Abstract
Coordination of the primary defense mechanisms against pathogens relies on the appropriate expression of pathogen recognition receptors (PRRs) triggering the early release of effector molecules of the innate immune system. To analyze the impact of this system on the counteraction of infections of the mammary gland (mastitis), we characterized the bovine gene encoding the key PRR Toll-like receptor 9 (TLR9) and mapped its precise position on chromosome BTA22. The sequence information was used to establish real-time PCR quantification assays to measure the mRNA abundances of TLR9, TLR2, and TLR4 together with those of β-defensin 5 (BNBD5), an early bactericidal effector molecule of the innate system, in healthy and infected mammary glands. Mastitis strongly increased (4- to 13-fold) the mRNA abundances of all of these genes except TLR9. Slight subclinical infections already caused a substantial increase in the copy numbers, though they did so the least for TLR9. Induction was not systemic, since mRNA abundance was low in uninfected control quarters of the udder but high in the severely infected quarters of the same animal. The number of TLR2 copies correlated well with those of TLR4, indicating coordinated regulation of these two PRRs during infection of the udder. Their coordinated regulation explains our unexpected observation that pure Staphylococcus aureus infections caused a strong increase also in TLR4 mRNA abundance. In situ hybridizations revealed that BNBD5 is expressed predominantly in the mammary epithelial cells (MEC) of the infected gland. Our data therefore suggest a significant contribution of the innate immune system to counteract mastitis and attribute a prominent effector function to the MEC.
Infections of the mammary gland (mastitis) are the most costly single trait diseases in dairy cattle housing (48). They are caused by a broad spectrum of bacterial and fungal pathogens. Invading pathogens activate the immune defense in the udder, which is manifest as an increase of somatic cells in the milk. Mainly polymorph nuclear neutrophil granulocytes are recruited to the gland (38, 40), and their numbers increase 10- to 50-fold during the first few hours after infection (22, 23). The etiology of the pathogens influences the severity of the inflammation. Contagious pathogens, like Staphylococcus aureus or Streptococcus agalactiae, tend to result in chronic, subclinical mastitis while the environmental coliform bacteria very often cause acute, clinical infections of the gland (17). The reasons for these pathogen-related differences in the ability of the host's immune defense system to cope with infections of the mammary gland are unknown. They might reside in factors contributing to the innate branch of the immune system, since the Toll-like receptor (TLR) pathogen receptors and the β-defensin-type bactericidal effector molecules of the innate immune system are both known to display some pathogen specificity. However, there are no systematic reports available on the mammary gland-specific or mastitis-related expression of any key factor of the innate immune system.
TLRs signal the presence of pathogens to the mammalian host (1, 25). They are prominently expressed in antigen-presenting cells, notably macrophages and dendritic cells (27), and are activated by pathogen-associated molecular patterns (PAMPs). They stimulate the innate immune system (5) as well as the adaptive system (32). Ten different TLRs (TLR1 to TLR10) have been identified in mammals (16, 41). Different TLRs are stimulated by different PAMPs (19), in part related to the species of the pathogen. TLR4 but not TLR2, for example, together with CD14, recognizes the presence of lipoprotein A, a component of the lipopolysaccharide (LPS) from gram-negative bacteria (24). TLR2 is activated inside the phagosome by peptidoglycans from gram-positive bacteria (7, 52).
TLR 9 is of pivotal importance to the mounting of the full immune defense. It is activated by unmethylated CpG dinucleotides of bacterial DNA (4, 20). Unmethylated CpG dinucleotides are known to act as strong adjuvants for TH1 and cytotoxic-T-cell responses as well as to promote the maturation of dendritic cells (49). Moreover, the demonstrated lack of TH1 helper function in TLR9-null mice (20) indicates a severe lesion in these animals to stimulate the adaptive immune response. TLR9s from humans and mice display species-specific CpG motif recognition (4). Prior to this report, the DNA sequence and gene structure of the bovine TLR9-encoding gene were unknown.
β-Defensins constitute a structural group of the broad variety of bactericidal peptides (44, 57). They are expressed in epithelial cells (14, 35, 43) and also expressed abundantly in polymorphonuclear cells (PMNs). Expression of the human β-defensin-2-encoding gene is triggered via TLR-mediated NF-κB activation (6). Likewise, LPS stimulates expression of the bovine tracheal antibacterial peptide-encoding gene via activation of NF-κB (13). The bactericidal activities of some β-defensins differ against various pathogens, as is known for humans (29, 42) and for some members of the 13 β-defensin-encoding genes in cattle (45).
β-Defensin 1 has recently been detected in the mammary glands of women (26), conceivably protecting the gland against infections. However, while a β-defensin-like bactericidal activity was isolated from bovine teats more than 30 years ago (21), no recent account has been given on the significance of β-defensin activity to counteract mastitis inside the udder. Thus, we set out to examine whether mastitis activates the innate immune system in the udder, as indicated by enhanced expression of TLRs and β-defensins in the gland. We chose to determine the expression of TLR2 and TLR4 as the likely receptors for PAMPs from gram-positive and -negative bacteria, respectively. TLR9 should complement the selection of TLRs, since it acts as a strong adjuvant for the mounting of the immune response. Hence, we characterized the bovine TLR9 cDNA sequence and mapped the gene in cattle. We selected the gene encoding β-defensin 5 (BNBD5) as a candidate for effector molecules of the innate system, since we had previously isolated the mRNA of this gene from infected udders and sequenced the entire cDNA and encoding gene. A detailed characterization of this gene will be given elsewhere (C. Pitra, W. Yang, and H. M. Seyfert, unpublished data). Samples from healthy and infected udders were collected, the pathogens were typed, and three questions were asked. (i) Would mastitis increase the abundance of factors contributing to the innate immune defense system in the udder of the cow? (ii) Would different species groups of mastitis bacteria exert a differential influence upon TLR2 versus TLR4 expression during true mastitis infections? (iii) Would gram-positive bacteria also induce the expression of BNBD5, shown by in vitro tests to be active against gram-negative bacteria only (45)?
MATERIALS AND METHODS
Collection of tissues and typing of pathogens.
Udder tissue samples were collected aseptically from slaughtered cows within 5 to 10 min after killing. Glands were pathologically and anatomically categorized as inflamed (presumptively infected; n = 19) and healthy (presumptively uninfected; n = 19) glands. Washed udder skin was heat sterilized and incised with a disinfected (70% alcohol) knife. A piece of tissue (5 by 5 by 5 cm) was removed from a deeper area of the udder quarter. A small section (0.5 by 0.5 by 0.5 cm) thereof was preserved in RNA later (Qiagen, Hilden, Germany) for RNA analysis. Bacterial contamination (grade and bacterial species) was determined from two fresh slices (duplicates) of the tissue sample after a smear test with standard bacteriological techniques. The degree of bacterial contamination was evaluated after cultivation on blood agar (24 h, 37°C) and categorized as uninfected if no colonies formed (0 CFU). Slight, moderate, and severe infections were assumed if samples gave rise to 1 to 10, 11 to 30, and >31 CFU, respectively. Substances inhibitory to bacterial growth (e.g., residuals from antibiotics) were not detected in any sample.
Isolation of the bovine TLR9 cDNA and genomic copy of the encoding gene. (i) cDNA isolation.
