Abstract
EGFR family members and c-Src are co-overexpressed in many cancers. The synergistic effect of EGFR and c-Src has been shown in the tumorigenesis of breast and other cancers. Reported mechanisms of synergy include transcriptional regulation by STAT5b and the regulation of cellular ATP production by mitochondrial protein COX II. Here we report a new mechanism of EGFR-c-Src synergy through choline kinase α (CHKA). The first enzyme of the phosphatidyl choline production pathway, CHKA is overexpressed in many cancers, and the product of the enzyme, phosphocholine, is also increased in tumor cells. In this report, we find that CHKA forms a complex with EGFR in a c-Src dependent manner. Endogenous CHKA and EGFR co-immunoprecipitated from a variety of breast cancer cell lines and immortalized mammary epithelial cells. CHKA interacted with the EGFR kinase domain upon c-Src co-overexpression and was phosphorylated in a c-Src-dependent manner on Y197 and Y333. Overexpression of EGFR and c-Src increased total cellular activity and protein levels of CHKA. Mutation of CHKA Y197 and Y333 reduced complex formation, EGFR-dependent activation of CHKA enzyme activity and EGF-dependent DNA synthesis. Furthermore, siRNA-mediated knockdown of CHKA in MCF-7 and MCF-10A cells reduced EGF-dependent cell proliferation. Together, these results strongly implicate a new c-Src-dependent link between CHKA and EGFR, which contributes to the regulation of cell proliferation and tumorigenesis.
Keywords: Choline kinase, proliferation, c-Src, EGFR, breast cancer, tyrosine phosphorylation
Introduction
Epidermal growth factor receptor (EGFR/HER1) is overexpressed in a wide variety of human cancers, including colon, lung, prostate, and breast (Biscardi et al 2000)(Biscardi et al 1999b), implicating its function in tumorigenesis. The non-receptor tyrosine kinase, c-Src, is also overexpressed in many of these same tumors, suggesting that the two tyrosine kinases may functionally interact. In breast tumors, for example, 70–100% overexpress c-Src, with the majority of these tumors co-overexpressing one of the EGFR family members (HER1–4) (reviewed in (Ishizawar and Parsons 2004)). Breast cancer cells or murine fibroblasts that co-overexpress EGFR and c-Src exhibit synergistic increases in anchorage-independent growth in cell culture and development of tumors in mouse xenograft models than those overexpressing only one of the pair (Maa et al 1995)(Biscardi et al 1998). In the immortalized mammary epithelial-derived cell lines, co-overexpression of EGFR and c-Src, but not EGFR or c-Src alone causes hyperproliferation, aberrant three dimensional acinar structures, enhancement of migration and invasion, and anchorage independent cell growth (Dimri et al 2007). Taken together, those results strongly suggest that co-overexpression of EGFR and c-Src is not just a by-product of tumorigenesis but a driver of the process in mammary epithelial cells.
Subsequent investigations have revealed several molecular mechanisms by which c-Src and EGFR cooperate. First, c-Src phosphorylates Y845 of the EGFR, a residue that resides in the activation loop of the kinase domain and is highly conserved among receptor tyrosine kinases (Biscardi et al 1999a)(Hunter and Cooper 1985). (Note that several reports indicate that Src-independent phosphorylation of Y845 can occur of (Qiu et al 2009)(Yang et al 2008)). Mutation of Y845 to phenylalanine (Y845F) has little to no affect on the ligand-activated catalytic activity of the EGFR but reduces EGF-induced DNA synthesis and growth in soft agar of murine fibroblasts and human breast cancer cell lines (Tice et al 1999) (Biscardi et al 1999a). These results indicate that phosphorylated Y845 (pY845) in EGFR plays a key role in anchorage-dependent and -independent cell proliferation. pY845 is required for activation of STAT5b, a cytosolic transcription factor whose action is critical for EGF-induced DNA synthesis (Kloth et al 2003). pY845 is also required for binding of the EGFR to a mitochondrial protein, cytochrome c oxidase subunit II (CoxII), a key component of the oxidative phosphorylation pathway (Boerner et al 2004). EGF-induced translocation of the EGFR to the mitochondria is accompanied by phosphorylation of Cox II and modulation of cellular ATP production (Demory et al 2009). Thus, to date, evidence suggests that the biological synergy between the EGFR and c-Src is mediated, at least in part, by pY845, which regulates cellular transcriptional programs through STAT5b and bioenergetics through Cox II. In this study we investigated additional mechanisms by which the EGFR and c-Src synergize to promote tumor progression.
Choline kinase α (CHKA), which converts choline to phosphocholine in the phosphatidylcholine synthesis (Kennedy) pathway (Aoyama et al 2004)(Wu and Vance 2010), is overexpressed in breast, lung, prostate, colorectal and bladder cancers (Ramirez de Molina et al 2002a)(Ramirez de Molina et al 2002c)(Ramirez de Molina et al 2002b)(Ramirez de Molina et al 2007)(Hernando et al 2009). The product of this enzyme, phosphocholine, is also increased in primary malignant tumors of the breast, brain, and prostate and in cancer cell lines (reviewed in (Glunde et al 2006)).
High levels of CHKA correlate with poor prognosis of non-small-cell lung cancers (Ramirez de Molina et al 2007), histological grade of breast cancer (Ramirez de Molina et al 2002a), and aggressiveness of bladder cancer (Hernando et al 2009). As a result of these studies, this enzyme is under consideration for therapeutic targeting. In pre-clinical studies, a small molecule choline kinase inhibitor (Ramirez de Molina et al 2004) or lentivirus shRNA-mediated knock-down of CHKA (Krishnamachary et al 2009) was found to suppress xenograft growth in vivo and growth factor-induced breast cancer cell growth in vitro (Cuadrado et al 1993)(Ramirez de Molina et al 2004), suggesting that CHKA plays a pivotal role in tumorigenesis. However, little is known about how CHKA is regulated and how high levels of the enzyme are achieved in cancer cells. In this study, we reveal one mechanism of regulation involving EGFR and c-Src. Specifically, we found that CHKA activity is increased upon overexpression of EGFR and c-Src and this increase requires c-Src kinase activity and complex formation of CHKA with EGFR and c-Src. c-Src-mediated phosphorylation enhances the association of CHKA with EGFR and is critical for EGF-induced DNA synthesis. Furthermore, EGF-induced breast epithelial cell proliferation is dependent upon CHKA. Together, these findings provide evidence for a third pathway that mediates the synergistic actions of EGFR-c-Src in breast cancer development.
Results
Yeast two-hybrid identification of an EGFR/choline kinase α2 interaction
To identify additional EGFR signaling pathways, we performed a yeast two-hybrid screen with the EGFR kinase domain (aa 672–960) as bait. In addition to MIG-6(Hackel et al 2001) and other clones, a single clone was identified that contained a splice variant or a splicing intermediate sequence of choline kinase α2 (CHKA2). Encoded within this clone were a 228-nucleotide (76 amino acid) sequence of an alternative reading-frame and a segment of an intron of CHKA2, followed by the C-terminal 343 aa (aa115–457) of CHKA2 in-frame. The resulting product of this clone activated transcription of yeast reporter genes in the two-hybrid system only when the EGFR-kinase domain bait was co-expressed. But neither the N-terminal 76 aa segment of the original clone, the CHKA aa115–457 segment, nor full length CHKA2 activated the reporter genes in the presence of the EGFR kinase domain, suggesting either that the full length splice variant was the only form of CHKA that could efficiently bind EGFR or that full-length wild type (wt) CHKA2 might require other co-factor(s) or some modification(s) to the protein for efficient binding. In this study we explored the latter possibility.
