396–8409. doi: 10.1128/MCB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saponaro M, et al. Cdk1 Targets Srs2 to Complete Synthesis-Dependent Strand Annealing and to Promote Recombinational Repair. Plos Genetics. 2010;6:e1000858. doi: 10.1371/journal.pgen.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 25.Burgess RC, et al. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J Cell Biol. 2009;185:969–981. doi: 10.1083/jcb.200810055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meindl A, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nature Gen. 2010;42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 27.Hu Y, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommers JA, et al. FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J Biol Chem. 2009;284:7502–7514. doi: 10.1074/jbc.M809019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fugger K, et al. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J Cell Biol. 2009;186:655–663. doi: 10.1083/jcb.200812138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