We followed the general procedures for RNA extraction and reverse transcription (RT)-PCR set-up as previously described (30). One microgram of total RNA extracted from an infected udder was primed in reverse with an oligo(dT) nucleotide primer. Initially, two different segments of the bovine TLR9 were amplified with two different sets of primer pairs. The primers had been placed on sequence elements well conserved between mice and humans. The primers TLR9 forward (5′-GCCTTCCTACCCTGTGAGCT) and reverse (5′-GMGGTCAGATTGGCCAGGTC[M, C, or A]) amplified 644 bp from the 5′-terminal half of the message. Primers TLR9E4f (5′-TGTTTGTGCTGGCCCACACGGAC) and TLR9E4r (5′-GCAGGATCACCAGCACTACGA) amplified a 114-bp segment from the 3′ terminus of the cDNA. Subcloning and sequencing revealed that the products contained the expected copies from the bovine TLR9 message. Subsequently, the oligonucleotides TLR9f and TLR9E4r were used to amplify the large segment of the bovine TLR9 cDNA.
(ii) RACE experiments.
We used the GeneRacer kit (Invitrogen, Karlsruhe, Germany) to establish the 5′ end of the cDNA sequence and the Marathon random amplification of cDNA ends (RACE) kit from Stratagene (La Jolla, Calif.) for the 3′ end.
(iii) Isolation of genomic copies from BAC libraries.
Genomic copies of the bovine TLR9-encoding gene were retrieved from the bovine bacterial artificial chromosome (BAC) library BBI-B750 (58) of the RZPD Resource Center (Berlin, Germany) based on PCR screenings. Primers 5′-CACCATCTTCAACGACCTGA and 5′-TCTCCAGGGACACCAGACTC amplified a 122-bp segment of the bovine TLR9-encoding gene from these BAC stocks, and the isolates were stored as our BAC clones 820 and 821. BAC 820 was used for further analyses.
Chromosome assignment of the bovine TLR9-encoding gene. (i) Chromosome preparation and FISH.
Metaphase spreads were prepared from fibroblast cultures by standard cytogenetic techniques. Chromosomes were G banded and Giemsa stained. For fluorescence in situ hybridization (FISH), G-banded metaphase spreads were digitized with MacKtype karyotype analysis software (Perceptive Scientific Instruments, Inc., League City, Tex.). Chromosomes were karyotyped according to the International System for Chromosome Nomenclature of Domestic Bovids for bovine G-banded chromosomes (11). BAC clone 820 harboring the bovine TLR9-encoding gene was used as a hybridization probe. It was labeled with biotin-16-dUTP with a nick translation system kit (Invitrogen). The DNA mixture was prepared with 0.5 μg of the probe, 5 μg of bovine C0t-1 DNA (prepared according to the method of Young and Anderson [56]), and 20 μg of salmon sperm DNA. Bound probe molecules were detected with fluorescein isothiocyanate-labeled anti-biotin antibody (Roche, Basel, Switzerland). Images were captured with a charge-coupled device camera system (Photometrics sensys; Roper Scientific, Inc., Trenton, N.J.). The analysis of fluorescein isothiocyanate fluorescence signals on propidium iodide-colored chromosomes was performed with FISH software MacProbe (Perceptive Scientific Instruments, Inc.).
Hybrid cell mapping.
PCR typing of TLR9 was performed in a cattle-mouse somatic-hybrid cell (SHC) panel (55), and in a cattle-hamster 5,000-rad whole-genome radiation hybrid panel WGRH5000 (54), essentially as described previously (18). The WGRH typing experiment was performed twice, and data were scored independently. PCR products were scored as present (1), absent (0), or ambiguous (2). Two-point linkage analysis was done by using the software RHMAPPER (47) to assign the TLR9 locus to a chromosome and a nearest marker of the bovine WGRH5000 gene map (3).
Real-time RT-PCR quantitation of mRNA copy numbers.
We used real-time PCR with the LightCycler Instrument (Roche) to determine the relative copy numbers of the different mRNA moieties, essentially as described previously (31). All RNA samples were extracted with the TRIZOL reagent (Invitrogen) and purified further with the RNase-free DNase set (Qiagen), as prescribed in the kit. For cDNA synthesis, 1 μg of total RNA was primed in reverse with a mixture of four different reverse oligonucleotide primers (25 pM of each per assay), with one specific for each of the different mRNAs. After first-strand synthesis (H− Superscript; Invitrogen) and cDNA purification, aliquots equivalent to 12.5 ng of total RNA were distributed to four different vials. Next, gene-specific primer pairs were added and segments of the cDNA were amplified by using the Fast-Start Sybr Green I kit (Roche). Gains in fluorescence intensity were assayed at temperatures previously optimized for each gene (between 78 and 86°C). Relative copy numbers of the individual mRNA moieties were calculated from a dilution series (106 to 10 copies) of the respective cDNA subclones. All samples were measured twice from two independent cDNA preparations. We resolved all Light-Cycler amplificates after each run on agarose gels to monitor their quality.
The following primers have been used for cDNA priming of the BNBD5 mRNA primer cDE8r (5′-CGTGTGTTTGCCTTCTTTTACCA), and 149 bp was amplified by combining cDe8f (5′-CTGGATCACCTGCTCCTCGT) with gD8ex2r (5′-GGTGCCAATCTGTCTCATGTTG). The latter primer was optimized against all available bovine β-defensin cDNA sequences so that the 3′-terminal nucleotides do not attach to any other of these sequences. Both BNBD5 primers are located on different exons, separated by 1,480 bp of intron sequence. TLR2 mRNA was primed in reverse with b_TLR2rn (5′-GATCTTCCGCAGCTTACAGAAG), and the primer combination b_TLR2f (5′-CATTCCCTGGCAAGTGGATTATC) and b_TLR2r2 (5′-GGAATGGCCTTCTTGTCAATGG) amplified 195 bp. The TLR4 cDNA was primed with b_TLR4rn (5′-TGCACACATCATTTGCTCAGCT), and 237 bp was amplified with the primers b_TLR4f (5′-CACTCCCTCTCTAGCCTTCAG) and b_TLR4r (5′-CCTTGACCCACTGCAGGAAAC). The cDNA of the TLR9 mRNA was primed with TLR9r2 (5′-GCAAGTGGTGGATGCGGTTG), and 84 bp was amplified with primer pair TLR9fn-TLR9r3 (5′-ACTGGCTGTTCCTGAAGTCTGT and 5′-GAGATTAAGGAGAGGCTGGTGA, respectively). The authenticity of all products amplified by applying this protocol was verified by subcloning and sequencing of the PCR products.
In situ hybridization.