EGFR and CHKA2 form a complex in a c-Src-dependent manner in mammalian cells
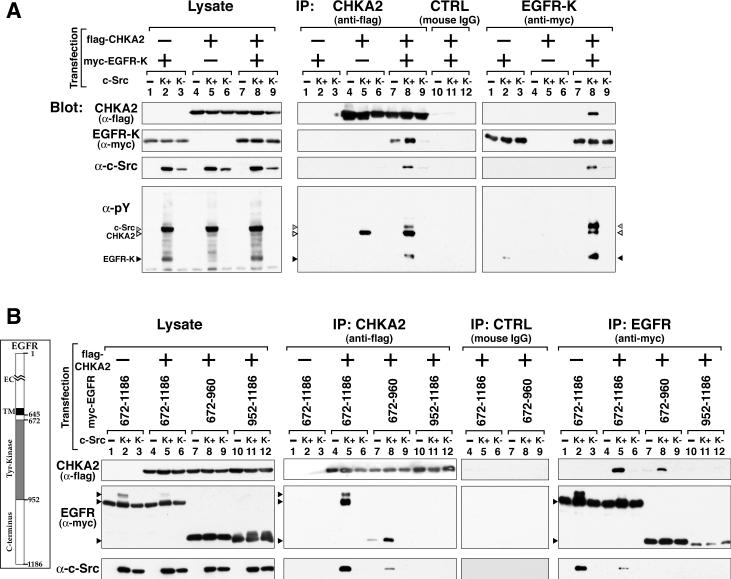
Since c-Src and EGFR have been shown to synergize in promoting tumorigenesis (Maa et al 1995)(Tice et al 1999), we hypothesized that c-Src could be a co-factor that facilitates the association of CHKA2 with the EGFR kinase domain in mammalian cells. To test this hypothesis, a co-immunoprecipitation assay of transiently transfected CHKA2 and EGFR kinase domain was performed in 293T cells that were co-overexpressing either wild type (wt, K+) or kinase-defective (K−) c-Src. Figure 1A shows that without any overexpression of c-Src, immunoprecipitates of flag-tagged CHKA2 contained only slightly detectable amounts of myc-tagged EGFR kinase domain (center panel, lane 7), which was significantly augmented by co-overexpression of wt c-Src (center panel, lane 8). The interaction was abolished by co-overexpression of K− c-Src (A430V, Fig. 1A, center panel, lane 9). The K+ c-Src-dependence of this association was confirmed in the reciprocal co-immunoprecipitation (right panel, lanes 7–9). These results suggest that the EGFR kinase domain and CHKA2 indeed make a complex in a c-Src-activity-dependent manner. Note that association of c-Src with this complex requires the expression of both EGFR kinase domain and CHKA2 (c-Src panel, lanes 8). Binding was observed with the K721A or Y845F mutants of the EGFR kinase domain (kinase inactive and c-Src-dependent phosphorylation site mutants, respectively) (data not shown), suggesting that neither pY845 nor EGFR kinase activity is required for association with CHKA2.
Figure 1. CHKA2 co-immunoprecipitates with EGFR kinase domain in a c-Src activity dependent manner in 293T cells.
A. Plasmids encoding flag-tagged CHKA2 and myc-tagged EGFR kinase domain (EGFR-K, aa 672–960) were co-transfected with those encoding wt (K+) or kinase-defective (K−) c-Src into 293T cells. Where indicated, the amount of total DNA for transfection was adjusted with the corresponding empty vectors (−). Lysates were examined for levels of specific proteins or used for co-immunoprecipitation as described in Materials and Methods. Western blotting of whole cell lysates (Lysate) is shown in the left panel and of CHKA2 and EGFR immunoprecipitates in the middle and right panels, respectively. The same amount of mouse IgG was used for negative control (CTRL) immunoprecipitation as for CHKA2 (lane 10–12). Immunprecipitated proteins were detected by Western blotting with the indicated antibodies. B. Plasmids encoding flag-tagged CHKA2 and various myc-tagged domains of the cytosolic region of EGFR (diagram, left) were co-transfected into 293T cells with those encoding wild type (K+) or kinase-defective (K−) c-Src and analyzed as in Panel A. Constructs containing the kinase domain of EGFR associated with CHKA2 in a c-Src-activity dependent manner.
We next asked whether any other region in the EGFR intracellular domain could support complex formation with CHKA2. Figure 1B shows that CHKA2 protein associated with the complete intracellular domain of EGFR (aa 672–1186) (center and right panels) and the isolated kinase domain (aa 672–960) in a K+ c-Src-dependent manner, but not to the C-terminal region (aa 952–1186). This result indicated that CHKA2 forms a complex with the EGFR cytosolic region mainly via its kinase domain and that the binding is dependent on c-Src activity.
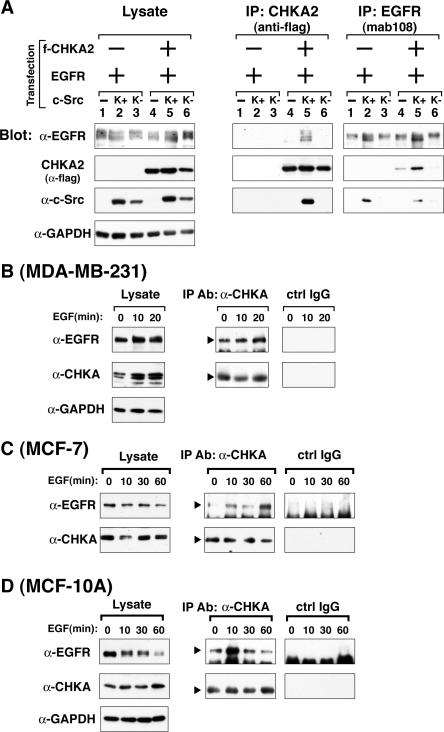
Figure 2A shows that full length EGFR also bound to CHKA in a c-Src-dependent manner, with enhanced association compared to the amount formed in the absence of overexpressed K+ c-Src. Confocal immunofluorescence highlighted the plasma membrane localization of CHKA when EGFR and c-Src were co-expressed in contrast to its cytosolic localization when expressed alone (SFig. 1), suggesting that the EGFR/c-Src complex recruited CHKA to the membrane. Similarly, association of endogenous EGFR with endogenous choline kinase α was examined before and after EGF stimulation. As shown in Figure 2B–D, in two breast cancer-derived cell lines, MDA-MB-231 and MCF-7, and in the immortalized mammary epithelial cell line, MCF-10A, antibody specific for CHKA co-precipitated EGFR with or without EGF stimulation. Association appeared to be slightly enhanced following EGF treatment, but with varying times of peak complex formation. In MCF-7 and MCF-10A cells the time of complex formation following EGF stimulation coincided with phosphorylation of EGFR at Y1068 and Y845 and c-Src at Y418 (SFig. 2). Pharmacological inhibition of c-Src but not of EGFR reduced CHKA tyrosine phosphorylation in response to EGF treatment of MCF-7 cells (SFig. 3), suggesting that tyrosine phosphorylation of CHKA in breast cancer cells is largely mediated by c-Src.