Ribonucleotide probes for the detection of the BNBD5-encoding messages were transcribed in vitro from a full-length cDNA cloned in opposite orientations into the vector pGEM-Teasy (Promega, Mannheim, Germany). We used the T7 polymerase to transcribe both sense and antisense probes and label them nonradioactively with the digoxigenin RNA labeling kit (Roche). Seven-micrometer sections were cut from paraformaldehyde (4%)-fixed and wax-embedded specimen. We followed the general procedures for the in situ hybridizations as described previously (33), with the following modifications. Slides were air dried after dewaxing, permeabilized with proteinase K (20 μg/ml), and acetylated. Next, the sections were denatured on a heat block (80°C, 2 min) to enhance the sensitivity of the hybridizations, cold shocked on ice, and subsequently maintained on a heating plate at 65°C. We applied (per section) 30 μl of hybridization mix containing 7 ng of the labeled probe, hybridized the mixture overnight at 58°C, washed it twice at 60°C (50% formamide, 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) and subsequently in 0.2× SSC at room temperature. Unbound probe molecules were removed by RNase digestion (37°C for 30 min with RNase A [10 μg] and T1 [100 U/ml] in 2× SSC). Signals were detected by applying anti-digoxigenin Fab fragments conjugated with the alkaline phosphatase and chromogenic substrates, as provided in the nucleic acid detection kit (Roche). Pilot experiments, applying all steps but excluding the probes, had revealed the necessity of including 5 mM levamisole in the substrate buffer for the development of the hybridization signals in mammary gland sections to prevent nonspecific background staining.
Statistical analyses.
All statistical analyses were conducted with the SigmaStat program package (Jandel Scientific, San Rafael, Calif.). Means were compared with the Mann-Whitney rank sum square test, and correlations were assessed with the Spearman rank order correlation.
Nucleotide sequence accession number.
Nucleotide sequences were deposited in National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/) and assigned the following accession numbers: TLR9 cDNA, NM_183081; TLR9 gene, AJ509824; BNBD5, A278799.
RESULTS
Characterization of the bovine TLR9 cDNA sequence and gene structure.
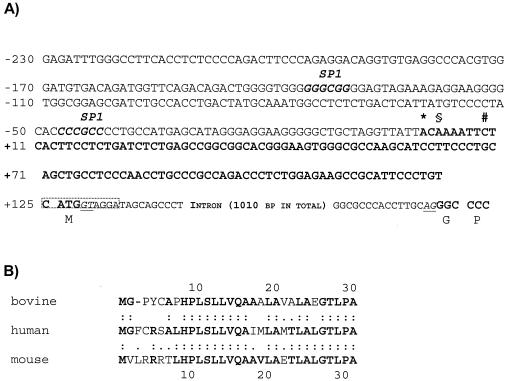
We have isolated and sequenced the gene encoding the entire bovine TLR9 and its cDNA equivalent. The cDNA sequence, as retrieved, comprises 123 bp of the 5′ untranslated region (UTR), an open reading frame of 3,090 bp, and 43 bp from the 3′ UTR. We conducted 5′ RACE experiments with RNA extracted from different tissues and cells to detect possible tissue-specific splice variants. However, the 5′ ends of the 19 different 5′ RACE clones sequenced from the different sources (mammary gland, brain, and activated macrophage suspensions) all clustered narrowly, and we define as +1 the 5′ nucleotide of our longest 5′ RACE clone (Fig. 1A). The 5′-adjacent sequence features two canonical SP-1 binding sites but no TATA box. The exact promoter definition awaits further analyses.
FIG. 1.
Nucleotide and amino acid sequences of the N terminus of the bovine TLR9-encoding gene. (A) 5′-Terminal sequence of the TLR9-encoding gene. The sequence is from BAC clone 820 and is also shown as retrieved from 5′-RACE clones (bold face letters). The most 5′-located nucleotides, as retrieved from the longest 5′-RACE clones, from various tissues are specified (*, mammary gland; §, brain; #, macrophages). Two SP-1 boxes are located in the presumed promoter sequence. An intron (splice donor and acceptor dinucleotides are underlined) separates the translational start codon from the rest of the coding sequence (codons are indicated). The sequence around the 5′ splice donor dinucleotide is precisely conserved between cattle, humans, and mice (boxed). (B) Comparison of the N-terminal amino acid sequence of TLR9 factors. The bovine signal peptide is shorter by 1 aa residue than those from humans and mice (:, identical residues; ., equivalent amino acid residues).
The architecture of the TLR9-encoding gene is conserved between cattle, humans, and mice. Sequence analysis of our BAC clone 820 reveals that exon 1 encodes the 5′ UTR and is 3′-terminated by the translational start codon ATG (Fig. 1A), similar to that in humans (16) and in mice (EMBL database file NW_000356). The intervening intron spans 1,010 bp in cattle but 1,215 bp in humans due to the insertion of an Alu repeat element (EMBL file NT_005986). Exon 2 encodes the entire body of the protein and the 3′ UTR. The 3′ UTR is conceivably longer than the short segment retrieved, but the adaptor-primers from the Marathon kit used for these experiments bound always to the GC-rich sequence element of the proximal 3′ UTR. No efforts were made to sequence further downstream on our BAC isolate 820.
The nucleotide sequence encoding the bovine TLR9 is 83.5 and 76.9% similar to the homologous genes from humans and mice, respectively. Conceptual translation of the bovine sequence shows that the bovine protein comprises 1,029 amino acid (aa) residues, and hence, is shorter by 3 aa residues than the homologous receptors from both other species. The entire protein sequence of the bovine receptor is 79.5 and 73.5% similar to those of humans and mice, respectively. However, the sequence of the N-terminal 7 aa of the TLR9 protein is divergent between all three species (Fig. 1B). Indeed, the signal peptide of the bovine receptor is shorter by 1 aa residue than that of the other two species.
The bovine TLR9-encoding gene is localized on BTA22.
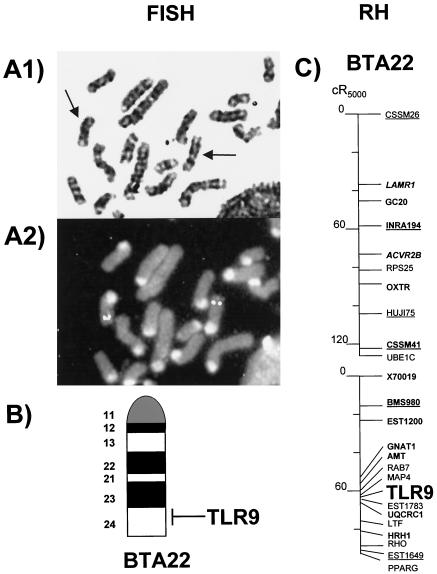
FISH with our BAC isolate 820 revealed that the TLR9-encoding gene is located on chromosome 22 (Fig. 2A). Using the G bands, we mapped TLR9 on band q24 (Fig. 2B). The assignment was confirmed by PCR typing on the SHC panel. TLR9 was assigned a position on BTA22 close to the reference marker HRH1, which had previously been mapped to the distal part of the chromosome (3). We found 97% concordance for the assigned gene in this assay, thus well above the widely accepted threshold value of 0.87 (9). PCR typing in the WGRH5000 panel was performed based on these SHC mapping results. Fourteen positive PCR signals in the 90 RH cell lines selected for PCR typing resulted in a retention frequency of 0.16 in an RHMAPPER two-point linkage analysis combined with the data of the cattle WGRH5000 gene map. This confirms the assignment of TLR9 to BTA22. TLR9 was placed between MAP4 and EST1649, with the strongest linkage to MAP4 (decimal log likelihood ratio score, 17.5) and a calculated distance of 3 cR5000 to MAP4 (Fig. 2C).
FIG. 2.
Mapping of TLR9 on cattle chromosome 22. (A) FISH of probe BAC 820 on a G-banded cattle metaphase spread prior to (A1, arrows specify chromosome 22) and after (A2) hybridization. Strong fluorescein signals can be seen in the telomeric region of chromosome BTA22. (B) TLR9 was assigned to position q24 of BTA22, according to the International System for Chromosome Nomenclature of Domestic Bovids 2000 for G-banded cattle chromosomes. (C) Assignment of TLR9 on the WGRH5000 map.
Infection of udder tissue enhances significantly the expression of BNBD5, TLR2, and TLR4 but not that of TLR9.
We wanted to examine the effect of mammary gland infection upon the abundance of the messages from these genes. Hence, samples were collected from pathologically and anatomically diseased (presumed infected) and healthy (presumed uninfected) glands. The bacteriologic typing of the latter samples revealed, however, that 9 of the 19 presumed uninfected glands gave rise to a few bacterial colonies in these evaluations, whereas all diseased quarters showed bacteriological contamination. Specifications of the individual samples and levels of expression of the respective genes are presented in Table 1.
TABLE 1.
Samples and mRNA copy numbers of the various genesa
| Animal no. | Comment | Pathogen(s) | Degree | No. of copies of:
|
|||
|---|---|---|---|---|---|---|---|
| BNBD5 | TLR2 | TLR4 | TLR9 | ||||
| 1 | Control | 0 | 30 | 65 | 41 | 43 | |
| 2 | Control | 0 | 72 | 999 | 2,167 | 366 | |
| 3 | Control | 0 | 95 | 253 | 449 | 469 | |
| 4 | Control | 0 | 288 | 1,666 | 3,176 | 2,653 | |
| 5 | Control | 0 | 20 | 2,373 | 1,435 | 381 | |
| 6 | Control | 0 | 124 | 1,292 | 3,959 | 1,204 | |
| 7 | Control | 0 | 12 | 14 | 10 | 19 | |
| 7 | S. aureus, A. pyogenes | 3 | 476 | 1,401 | 3,113 | 1,777 | |
| 8 | Control | 0 | 1,582 | 2,269 | 5,795 | 342 | |
| 8 | S. dysgalactiae | 1 | 1,916 | 6,797 | 8,348 | ND | |
| 9 | Control | 0 | 25 | 4,398 | 1,343 | 216 | |
| 9 | E. coli, A. pyogenes | 3 | 182 | 7,643 | 7,360 | 616 | |
| 10 | Control | 0 | 19 | 534 | 396 | 116 | |
| 10 | S. canis | 3 | 438 | 18,184 | 4,979 | 1,132 | |
| 11 | Control | E. coli | 1 | 37 | 593 | 2,296 | 394 |
| 12 | Control | S. aureus | 1 | 504 | 7,354 | 10,568 | 1,319 |
| 12 | Enter | 2 | 958 | 2,458 | 8,148 | 2,548 | |
| 13 | Control | S. aureus | 1 | 129 | 1,107 | 2,040 | 2,556 |
| 13 | S. aureus | 3 | 5,399 | 6,553 | 5,676 | 456 | |
| 14 | Control | Unspec | 1 | 3,151 | 4,754 | 4,905 | 4,254 |
| 14 | E. coli | 3 | 5,425 | 4,712 | 4,580 | 515 | |
| 15 | Control | Unspec | 1 | 311 | 6,348 | 7,194 | 691 |
| 15 | E. coli, A. pyogenes | 3 | 74 | 3,813 | 5,871 | 492 | |
| 16 | RF, control | S. uberis | 1 | 462 | 547 | 758 | 17 |
| 16 | RB, control | S. uberis | 1 | 1,510 | 2,783 | 8,919 | 270 |
| 16 | LF, infected | S. aureus | 3 | 4,160 | 7,149 | 16,039 | 420 |
| 17 | RF, control | Cory | 1 | 716 | 2,591 | 14,324 | 892 |
| 17 | RB, infected | Cory | 2 | 3,001 | 2,670 | 6,121 | 250 |
| 18 | LF, chronic SC | S. aureus | 2 | 121 | 10,919 | 5,889 | 175 |
| 18 | LB, C, infected | S. aureus | 3 | 6,617 | 75,311 | 63,230 | 1,608 |
| 18 | LB, A, infected | S. aureus | 3 | 2,022 | 21,722 | 17,299 | 362 |
| 19 | S. dysgalactiae | 2 | 71 | 152 | 294 | 239 | |
| 20 | S. aureus | 2 | 1,860 | 4,726 | 10,401 | 400 | |
| 21 | S. uberis | 3 | 29,383 | 13,859 | 7,877 | 209 | |
| 22 | A. pyogenes | 3 | 2,255 | 2,493 | 7,352 | 339 | |
| 23 | A. pyogenes | 3 | 2,262 | 3,094 | 7,325 | 247 | |
| 24 | S. uberis | 3 | 7,500 | 3,795 | 5,141 | 911 | |
| 25 | S. dysgalactiae | 3 | 1,861 | 3,985 | 5,787 | 647 | |
Abbreviations: LF, RF, and RB, left front, right front, and right back quarters of the udder, respectively; C and A, cisternal and alveolar area of the same udder quarter; unspec, unspecified bacteria; chronic SC, chronic subclinical infection; Cory, corynebacterium; Enter, enterococcal species; ND, not determined, due to consistently emerging false PCR products in this sample with these primers. Control, presumptive uninfected sample. Degree indicates the number of CFU derived from the samples (0, none; 1, <11; 2, 11 to 30; 3, >30).
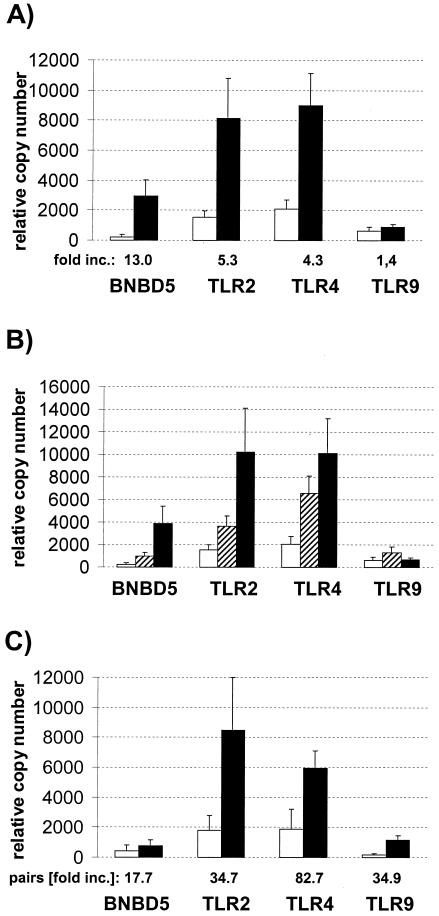
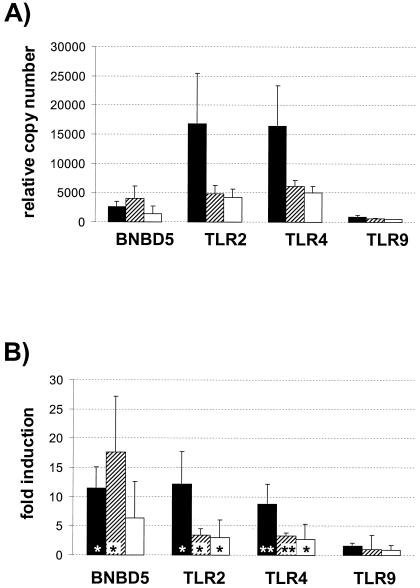
The average relative mRNA copy numbers of all four genes were lower in the truly uninfected glands than those measured in those samples containing bacteria (Fig. 3A). The average concentration of the BNBD5 messages was 13 times higher in the infected glands than in the uninfected samples. The abundance of TLR2 and TLR4 messages was also increased in the infected samples, about four- to fivefold. These differences are significant for BNBD5 and TLR2 (P, <0.02) and are highly significant for TLR4 (P, <0.001). The concentration of the TLR9 messages, on the other hand, was only slightly increased by mastitis, and the observed 1.4-fold increase in the abundance of the TLR9 messages in the infected samples is not statistically significant.