Figure 2. CHKA2 co-immunoprecipitates with exogenous or endogenous full length EGFR.
A. Plasmids encoding full length EGFR and flag-tagged CHKA2 (f-CHKA2) were cotransfected into 293T cells along those encoding K+ or K− c-Src and analyzed as in Figure 1A, except that mab108 was used instead of myc Ab for immunoprecipitation of EGFR. B–D. Indicated cells were serum starved overnight and stimulated with 50 (B), 10 (C) or 5 ng/mL (D) EGF for the indicated times. CHKA was immunoprecipitated with rabbit anti-CHKA antibody and rabbit IgG was used as a negative control (crtl IgG). Immunoblotting was carried out as described in Materials and Methods.
CHKA is required for maximum EGF-dependent cell growth in mammary epithelium-derived cell lines
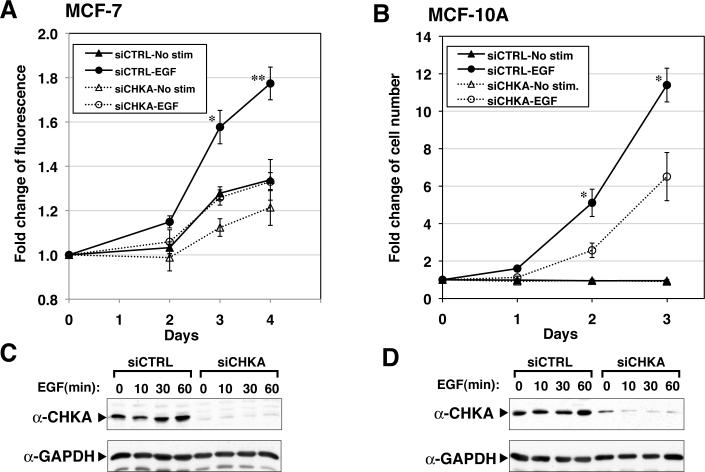
Since complex formation between endogenous EGFR and CHKA was observed, we examined the role of CHKA in EGF-dependent cell growth. Figure 3 shows the effects of silencing CHKA on cell growth of MCF-7 and MCF-10A cells. In an alamarBlue assay, clear suppression of EGF-dependent growth was seen in siCHKA transfected MCF-7 cells compared to control siRNA transfected cells at day 4 (p=0.0062) (Panel A). Cells undergoing control siRNA transfection without EGF stimulation and CHKA siRNA transfection with or without EGF stimulation were statistically undistinguishable, suggesting that CHKA is required for EGF-dependent cell growth in the MCF-7 cell line. Knock-down of CHKA protein was confirmed by western blotting (Panel C).
Figure 3. Silencing of endogenous CHKA reduces EGF-stimulated growth of MCF-7 and MCF-10A cells.
Cells were transfected with control (siCTRL) or CHKA (siCHKA) siRNA and treated as described in Materials and Methods. A. After starvation, MCF-7 cells were incubated with or without 10 ng/mL EGF, and cell viability was measured by the alamarBlue assay (n=3). B. Growth of MCF-10A cells in response to 5 ng/mL EGF was monitored by cell counting (n=3). Similar results were observed using the MTS assay (data not shown). Statistical significance between asterisk marked treatment and other treatments was determined by Student's t test. *: p < 0.05, **: p <0.01. The results are expressed as the mean ± SEM for three experiments. C, D. The same siRNA transfected samples in A and B were harvested at different time points of EGF stimulation and Western blotted to assess levels of CHKA along with control GAPDH. C: MCF-7. D: MCF-10A.
Cell growth of MCF-10A cells was monitored by counting cell number (Panel B) and the MTS assay (data not shown). Results from both assays were similar. At days 2 and 3, silencing of CHKA significantly reduced EGF-dependent cell proliferation compared to control siRNA (p = 0.037 and 0.036, respectively). Knock-down of CHKA in MCF-10A was confirmed (Panel D). Overall, these results strongly support a role for CHKA in the EGF-dependent cell growth of mammary epithelial-derived cell lines.
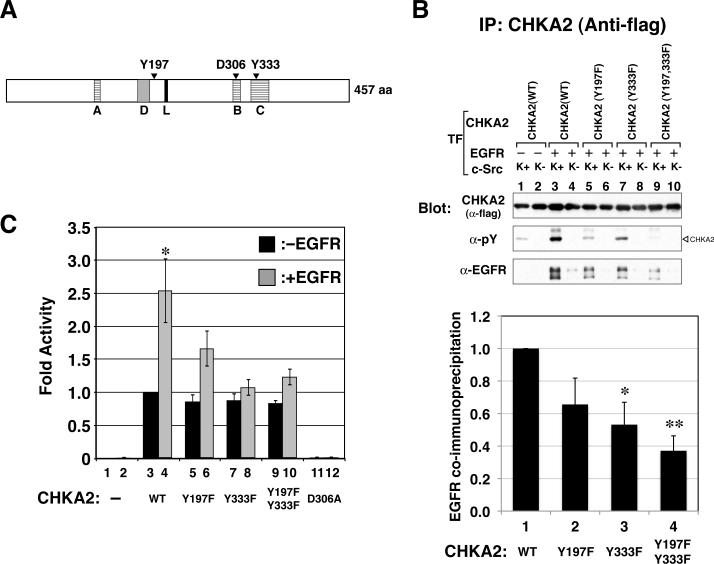
Two newly identified c-Src-dependent phosphorylation sites of CHKA2 protein are required for EGFR-dependent cell proliferation
To characterize the functional relationship between CHKA, EGFR and c-Src, we investigated the phosphorylation sites on CHKA. We had previously noticed that CHKA2 became tyrosine phosphorylated in a c-Src-dependent manner with or without EGFR kinase domain overexpression (Fig. 1A, middle panel). We hypothesized that phosphorylation of CHKA2 could contribute to the complex formation and/or activation of CHKA2. To identify phosphorylation sites on CHKA2, flag-tagged CHKA2 was overexpressed along with c-Src in 293T cells, purified and sequenced by mass spectrometry. With >90% coverage of the protein sequence (which included all the tyrosine residues in the protein), two phospho-tyrosines (pY), pY197 and pY333, were identified. The Y197 site is located just C-terminal to the dimer interface, and the Y333 site is in the choline kinase motif (Peisach et al 2003)(Malito et al 2006)(Fig. 4A). Mutagenesis of either of these residues partially reduced the total tyrosine phosphorylation of CHKA2 (Panel B, anti-pY blot), while mutation of both residues reduced phosphorylation to almost undetectable levels (lane 9), suggesting that Y197 and Y333 are the major tyrosine phosphorylation sites on the protein. The absence of CHKA2 tyrosine phosphorylation in the presence of kinase defective c-Src indicated that phosphorylation of both residues is c-Src mediated, although it is not clear whether this occurs in a direct or indirect manner. In the same context, the binding of various CHKA2 mutants to full length EGFR was examined. Co-immunoprecipitated EGFR with wt or mutant forms of CHKA2 was detected with anti-EGFR antibody and quantified (Fig. 4B). The two single tyrosine mutants showed reduced binding to EGFR relative to wt, while the double mutant exhibited an even further reduction (37%) compared to wt (Panel B bottom, bar 1 vs. 4, p=0.00048). These results indicate that two c-Src-dependent phosphorylation sites are important for formation of the CHKA-EGFR complex.