FIG. 3.
Relationship between health status and mRNA copy numbers. (A) Mean relative copy numbers from the indicated genes, as measured from proven uninfected (open columns; n = 10) or infected (filled columns; n = 28) quarters of mammary glands. Error bars indicate standard errors of the means. (B) Same as shown in panel A; however, separating the group of infected quarters into slight subclinical infections (hatched columns; n = 9) and moderate or severe infections (black columns; n = 19). (C) Differences in mRNA abundances between healthy control and infected quarters of the same animal (control) (open columns; n = 4). The average magnitude of increased concentration from pairwise comparisons of control and infected quarters of the same animal is given below the graph (pairs [fold increase {inc.}]).
Severity of infection correlates with increased abundance of messages for BNBD5, TLR2, and TLR4 but not for TLR9.
Slight infections already result in a substantial increase of the mRNA concentrations of the former three genes (Fig. 3B). Subgrouping the values of all samples with attributable bacterial counts into slightly infected materials (Table 1, degree 1) and pathologically and anatomically diseased (e.g., moderately or severely infected) (Table 1, degrees 2 and 3) reveals that the samples harboring only a few bacteria already had clearly higher copy numbers of these three genes than those found in the uninfected samples. The increased mRNA concentrations of the TLR2- and TLR4-encoding genes, as measured in the slightly infected samples, is statistically significant (P, <0.05), and that for BNBD5 is close to significance (P, 0.07). However, the average mRNA concentration of all three genes is further increased in the more severely infected glands. The average abundance of TLR9 messages would appear doubled in the slightly infected samples, but this increase is statistically insignificant.
Induction of these genes is not systemic but is confined to the infected quarter of the udder.
Nonsystemic induction of increased mRNA abundance in the udder becomes evident when comparing mRNA abundances in proven uninfected control quarters to those from the infected quarters of the same animal (Fig. 3C; Table 1, animals 7 to 10). Infection increases the mRNA copy numbers of all four genes, including TLR9, over the level detected in the proven uninfected quarters of the same gland. The extent of increase in the mRNA abundances, as associated with infection, becomes even clearer in pairwise comparisons, expressing the mRNA concentration of the infected quarter as a multiple of the level measured in the uninfected control quarter of the same animal. All four mRNAs, including TLR9, are 17- to ∼80-fold more abundant in the infected quarters than in the proven uninfected controls. However, the magnitude of this factor may be somewhat exaggerated, since its calculation depends heavily on the absolute very low copy numbers measured in the uninfected glands, which are eventually close to the background.
Coordinated increase of TLR2, TLR4, and BNBD5 mRNA abundance.
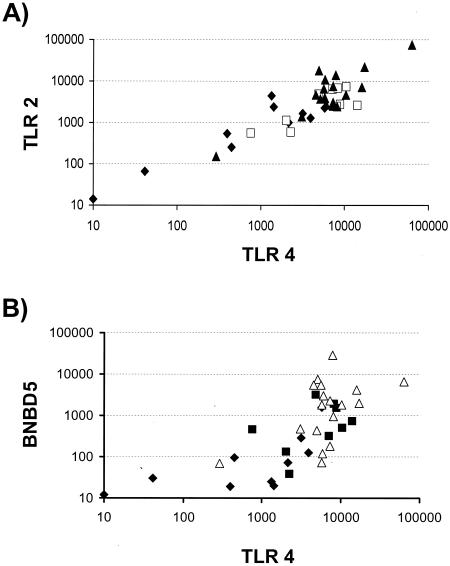
The reciprocal plot of the individual TLR2 versus TLR4 copy numbers measured from the same samples suggests a linear correlation between both values (Fig. 4A). A clear positive and significant correlation exists between all values of these two pathogen recognition receptors (r, 0.75; P, <0.001) and between those from any subgroups of data, as from the moderately and severely infected samples alone (r, 0.47; P, <0.05), slightly infected samples (r, 0.63; P, 0.058) or proven uninfected samples (r, 0.67; P, <0.05).
FIG. 4.
Correlated levels of TLR2, TLR4, and BNBD5 mRNA abundance. (A) Reciprocal plot of individual TLR2 and TLR4 copy numbers from healthy (carets), slight (squares), or moderately and severely (triangles) infected quarters. Filled symbols represent subgroups with P values of <0.05 as the level of significance for the correlation between both parameters. (B) Relationship between TLR4 and BNBD5 mRNA concentrations (symbols are as defined for panel A). A similar plot is obtained if the BNBD5 data are plotted against those for TLR2 (data not shown).
The concentrations of the BNBD5 messages correlate with those of TLR2 (r, 0.60;, P, <0.001) and TLR4 (r, 0.66; P, <0.001) (Fig. 4B). The statistical significance stems in both of these comparisons from the 19 uninfected or slightly infected samples (r, 0.64; P, <0.003; r, 0.79; P, <0.001) for TLR2 and TLR4, respectively. These correlations are statistically insignificant in the moderately and severely infected samples.
The concentrations of TLR9-encoding messages, on the other hand, are not correlated with any one of those from the other genes examined.
S. aureus increases strongly the abundances of all mRNAs examined except TLR9.
The bacteriologic evaluation of the infected quarters had revealed a variety of pathogenic bacteria. S. aureus was the sole pathogen in eight samples. These revealed much higher concentrations (8- to 12-fold) of the mRNAs from BNBD5, TLR2, and TLR4 than measured in the healthy quarters (Fig. 5). The level of TLR9 expression in these samples was almost similar (1.6-fold) to those in the controls. The rest of the infected samples were compiled into two other groups. One of these surmises all samples harboring all other bacteria (Streptococcus dysgalactiae, Streptococcus uberis, Streptococcus canis, Actinomyces pyogenes, and Corynebacterium) except Escherichia coli. Samples containing E. coli form the last group. BNBD5 abundance is greatly increased in both of the latter groups. All other pathogens stimulated TLR2 and TLR4 levels in the infected glands to a lesser extent than S. aureus. Levels of TLR9-encoding messages, however, are not significantly modulated by any of the pathogens. The values from three samples have not been incorporated into this comparison: the one containing enterococci and the two samples with unspecified bacteria (animals 12, 14, and 15) (Table 1).