Figure 4. c-Src-dependent phosphorylation of tyrosines 197 and 333 in CHKA2.
A. Schematic of CHKA2 structure indicating positions of Y197 and Y333, two c-Src-dependent phosphorylation sites, as well as D306, a catalytically critical residue. A: ATP binding loop; B: Brenner's motif (conserved in many phosphotransferases); C: choline kinase motif; D: dimer interface; L: linker (Malito et al 2006). B. Wild-type and mutant forms of flag-CHKA2 mutated at c-Src phosphorylation sites were exogenously expressed along with EGFR and K+ or K− c-Src in 293T cells. Flag-CHKA2 immunoprecipitate was western blotted with the indicated antibodies. Graph: Mutant forms of CHKA2 showed reduced binding to EGFR. Quantification of binding measured by the amount of EGFR co-immunoprecipitated with anti-flag antibody is shown in the bottom graph. The value of wt is set at 1. The results are expressed as the mean ± SEM for four experiments. Asterisk indicates statistical significance (*: p < 0.05, **: p <0.01) to wt. C. Relative in vitro choline kinase activity of 293T lysates expressing exogenous wt or mutant forms of CHKA with or without exogenously expressed full length EGFR. Black and grey bars indicate CHKA alone or in combination with EGFR, respectively. D306A is a catalytically inactive mutant of CHKA (Liao et al 2006). Wild-type activity without EGFR overexpression is set at 1. The results are expressed as the mean ± SEM for three experiments. Asterisk-marked bar has statistical significance (p < 0.05) with all samples except the Y197F mutant with EGFR cotransfected.
To address whether c-Src-mediated phosphorylation of CHKA2 affected its activity, the in vitro choline kinase activity of wt and mutant, ectopically-expressed CHKA2 was assayed in extracts of 293T cells in the presence or absence of co-expressed full-length EGFR. Figure 4C shows no difference in activity of the various forms of CHKA2 in the absence of overexpressed EGFR. However, in the presence of co-transfected EGFR, the activity of wt CHKA2 was increased 2.5 fold (p=0.0261). Y197F and Y333F single mutants and the Y197/333F double mutant showed reduced activity in the presence of EGFR, with the greatest reductions seen with the Y333F and Y197F/Y333F variants (p=0.0249 and 0.0225, respectively). As a control, an inactivating mutation in the Brenner's motif (Liao et al 2006), D306A of the human clone, completely eliminated CHKA2 activity.
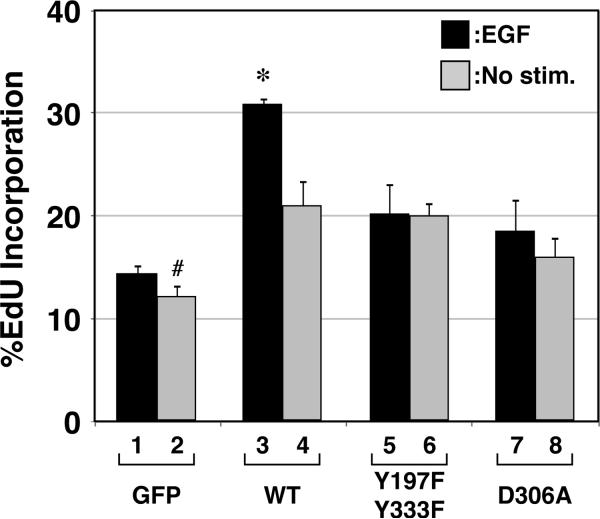
Further characterization of the Y197/333F double mutant revealed that it failed to support the enhanced EGF-induced DNA synthesis seen with overexpressed wt CHKA2, which was 2.2 fold above the EGF-treated and estrogen-starved controls in MCF-7 cells (Fig. 5, compare bar 1 with 3). Wild-type CHKA2 enhanced basal DNA synthesis 1.7 fold above control (compare bars 2 and 4, p=0.0050), indicating that overexpression of CHKA2 alone had a stimulatory effect on cell cycle progression. A similar level of DNA synthesis was maintained by the Y197/333F double mutant and the catalytically inactive D306A mutant, suggesting that CHKA2 has a small but positive effect on DNA synthesis that is independent of its tyrosine phosphorylation or catalytic activity (Compare bar 2 with bars 6 and 8). The D306A mutant, like the double Y197/333F mutant, failed to support an EGF-dependent increase in DNA synthesis (compare bar 1 with bars 5 and 7), suggesting that the biological activity of the Y197/333F mutant is similar to that of the catalytically inactive mutant. Taken together, these studies indicate that c-Src-dependent phosphorylation of CHKA2 is critical for EGFR-dependent activation of CHKA2.
Figure 5. Overexpression of wt CHKA2 enhances EGF-dependent and -independent MCF-7 cell proliferation, while CHKA2-Y197/333F abolishes the EGF-dependent enhancement.
MCF-7 cells transfected with plasmids encoding wt or mutant forms of flag-tagged CHKA2, were estrogen-deprived and incubated with or without 50 ng/mL EGF for 15 hrs. DNA synthesis was measured by EdU incorporation and quantified by immunofluorescence microscopy as percent flag-positive cells that were positive for EdU incorporation as described in Materials and Methods. The asterisk indicates statistical significance (p< 0.03) as compared to all other bars. The results are expressed as the mean ± SEM for three experiments. Bar 2 (#) is statistically significantly different (p < 0.05) from all other conditions.