FIG. 5.
Influence of pathogen species on mRNA abundance. (A) Relative copy numbers, as measured in samples infected with S. aureus (filled bars; n = 8), a spectrum of streptococcal and coryneform bacteria (hatched bars; n = 13), and those containing E. coli (white bars; n = 4, two of these had E. coli as the sole pathogen, and the others contained A. pyogenes as a second pathogen). (B) Magnitude of increased copy numbers in severely infected samples, expressed as multiples from the levels found in uninfected quarters (grouping of values is as in panel A) *, P < 0.05; **, P < 0.001.
BNBD5 is expressed in MEC.
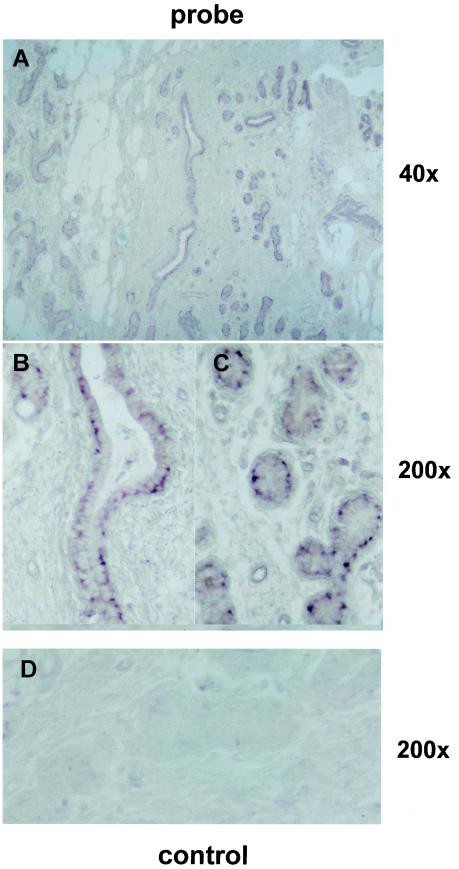
In situ hybridizations towards serial sections of infected mammary glands were used to identify the relevant cell type expressing the bactericidal effector gene BNBD5 within the gland. The antisense probe hybridized towards mammary epithelial cells (MEC) of alveoli and cells lining the milk ducts Fig. 6, whereas the sense control did not bind specifically to these cells. The low magnification overview shows that MEC are the predominant sites for the expression of this gene in the infected udder. Sections from proven uninfected glands did not reveal any specific hybridization signal (data not shown). These data together suggest that the pathogenic signal is transduced to MEC within the udder to stimulate the expression of this effector gene of the innate immune defense.
FIG.6.
BNBD5 is expressed in MEC and cells lining the milk ducts. Mammary gland sections of an udder were hybridized with either the antisense probe (A to C) or the sense control (D) of the BNBD5-encoding gene. The signal was developed for 20 h. The reddish label is found in MEC lining the milk ducts and in collapsed alveoli, and it is in its highest concentrations at basolateral locations inside the cells. The samples were taken from a slaughtered cow which had not been milked for 2 weeks, due to the infection. Parallel samples revealed high levels of BNBD5 in real-time RT-PCR quantification. Magnification, ×40 (A) and ×200 (B to D).
DISCUSSION
A TH1-driven secretion of cytokines and chemokines is necessary to mount a successful defense against invading pathogens of the udder (15). Bias towards a TH2-driven response results in malfunction of the immune response, as frequently encountered during early lactation and conceivably caused by the recent pregnancy (39). Activation of TLRs influences the polarization of the proinflammatory immune reaction towards either a TH1 or TH2 response (12), and the outcome of this polarization depends heavily upon the activity of subsets from dendritic cells (34). These subsets of dendritic cells, in turn, are characterized by different patterns of TLR expression (27). This background prompted us to examine the possible involvement of differential abundance of TLRs to counteract infection of the udder in the cow. Levels of TLR9 expression should be included in the survey due to its pivotal importance in mounting a TH1 response (20).
Characterization and mapping of the bovine TLR9-encoding gene.
We had to sequence the bovine gene encoding TLR9 to allow for the measurement of the respective mRNA abundance in real-time PCR assays. Moreover, characterization of the TLR9 gene from the bovine species is a necessary prerequisite for its eventual use in augmenting the strength of the immune response in cattle, since relevant PAMPs for TLR9 are species specific (4). Our comparison of the protein sequences of TLR9 factors from various species reveals considerable heterogeneity of the extreme N terminus. N-terminal heterogeneity of the TLR9 factors might be relevant for the species specificity of TLR9 signaling, since it was shown that the N-terminal 58 aa residues are indispensable for TLR9 function (50). Assignment of TLR9 to the bovine chromosome 22 and its fine mapping provides the basis for a detailed evaluation of an eventual trait-related genetic variance as associated with this gene. Regarding mastitis, no quantitative trait loci for clinical mastitis have been found on BTA 22 in genome-wide scans (28), though such studies are rare. Earlier studies, however, had shown a quantitative trait locus on BTA22 for the somatic cell scores trait on this chromosome (2). This trait is also related to udder health (46). Our mapping information allows for an efficient examination of the contribution of TLR9 to either of these traits.
Influence of mastitis on the expression of the four genes.
Our key observation is that mastitis increases the abundances of messages encoding the pathogen-specific receptors TLR2 and TLR4 in the udder together with the bactericidal peptide BNBD5. While the demonstration of enhanced synthesis of the encoded proteins awaits the establishment of suitable antibodies against them, our data suggest conclusively that the innate branch of the immune system contributes significantly to the immune surveillance of the mammary gland and very probably, to the counteraction of mastitis.
Most of the samples (except those from animals 16, 17, and 18) were collected from a commercial slaughterhouse, irrespective of breed and lactation status. Hence, we have no records regarding the clinical history of the animals or other parameters reflecting the degree of inflammation or progressiveness of mastitis in the respective udder quarters, e.g., cell counts of somatic cells in the milk. However, all udder quarters scored on the basis of bacterial counts as moderately or severely infected had shown pathological alterations of the tissue, indicating ongoing mastitis at the time of sampling. Somewhat unclear, however, is the degree of inflammation in those nine presumptively uninfected control udders, whose samples gave rise to a few CFU. While we cannot exclude bacterial contamination during sampling, it is obvious that some of these revealed considerably higher expression of BNBD5, TLR2, and TLR4 and hence may have been inflamed at the time of sampling (Table 1, degree 1; Fig. 3B). Clearly, a decisive analysis of the interdependence between the onset of inflammation and increased abundance of the factors of the innate immune system inside the udder requires a different experimental approach.
We target the MEC as a highly relevant effector cell of the immune surveillance in the mammary gland of the cow, since the MEC is the predominant source of β-defensin 5 expression in the infected gland. This settles for the cow ambiguities which arose in humans concerning the role of inducible defensin expression in mammary glands. While the constitutively active human hBD-1-encoding gene was found to be expressed in the human MEC (51), the authors failed to find expression of the inducible hBD-2 gene in their mammary samples. Our data show that the bovine BNBD5 gene is expressed in MEC and that expression of this gene is induced by pathogenic bacteria. Hence, MEC are highly relevant effector cells of the innate immune defense for the cow.