CHKA2 total cellular activity and protein levels are regulated by EGFR and c-Src
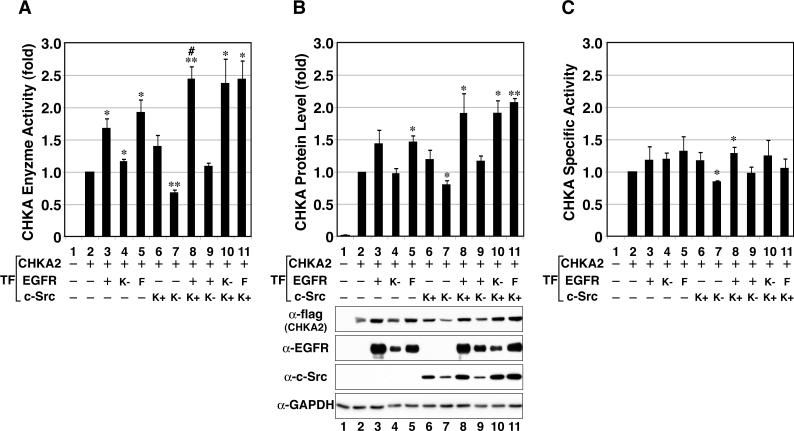
To determine the relative contributions of EGFR and c-Src to the activity of wt CHKA2, extracts of MCF-7 cells overexpressing full-length wt EGFR and/or c-Src were assayed for CHKA2 activity. Figure 6A shows that when either EGFR or c-Src was co-transfected with CHKA2, in vitro choline kinase activity was increased 1.68-fold or 1.40-fold, respectively, compared to control cells transfected with CHKA2 alone (lanes 3 & 6 vs. 2), while co-overexpression of both EGFR and c-Src significantly increased this level to 2.45 fold (lane 8 vs. 2). Co-overexpression of EGFR with kinase-defective c-Src instead of wt c-Src reduced the CHKA2 activity to that seen with the CHKA2 alone control (1.10-fold, lane 9 vs. 2), suggesting that c-Src activity was required for EGFR-dependent activation of CHKA2. In addition, kinase-negative (K−) c-Src significantly reduced CHKA activity in the absence of EGFR (lane 7 vs. 2), but K− EGFR alone had no effect on CHKA activity (lane 4), suggesting that the endogenous kinase activity of c-Src but not of EGFR is critical for regulating CHKA activity. It is also noteworthy that co-expression of K− EGFR with wt c-Src fully enhanced CHKA activity (lane 10 vs. 8), suggesting that EGFR protein but not activity is required for activation of CHKA by EGFR and c-Src. The Y845F mutant of the EGFR had nearly identical effects on CHKA as wt EGFR (lanes 5 vs. 3, 11 vs. 8), indicating that phosphorylation of this site is dispensable for the effects of EGFR on CHKA. Under the conditions of this experiment, we found negligible effect of EGF stimulation on CHKA2 in vitro enzyme activity (data not shown). Interestingly, protein levels of transfected CHKA2, as measured by immunoblotting with a flag monoclonal antibody, correlated well with total choline kinase activity (Panels A and B), and the specific activities of CHKA2 from each transfection group (Panel C) were found to be quite similar, varying between 0.85 and 1.33. Comparable results were also seen in 293T cells (data not shown). These results suggest that total choline kinase activity is cooperatively enhanced by EGFR and c-Src co-overexpression primarily through an increase in total CHKA2 protein levels. Thus, one mechanism by which total CHKA2 activity can be increased in cancer cells co-overexpressing EGFR and c-Src is by c-Src-dependent phosphorylation of the enzyme and association with EGFR.
Figure 6. Choline kinase activity and protein level are enhanced by co-overexpression of EGFR and/or c-Src.
Flag-CHKA2 plasmid was co-transfected with plasmids expressing full-length wt (+), K721A (K−) or Y845F (F) variants of EGFR and/or wt c-Src or kinase defective c-Src into MCF-7 cells, and CHKA activity and protein level were assayed. The results are expressed as the mean ± SEM for three experiments. Statistical significance between the CHKA2 alone (bar 2) and other groups was determined by Student's t test, *: p < 0.05, **: p < 0.01. A. In vitro choline kinase activity was measured from equal amounts of sample lysate as described in Materials and Methods. The value of the CHKA2 alone sample was set at 1. Bar 8 is statistically significantly different (#: p <0.05) from all other treatments except bars 10 and 11. B. Relative flag-CHKA2 protein levels were measured by Western blotting using anti-flag monoclonal antibody and quantified by densitometric analysis. GAPDH served as a loading control. Examples of Western blotting are shown below the graph. C. Relative specific activities of flag-CHKA2 in the various samples were calculated by dividing the relative activity by the relative protein amount.
Discussion
In addition to regulating transcription through STAT5b (Kloth et al 2003) and cancer cell bioenergetics through the electron transport enzyme, Cox II (Boerner et al 2004)(Demory et al 2009), this study describes a third mechanism by which c-Src can synergize with the EGFR to promote tumorigenesis (Maa et al 1995), namely, by increasing CHKA protein levels resulting in increased total cellular CHKA activity. High levels of CHKA and elevation of phosphocholine and tCho have been found in many cancers, including breast cancer (Glunde et al 2006)(Wu and Vance 2010), suggesting that it plays a critical role in oncogenesis. tCho can be detected by magnetic resonance spectroscopy, a powerful non-invasive diagnostic method for breast and other cancers (Glunde et al 2006). Among the tCho compounds, high-resolution NMR studies revealed that the level of phosphocholine, the product of choline kinase, is most closely correlated with tumorigenesis (Glunde et al 2006)(Morse et al 2009).
How phosphocholine contributes to tumorigenesis is not clear at this point, but it could be due to its role as an essential phospholipid in membrane biogenesis, or alternatively, it has been speculated that phosphocholine could serve as a second messenger, regulating cell cycle progression (Cuadrado et al 1993). Indeed, shRNA-mediated depletion or pharmacological inhibition of CHKA reduces MDA-MB-231 breast cancer cell proliferation in vitro and tumor size in xenograft models (Ramirez de Molina et al 2004)(Krishnamachary et al 2009)(Chua et al 2009) providing evidence for a crucial role in tumorigenesis. It is noteworthy that MDA-MB-231 cells express high levels of EGFR and c-Src (Belsches-Jablonski et al 2001), thus serving as one example where EGFR/c-Src-mediated tumorigenesis can be suppressed by choline kinase inhibition. Several reports(Chua et al 2009)(Yalcin et al 2010) also show that inhibition of choline kinase attenuates PI3K/AKT activation, a major component of survival signaling, thus providing another explanation for the reduction in cell proliferation we observed (Fig. 3).
Cellular phosphocholine levels are enhanced by other factors (such as insulin, PDGF, heregulin and bFGF) in addition to EGF (Cuadrado et al 1993)(Uchida 1996)(Ramirez de Molina et al 2004). Interestingly, signaling downstream of these receptors has also been linked to c-Src (Biscardi et al 1999b), raising the possibility that CHKA may be regulated by these growth factor receptors in a similar fashion as EGFR and c-Src. It has been shown that heregulin, a ligand of Her3/ErbB3, enhances phosphocholine production and DNA synthesis in MCF-7 cells. Conversely, a choline kinase inhibitor abolishes both the basal and heregulinstimlated DNA synthesis in these cells (Ramirez de Molina et al 2004). Our laboratory has also found that siRNA knockdown of CHKA reduced heregulin-dependent cell growth of MCF-7 cells, and furthermore, Her2/ErbB2 was co-immunoprecipitated with CHKA (T. Miyake and S.J. Parsons, unpublished data), suggesting that CHKA could complex with and serve as a downstream mediator of ErbB2/3 signaling. c-Src has also been shown to regulate ErbB2/3 complex formation and subsequent signaling (Ishizawar et al 2007), raising the question of the involvement of c-Src in phosphocholine production following heregulin treatment.
Our results demonstrate that complex formation between EGFR and CHKA occurs in a c-Src dependent manner, which results in enhanced protein level and total activity of CHKA. Complex formation could recruit CHKA to the membrane (SFig.1), and/or affect conformations or activities of EGFR, c-Src and/or CHKA, which in turn could change their downstream signaling. c-Src-mediated phosphorylation of CHKA2 did not require the EGFR kinase domain (Fig. 1A, IP:CHKA, lane 5), but CHKA phosphorylation was enhanced by co-overexpression of full length EGFR and c-Src (Fig. 4B, α-pY, blot lane 1 vs. 3). Also, the Y197/333F mutant of CHKA reduced, but still retained association with EGFR. These results suggest that complex formation of EGFR and CHKA enhances the c-Src-mediated phosphorylation of CHKA and that association of EGFR and CHKA may be regulated by other factor(s) besides phosphorylation of Y197 and Y333. Note that the possibility that c-Src-mediated phosphorylation of CHKA is not direct but carried out by another tyrosine kinase activated by c-Src cannot be ruled out at this point. The exact nature of the EGFR/CHKA complex is also not clear. One key question is whether the binding between EGFR and CHKA is direct. Currently, we cannot exclude the possibility that other factors regulated by c-Src are required for a stable interaction between EGFR and CHKA. Such factors, in addition to the catalytic activity of c-Src itself could be potential therapeutic targets for inhibiting formation and function of the EGFR-CHKA2 complex.