The correlated increase in the abundance of TLR2 and TLR4 messages suggests that expressions of these two receptors are regulated in concert. Both receptors are known to form eventually heterodimers (37), supporting our observation. It has recently been shown that their different agonists (peptidoglycan and LPS, respectively) elicit different effector functions in macrophages (8, 36), suggesting a possible differential pathogen-specific effect upon their regulation. Our data, however, indicate clearly that they are regulated alike during the complex events of mounting the immune response in the udder.
The cell types expressing the TLRs in the mammary gland await further analyses. The sensitivity of our in situ hybridizations was not sufficient to detect these messages in our mammary gland sections with this method. Conceivably, a substantial proportion of the increased abundance of these mRNAs may be attributed to PMNs which are copiously recruited into the infected udder quarter as a consequence of infection (23, 38). Assuming coordinated synthesis of these two TLRs in PMNs could explain the unexpected result that pure S. aureus infections increase the TLR4 mRNA abundance in the infected udder. Otherwise, it might seem paradoxical that PAMPs from a gram-positive pathogen increase the abundance of TLR4, the proven receptor for lipid A, a relevant PAMP from gram-negative bacteria (24). On the other hand, it would not be surprising to find TLR2 and TLR4 expressed in MEC as well. TLR2 was shown to drive hBD2 expression in model cells (6), and inflammation-induced expression of TLR2 and TLR4 has been demonstrated in renal epithelial cells (53). Given our demonstration that MEC express BNBD5, these cells might well harbor the entire cascade of factors necessary to transduce the signal from the pathogen to the bactericidal effector gene.
TLR9 abundance in the mammary gland is regulated differently from the other two TLRs. We did not observe any indication for coordinated expression of this pathogen-specific receptor neither with other TLRs nor the BNBD5-encoding gene. The insignificant small extent of mastitis-related increase in the abundance of this mRNA moiety in the udder together with the lacking correlation of the TLR9 mRNA abundance with any of the other parameters examined suggests that this receptor is expressed in a different compartment of the udder. Mastitis does not enhance the expression of this factor nor does it recruit such cells into the gland expressing it in high levels. TLR9 is known to be expressed in professional antigen-presenting cells, such as dendritic cells (27) and immune cell-rich tissues (10).
Acknowledgments
We thank Liz Glass and Kirsty McGuire of the Roslin Institute (Edinburgh, United Kingdom) for providing the RNA from activated macrophages. We are grateful for the dedicated technical assistance of A. Deike and K. Oden.
This work was supported by the Deutsche Forschungsgemeinschaft (Se 326/12-1) and the Research Council of New Zealand.
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. [DOI] [PubMed] [Google Scholar]
- 2.Ashwell, M. S., C. E. Rexroad, Jr., R. H. Miller, and P. M. VanRaden. 1996. Mapping economic trait loci for somatic cell score in Holstein cattle using microsatellite markers and selective genotyping. Anim. Genet. 27:235-242. [DOI] [PubMed] [Google Scholar]
- 3.Band, M. R., J. H. Larson, M. Rebeiz, C. A. Green, D. W. Heyen, J. Donovan, R. Windish, C. Steining, P. Mahyuddin, J. E. Womack, and H. A. Lewin. 2000. An ordered comparative map of the cattle and human genomes. Genome Res. 10:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, S., C. J. Kirschning, H. Hacker, V. Redecke, S. Hausmann, S. Akira, H. Wagner, and G. B. Lipford. 2001. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA 98:9237-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendelac, A., and D. T. Fearon. 2000. Innate immunity: receptors and effectors of innate immunity. Curr. Opin. Immunol. 12:11-12. [Google Scholar]
- 6.Birchler, T., R. Seibl, K. Buchner, S. Loeliger, R. Seger, J. P. Hossle, A. Aguzzi, and R. P. Lauener. 2001. Human Toll-like receptor 2 mediates induction of the antimicrobial peptide human beta-defensin 2 in response to bacterial lipoprotein. Eur. J. Immunol. 31:3131-3137. [DOI] [PubMed] [Google Scholar]
- 7.Campos, M. A., I. C. Almeida, O. Takeuchi, S. Akira, E. P. Valente, D. O. Procopio, L. R. Travassos, J. A. Smith, D. T. Golenbock, and R. T. Gazzinelli. 2001. Activation of Toll-like receptor-2 by glycosylphosphatidylinositol anchors from a protozoan parasite. J. Immunol. 167:416-423. [DOI] [PubMed] [Google Scholar]
- 8.Carl, V. S., K. Brown-Steinke, M. J. Nicklin, and M. F. Smith, Jr. 2002. Toll-like receptor 2 and 4 (TLR2 and TLR4) agonists differentially regulate secretory interleukin-1 receptor antagonist gene expression in macrophages. J. Biol. Chem. 277:17448-17456. [DOI] [PubMed] [Google Scholar]
- 9.Chevalet, C., and F. Corpet. 1986. Statistical decision rules concerning synteny or independence between markers. Cytogenet. Cell Genet. 43:132-139. [DOI] [PubMed] [Google Scholar]
- 10.Chuang, T. H., and R. J. Ulevitch. 2000. Cloning and characterization of a sub-family of human Toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur. Cytokine Netw. 11:372-378. [PubMed] [Google Scholar]
- 11.Cribiu, E. P., D. Di Berardino, G. P. Di Meo, A. Eggen, D. S. Gallagher, I. Gustavsson, H. Hayes, L. Iannuzzi, C. P. Popescu, J. Rubes, S. Schmutz, G. Stranzinger, A. Vaiman, and J. Womack. 2001. International System for Chromosome Nomenclature of Domestic Bovids (ISCNDB 2000). Cytogenet. Cell Genet. 92:283-299. [DOI] [PubMed] [Google Scholar]
- 12.Dabbagh, K., and D. B. Lewis. 2003. Toll-like receptors and T-helper-1/T-helper-2 responses. Curr. Opin. Infect. Dis. 16:199-204. [DOI] [PubMed] [Google Scholar]
- 13.Diamond, G., V. Kaiser, J. Rhodes, J. P. Russell, and C. L. Bevins. 2000. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect. Immun. 68:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond, G., M. Zasloff, H. Eck, M. Brasseur, W. L. Maloy, and C. L. Bevins. 1991. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc. Natl. Acad. Sci. USA 88:3952-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dosogne, H., F. Vangroenweghe, and C. Burvenich. 2002. Potential mechanism of action of J5 vaccine in protection against severe bovine coliform mastitis. Vet. Res. 33:1-12. [DOI] [PubMed] [Google Scholar]
- 16.Du, X., A. Poltorak, Y. Wei, and B. Beutler. 2000. Three novel mammalian toll-like receptors: gene structure, expression, and evolution. Eur. Cytokine Netw. 11:362-371. [PubMed] [Google Scholar]