The role of c-Src in the CHKA-EGFR complex formation is crucial, since dominant negative c-Src can completely abolish this association. Our study revealed two c-Src-dependent phosphorylation sites (Y197 and Y333) on CHKA2 that are important for efficient complex formation, EGFR-dependent increase of CHKA2 protein levels and enhancement of EGF-dependent cell growth upon CHKA2 overexpression (Figs. 3, 4, and 5). Mutation of each of the single phosphorylation sites reduced complex formation between CHKA and EGFR, while the combination was even more effective, suggesting that both sites are important for complex formation and its subsequent consequences. Since the Y197 site is located proximal to the dimer interface and Y333 is within the choline kinase motif, phosphorylation of these sites could also regulate dimerization of the enzyme and/or enzyme activity, respectively.
Since EGF stimulation induces c-Src binding to the EGFR and activation (Stover et al 1995)(Mao et al 1997), it is likely that in normal cells the binding of EGF ligand activates EGFR and c-Src (Ishizawar and Parsons 2004), which in turn enhances the association of EGFR with and activation and/or increasing protein levels of CHKA2. Figure 2B–D supports this hypothesis, since greater amounts of endogenous EGFR were found in association with endogenous CHKA following EGF stimulation, although basal association between the two molecules was also observed. Our transient transfection experiment may mimic the condition of some of the tumor cells in the sense that CHKA protein levels and total activity are basally increased by co-overexpression of EGFR and c-Src.
In summary, this study describes a new mechanism of biological synergy between EGFR and c-Src, namely regulation of total cellular CHKA activity through increasing protein levels, which in turn enhances lipid metabolism or phosphocholine–mediated signaling. Identification of CHKA as a downstream mediator of EGFR/c-Src signaling suggests that CHKA may be an effective target of therapeutic intervention, especially in tumors co-overexpressing EGFR and c-Src. Indeed, the combination of molecules raises the possibility of targeting all three enzymes in the complex, since they function cooperatively in regulating pathways critical to cell proliferation. Additionally, identification of other molecules in the complex is likely to lead not only to a greater understanding of the mechanisms of CHKA regulation by EGFR complex formation and c-Src dependent phosphorylation, but may also reveal new therapeutic targets for breast and other cancer types that overexpress EGFR, c-Src and/or choline kinase.
Materials and Methods
Molecular biology
Two hybrid screening was performed using MATCHMAKER GAL4 Two-Hybrid System 3 from Clontech (Palo Alto, CA) according to the manufacturer's instructions. The coding region of EGFR kinase domain (aa 672–960) was cloned into pGBK-T7 and used as a bait-expressing plasmid. Human mammary gland MATACHMAKER library was purchased from Clontech. Plasmids containing full length CHKA2 cDNA were obtained from Invitrogen (Carlsbad, CA), and the coding sequences of each were confirmed. Full length CHKA2 was then amplified by Platinum Pfx DNA polymerase (Invitrogen) and cloned into a pCMV-flag vector in which the HA-tag sequence of pCMV-HA (Clontech) was replaced with the flag-tag sequence. Y197F, Y333F and D306A mutations in CHKA2 were introduced by PCR using the method described in Ho et al. (Ho et al 1989). PCR-amplified regions of human EGFR [EGFR kinase domain (aa 672–960), C-terminal region (aa 952–1186) and cytosolic region (aa 672–1186)] were cloned into a pCMV-myc vector (Clontech), and sequences were confirmed. Expression plasmids for full length EGFR and wt and kinase defective c-Src were described previously (Boerner et al 2004).
Cell culture and transfection
293T, MCF-7 and MDA-MB-231 cell lines were obtained from American Type Culture Collection (Manassas, VA) and cultured in DMEM, Penicillin/Streptomycin and 10% FBS (Invitrogen) at 37°C in a humidified environment. MCF-10A cells were cultured as previously described (Debnath et al 2003). For plasmid transfection of 293T and MCF-7 cells, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer's protocol. When the presence or absence of an encoding transfected cDNA was tested, the control transfection was carried out with the corresponding empty vector. Dasatinib and Gefitinib(LC laboratories, Woburn, MA), as well as PP2 (EMD, Rockland, MA) were used at the indicated concentrations.
Antibody production and purification
Rabbit anti-human CHKA2 antibody was raised by Covance (Denver, PA). A peptide containing the sequence of CHKA2 aa 265–280 was conjugated with KLH and injected subcutaneously with adjuvant. Antisera were cleared of antibodies cross-reacting with carrier and non-specific peptides, then further purified with SulfoLink Immobilization Kit from Thermo Fisher Scientific (Rockford, IL) conjugated with the immunizing peptide.
Immunoblotting and immunoprecipitation
Cell lysates were prepared in NP40 buffer (50mM HEPES pH7.4, 1% NP-40, 150mM NaCl). Lysates used to immunoprecipitate full-length EGFR were prepared in CHAPS buffer (50mM HEPES pH7.4, 0.65% CHAPS, 150mM NaCl). Both buffers contained 2mM EDTA, 1mM sodium orthovanadate, and Protease Inhibitor Complete from Roche Applied Science (Indianapolis, IN). Protein concentration was determined by the BCA assay (Thermo Fisher Scientific). For immunoprecipitation, 100 or 500μg protein lysate was incubated with either flag-M2-antibody-conjugated agarose (Sigma-Aldrich, St.Louis, MO), mouse-anti-myc (Cell Signaling Technology, Danvers, MA), EGFR mouse monoclonal antibody 108 (Wright et al 1996), rabbit anti-CHKA, or appropriate species control IgG (Jackson ImmunoResearch, West Grove, PA) overnight at 4°C. For non-conjugated antibodies, Protein A agarose (Millipore, Billerica, MA) was added and incubated for one hour. Agarose beads were then washed four times with the cognate lysis buffer, and immunoprecipitated proteins were extracted with hot 2× sample buffer. Immunoblotting was performed using standard methods with the following antibodies: rabbit anti-myc-tag, EGFR, and c-Src (Cell Signaling), rabbit anti-flag-tag (Sigma-Aldrich), GAPDH (Millipore), and HRP-conjugated pY99, goat anti-rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA), sheep anti-mouse IgG (GE Healthcare, Piscataway, NJ) and native IgG detection reagent (HRP) (Thermo Fisher Scientific). Phospho-EGFR and phospho-Src antibodies were described previously (Boerner et al 2004).
CHKA siRNA knockdown and cell proliferation assays
On-Target plus SMART pool CHKA2 siRNA or Non-targeting pool for siRNA control (Thermo Fisher Scientific) was transfected into indicated cells using Lipofectamine RNAiMax (Invitrogen) according to manufacturer's directions. Single siRNAs and luciferase siRNA were also tested to ensure specificity (SFig. 4). After transfection MCF-10A cells were incubated in EGF-depleted medium overnight, then stimulated with or without 5ng/mL EGF. Cell growth was monitored by counting trypan blue negative cells, thus representing only live cells. MCF-7 cells were incubated in full growth media 24 hrs following transfection, starved of serum in DMEM+0.1% BSA overnight, and stimulated with 10 ng/mL EGF for the indicated times. Cell growth was monitored by the alamarBlue assay (Invitrogen).
To examine the effect of transfected CHKA2 on MCF-7 cell DNA synthesis, the Click-iT EdU Imaging assay (Invitrogen) was carried out. Expression plasmids encoding various forms of flag-tagged-CHKA2, or flag-tagged GFP were transfected into MCF-7 cells. The day after transfection, cells were cultured in PRF medium (Phenol Red-Free DMEM + 5% charcoal-stripped FBS) for 24 hrs and then incubated in fresh PRF medium with or without 50 ng/mL EGF for 15 hrs. Ten μM EdU was added and incubated for an additional 9 hrs. Cells were fixed with Buffered Formalde-Fresh (Thermo Fisher Scientific), stained with Click-iT EdU imaging reagents, mouse anti-flag antibody, and AlexaFluor-conjugated anti-mouse antibody (Invitrogen) and scored by immunofluorescence microscopy for anti-flag positive cells that were EdU positive.
In vitro choline kinase assay
Cell lysates were prepared one day after transfection of 293T cells with the indicated plasmid(s). Transfected MCF-7 cells were cultured for two days in PRF medium before cell lysates were prepared. In both cases, protein concentration was measured by the BCA assay, and samples used for further assay contained equal amounts of protein lysate. In vitro choline kinase activity was measured as described previously (Ishidate and Nakazawa 1992) with minor modifications. Aliquots of cell lysate were incubated in the reaction buffer containing 0.1M Tris HCl pH 8.0, 12 mM MgCl2, 10 mM ATP, [methyl-14C]-Choline chloride 0.25 mM (MP Biomedicals, Solon, OH), 1% NP-40 for 15 min at 37°C. Phosphocholine was isolated using AG 1-x8 Resin (Bio-Rad) as described in (Ishidate and Nakazawa 1992), and radioactivity was measured by scintillation counting. An appropriate lysate concentration for optimal linear activity was predetermined using extract expressing flag-CHKA2. Note that endogenous choline kinase activity was less than 1% of transfected choline kinase activity.
Protein purification and mass spectrometry identification of CHKA2 tyrosine phosphorylation
Flag-tagged CHKA2 constructs were transfected into 293T cells along with wt c-Src. Flag-tagged CHKA2 was purified following the method described in Grigera et al.(Grigera et al 2005). The sample was reduced with DDT, alkylated with iodoacetamide, and digested with ~0.5ug Arg-C (25%) and ~0.5ug Glu-C (25%) overnight at room temperature. After digestion, each sample was acidified to 5% with acetic acid, and 5% of each sample was loaded for LC-MS mass spectrometry analysis. The data were analyzed by the Sequest search algorithm against the CHKA2 protein sequence.
Statistical Analysis
Statistical differences between groups were determined by Student's t test. Values p <0.05 were taken as significant.
Supplementary Material
Acknowledgments
We thank Dr. Nicholas E. Sherman in the W.M. Keck Biomedical Mass Spectrometry Laboratory for analysis of CHKA2 phosphorylation sites, and Drs. Jill Slack-Davis and John DaSilva for their critical comments. We also thank members of Sarah J. Parsons' laboratory and the Women's Oncology Group in the University of Virginia Cancer Center for helpful discussions. This work was supported by NIH-NCI grant R01CA123037(S.J.P.).
This work was supported by NIH-NCI grant R01CA123037(S.J.P.).
Footnotes
Conflict of interest The authors declare no conflict of interest.
Supplementary information is available at Oncogene's website.
References
- Aoyama C, Liao H, Ishidate K. Structure and function of choline kinase isoforms in mammalian cells. Prog Lipid Res. 2004;43:266–281. doi: 10.1016/j.plipres.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Belsches-Jablonski AP, Biscardi JS, Peavy DR, Tice DA, Romney DA, Parsons SJ. Src family kinases and HER2 interactions in human breast cancer cell growth and survival. Oncogene. 2001;20:1465–1475. doi: 10.1038/sj.onc.1204205. [DOI] [PubMed] [Google Scholar]
- Biscardi JS, Belsches AP, Parsons SJ. Characterization of human epidermal growth factor receptor and c-Src interactions in human breast tumor cells. Mol Carcinog. 1998;21:261–272. doi: 10.1002/(sici)1098-2744(199804)21:4<261::aid-mc5>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem. 1999a;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- Biscardi JS, Tice DA, Parsons SJ. c-Src, receptor tyrosine kinases, and human cancer. Adv Cancer Res. 1999b;76:61–119. doi: 10.1016/s0065-230x(08)60774-5. [DOI] [PubMed] [Google Scholar]
- Biscardi JS, Ishizawar RC, Silva CM, Parsons SJ. Tyrosine kinase signalling in breast cancer: epidermal growth factor receptor and c-Src interactions in breast cancer. Breast Cancer Res. 2000;2:203–210. doi: 10.1186/bcr55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner JL, Demory ML, Silva C, Parsons SJ. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol Cell Biol. 2004;24:7059–7071. doi: 10.1128/MCB.24.16.7059-7071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua BT, Gallego-Ortega D, Ramirez de Molina A, Ullrich A, Lacal JC, Downward J. Regulation of Akt(ser473) phosphorylation by choline kinase in breast carcinoma cells. Mol Cancer. 2009;8:131. doi: 10.1186/1476-4598-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Carnero A, Dolfi F, Jimenez B, Lacal JC. Phosphorylcholine: a novel second messenger essential for mitogenic activity of growth factors. Oncogene. 1993;8:2959–2968. [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Demory ML, Boerner JL, Davidson R, Faust W, Miyake T, Lee I, et al. Epidermal growth factor receptor translocation to the mitochondria: Regulation and effect. J Biol Chem. 2009 doi: 10.1074/jbc.M109.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri M, Naramura M, Duan L, Chen J, Ortega-Cava C, Chen G, et al. Modeling breast cancer-associated c-Src and EGFR overexpression in human MECs: c-Src and EGFR cooperatively promote aberrant three-dimensional acinar structure and invasive behavior. Cancer Res. 2007;67:4164–4172. doi: 10.1158/0008-5472.CAN-06-2580. [DOI] [PubMed] [Google Scholar]
- Glunde K, Ackerstaff E, Mori N, Jacobs MA, Bhujwalla ZM. Choline phospholipid metabolism in cancer: consequences for molecular pharmaceutical interventions. Mol Pharm. 2006;3:496–506. doi: 10.1021/mp060067e. [DOI] [PubMed] [Google Scholar]
- Grigera PR, Jeffery ED, Martin KH, Shabanowitz J, Hunt DF, Parsons JT. FAK phosphorylation sites mapped by mass spectrometry. J Cell Sci. 2005;118:4931–4935. doi: 10.1242/jcs.02696. [DOI] [PubMed] [Google Scholar]
- Hackel PO, Gishizky M, Ullrich A. Mig-6 is a negative regulator of the epidermal growth factor receptor signal. Biol Chem. 2001;382:1649–1662. doi: 10.1515/BC.2001.200. [DOI] [PubMed] [Google Scholar]
- Hernando E, Sarmentero-Estrada J, Koppie T, Belda-Iniesta C, Ramirez de Molina V, Cejas P, et al. A critical role for choline kinase-alpha in the aggressiveness of bladder carcinomas. Oncogene. 2009;28:2425–2435. doi: 10.1038/onc.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hunter T, Cooper JA. Protein-tyrosine kinases. Annu Rev Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- Ishidate K, Nakazawa Y. Choline/ethanolamine kinase from rat kidney. Methods Enzymol. 1992;209:121–134. doi: 10.1016/0076-6879(92)09016-v. [DOI] [PubMed] [Google Scholar]
- Ishizawar R, Parsons SJ. c-Src and cooperating partners in human cancer. Cancer Cell. 2004;6:209–214. doi: 10.1016/j.ccr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Ishizawar RC, Miyake T, Parsons SJ. c-Src modulates ErbB2 and ErbB3 heterocomplex formation and function. Oncogene. 2007;26:3503–3510. doi: 10.1038/sj.onc.1210138. [DOI] [PubMed] [Google Scholar]
- Kloth MT, Laughlin KK, Biscardi JS, Boerner JL, Parsons SJ, Silva CM. STAT5b, a Mediator of Synergism between c-Src and the Epidermal Growth Factor Receptor. J Biol Chem. 2003;278:1671–1679. doi: 10.1074/jbc.M207289200. [DOI] [PubMed] [Google Scholar]
- Krishnamachary B, Glunde K, Wildes F, Mori N, Takagi T, Raman V, et al. Noninvasive detection of lentiviral-mediated choline kinase targeting in a human breast cancer xenograft. Cancer Res. 2009;69:3464–3471. doi: 10.1158/0008-5472.CAN-08-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Aoyama C, Ishidate K, Teraoka H. Deletion and alanine mutation analyses for the formation of active homo- or hetero-dimer complexes of mouse choline kinase-alpha and -beta. Biochim Biophys Acta. 2006;1761:111–120. doi: 10.1016/j.bbalip.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Maa MC, Leu TH, McCarley DJ, Schatzman RC, Parsons SJ. Potentiation of epidermal growth factor receptor-mediated oncogenesis by c-Src: implications for the etiology of multiple human cancers. Proc Natl Acad Sci U S A. 1995;92:6981–6985. doi: 10.1073/pnas.92.15.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malito E, Sekulic N, Too WC, Konrad M, Lavie A. Elucidation of human choline kinase crystal structures in complex with the products ADP or phosphocholine. J Mol Biol. 2006;364:136–151. doi: 10.1016/j.jmb.2006.08.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao W, Irby R, Coppola D, Fu L, Wloch M, Turner J, et al. Activation of c-Src by receptor tyrosine kinases in human colon cancer cells with high metastatic potential. Oncogene. 1997;15:3083–3090. doi: 10.1038/sj.onc.1201496. [DOI] [PubMed] [Google Scholar]
- Morse DL, Carroll D, Day S, Gray H, Sadarangani P, Murthi S, et al. Characterization of breast cancers and therapy response by MRS and quantitative gene expression profiling in the choline pathway. NMR Biomed. 2009;22:114–127. doi: 10.1002/nbm.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisach D, Gee P, Kent C, Xu Z. The crystal structure of choline kinase reveals a eukaryotic protein kinase fold. Structure. 2003;11:703–713. doi: 10.1016/s0969-2126(03)00094-7. [DOI] [PubMed] [Google Scholar]
- Qiu C, Tarrant MK, Boronina T, Longo PA, Kavran JM, Cole RN, et al. In vitro enzymatic characterization of near full length EGFR in activated and inhibited states. Biochemistry. 2009;48:6624–6632. doi: 10.1021/bi900755n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez de Molina A, Gutierrez R, Ramos MA, Silva JM, Silva J, Bonilla F, et al. Increased choline kinase activity in human breast carcinomas: clinical evidence for a potential novel antitumor strategy. Oncogene. 2002a;21:4317–4322. doi: 10.1038/sj.onc.1205556. [DOI] [PubMed] [Google Scholar]
- Ramirez de Molina A, Penalva V, Lucas L, Lacal JC. Regulation of choline kinase activity by Ras proteins involves Ral-GDS and PI3K. Oncogene. 2002b;21:937–946. doi: 10.1038/sj.onc.1205144. [DOI] [PubMed] [Google Scholar]
- Ramirez de Molina A, Rodriguez-Gonzalez A, Gutierrez R, Martinez-Pineiro L, Sanchez J, Bonilla F, et al. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochem Biophys Res Commun. 2002c;296:580–583. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- Ramirez de Molina A, Banez-Coronel M, Gutierrez R, Rodriguez-Gonzalez A, Olmeda D, Megias D, et al. Choline kinase activation is a critical requirement for the proliferation of primary human mammary epithelial cells and breast tumor progression. Cancer Res. 2004;64:6732–6739. doi: 10.1158/0008-5472.CAN-04-0489. [DOI] [PubMed] [Google Scholar]
- Ramirez de Molina A, Sarmentero-Estrada J, Belda-Iniesta C, Taron M, Ramirez de Molina V, Cejas P, et al. Expression of choline kinase alpha to predict outcome in patients with early-stage non-small-cell lung cancer: a retrospective study. Lancet Oncol. 2007;8:889–897. doi: 10.1016/S1470-2045(07)70279-6. [DOI] [PubMed] [Google Scholar]
- Stover DR, Becker M, Liebetanz J, Lydon NB. Src phosphorylation of the epidermal growth factor receptor at novel sites mediates receptor interaction with Src and P85 alpha. J Biol Chem. 1995;270:15591–15597. doi: 10.1074/jbc.270.26.15591. [DOI] [PubMed] [Google Scholar]
- Tice DA, Biscardi JS, Nickles AL, Parsons SJ. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc Natl Acad Sci U S A. 1999;96:1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T. Stimulation of phospholipid synthesis in HeLa cells by epidermal growth factor and insulin: activation of choline kinase and glycerophosphate acyltransferase. Biochim Biophys Acta. 1996;1304:89–104. doi: 10.1016/s0005-2760(96)00109-9. [DOI] [PubMed] [Google Scholar]
- Wright JD, Reuter CW, Weber MJ. Identification of sites on epidermal growth factor receptors which are phosphorylated by pp60src in vitro. Biochim Biophys Acta. 1996;1312:85–93. doi: 10.1016/0167-4889(96)00027-4. [DOI] [PubMed] [Google Scholar]
- Wu G, Vance DE. Choline kinase and its function. Biochem Cell Biol. 2010;88:559–564. doi: 10.1139/O09-160. [DOI] [PubMed] [Google Scholar]
- Yalcin A, Clem B, Makoni S, Clem A, Nelson K, Thornburg J, et al. Selective inhibition of choline kinase simultaneously attenuates MAPK and PI3K/AKT signaling. Oncogene. 2010;29:139–149. doi: 10.1038/onc.2009.317. [DOI] [PubMed] [Google Scholar]
- Yang S, Park K, Turkson J, Arteaga CL. Ligand-independent phosphorylation of Y869 (Y845) links mutant EGFR signaling to stat-mediated gene expression. Exp Cell Res. 2008;314:413–419. doi: 10.1016/j.yexcr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.