- 17.Erskine, R. J., R. J. Eberhart, L. J. Hutchinson, S. B. Spencer, and M. A. Campbell. 1988. Incidence and types of clinical mastitis in dairy herds with high and low somatic cell counts. J. Am. Vet. Med. Assoc. 192:761-765. [PubMed] [Google Scholar]
- 18.Goldammer, T., S. R. Kata, R. M. Brunner, U. Dorroch, H. Sanftleben, M. Schwerin, and J. E. Womack. 2002. A comparative radiation hybrid map of bovine chromosome 18 and homologous chromosomes in human and mice. Proc. Natl. Acad. Sci. USA 99:2106-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 20.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 21.Hibbitt, K. G., J. Brownlie, and C. B. Cole. 1971. The antimicrobial activity of cationic proteins isolated from the cells in bulk milk samples. J. Hyg. (London) 69:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, A. W. 1981. Factors influencing the outcome of Escherichia coli mastitis in the dairy cow. Res. Vet. Sci. 31:107-112. [PubMed] [Google Scholar]
- 23.Hill, A. W., A. L. Shears, and K. G. Hibbitt. 1979. The pathogenesis of experimental Escherichia coli mastitis in newly calved dairy cows. Res. Vet. Sci. 26:97-101. [PubMed] [Google Scholar]
- 24.Hirschfeld, M., Y. Ma, J. H. Weis, S. N. Vogel, and J. J. Weis. 2000. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J. Immunol. 165:618-622. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, and R. A. Ezekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 284:1313-1318. [DOI] [PubMed] [Google Scholar]
- 26.Jia, H. P., T. Starner, M. Ackermann, P. Kirby, B. F. Tack, and P. B. J. McCray. 2001. Abundant human beta-defensin-1 expression in milk and mammary gland epithelium. J. Pediatr. 138:109-112. [DOI] [PubMed] [Google Scholar]
- 27.Kadowaki, N., S. Ho, S. Antonenko, R. W. Malefyt, R. A. Kastelein, F. Bazan, and Y. J. Liu. 2001. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 194:863-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klungland, H., A. Sabry, B. Heringstad, H. G. Olsen, H. Gomez-Raya, D. I. Vage, I. Olsaker, J. Odegard, G. Klemetsdal, N. Schulman, J. Vikki, J. Ruane, M. Aasland, K. Ronningen, and S. Lien. 2001. Quantitative trait loci affecting clinical mastitis and somatic cell count in dairy cattle. Mamm. Genome 12:837-842. [DOI] [PubMed] [Google Scholar]
- 29.Liu, A. Y., D. Destoumieux, A. V. Wong, C. H. Park, E. V. Valore, L. Liu, and T. Ganz. 2002. Human beta-defensin-2 production in keratinocytes is regulated by interleukin-1, bacteria, and the state of differentiation. J. Investig. Dermatol. 118:275-281. [DOI] [PubMed] [Google Scholar]
- 30.Mao, J., S. Marcos, S. K. Davis, J. Burzlaff, and H. M. Seyfert. 2001. Genomic distribution of three promoters of the bovine gene encoding acetyl-CoA carboxylase alpha and evidence that the nutritionally regulated promoter I contains a repressive element different from that in rat. Biochem. J. 358:127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao, J., A. J. Molenaar, T. T. Wheeler, and H.-M. Seyfert. 2002. Stat5 binding contributes to lactational stimulation of promoter III expressing the bovine acetyl-CoA-carboxylase α-encoding gene in the mammary gland. J. Mol. Endocrinol. 29:73-88. [DOI] [PubMed] [Google Scholar]
- 32.Medzhitov, R., P. Preston-Hurlburt, and C. A. Janeway, Jr. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394-397. [DOI] [PubMed] [Google Scholar]
- 33.Molenaar, A. J., S. R. Davis, and R. J. Wilkins. 1992. Expression of α-lactalbumin, α-S1-casein, and lactoferrin genes is heterogeneous in sheep and cattle mammary tissue. J. Histochem. Cytochem. 40:611-618. [DOI] [PubMed] [Google Scholar]
- 34.Moser, M., and K. M. Murphy. 2000. Dendritic cell regulation of TH1-TH2 development. Nat. Immunol. 1:199-205. [DOI] [PubMed] [Google Scholar]
- 35.O′Neil, D. A., E. M. Porter, D. Elewaut, G. M. Anderson, L. Eckmann, T. Ganz, and M. F. Kagnoff. 1999. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163:6718-6724. [PubMed] [Google Scholar]
- 36.O′Neill, L. A. J. 2002. Toll-like receptor signal transduction and the tailoring of innate immunity: a role for Mal? Trends Immunol. 23:296-300. [DOI] [PubMed] [Google Scholar]
- 37.Ozinsky, A., D. M. Underhill, J. D. Fontenot, A. M. Hajjar, K. D. Smith, C. B. Wilson, L. Schroeder, and A. Aderem. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc. Natl. Acad. Sci. USA 97:13766-13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paape, M. J., K. Shafer-Weaver, A. V. Capuco, K. Van Oostveldt, and C. Burvenich. 2000. Immune surveillance of mammary tissue by phagocytic cells. Adv. Exp. Med. Biol. 480:259-277. [DOI] [PubMed] [Google Scholar]
- 39.Raghupathy, R. 1997. Th1-type immunity is incompatible with successful pregnancy. Immunol. Today 18:478-482. [DOI] [PubMed] [Google Scholar]
- 40.Riollet, C., P. Rainard, and B. Poutrel. 2000. Cells and cytokines in inflammatory secretions of bovine mammary gland. Adv. Exp. Med. Biol. 480:247-258. [DOI] [PubMed] [Google Scholar]
- 41.Rock, F. L., G. Hardiman, J. C. Timans, R. A. Kastelein, and J. F. Bazan. 1998. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA 95:588-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schibli, D. J., H. N. Hunter, V. Aseyev, T. D. Starner, J. M. Wiencek, P. B. McCray, Jr., B. F. Tack, and H. J. Vogel. 2002. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J. Biol. Chem. 277:8279-8289. [DOI] [PubMed] [Google Scholar]
- 43.Schonwetter, B. S., E. D. Stolzenberg, and M. A. Zasloff. 1995. Epithelial antibiotics induced at sites of inflammation. Science 267:1645-1648. [DOI] [PubMed] [Google Scholar]
- 44.Schroder, J. M. 1999. Epithelial antimicrobial peptides: innate local host response elements. Cell. Mol. Life Sci. 56:32-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selstedt, M. E., Y.-Q. Tang, W. L. Morris, P. A. McGuire, M. J. Novotny, A. H. Henschen, and J. S. Cullor. 1993. Purification, primary structures, and antibacterial activities of β-defensins, a new family of antibacterial peptides from bovine neutrophils. J. Biol. Chem. 268:6641-6648. [PubMed] [Google Scholar]
- 46.Shook, G. E. 1993. Genetic improvement of mastitis through selection on somatic cell count. Vet. Clin. N. Am. 9:563-577. [DOI] [PubMed] [Google Scholar]
- 47.Slonim, D., L. Kruglyak, L. Stein, and E. Lander. 1997. Building human genome maps with radiation hybrids. J. Comput. Biol. 4:487-504. [DOI] [PubMed] [Google Scholar]
- 48.Sordillo, L. M., and K. L. Streicher. 2002. Mammary gland immunity and mastitis susceptibility. J. Mammary Gland Biol. Neoplasia 7:135-146. [DOI] [PubMed] [Google Scholar]
- 49.Sparwasser, T., E. S. Koch, R. M. Vabulas, K. Heeg, G. B. Lipford, J. W. Ellwart, and H. Wagner. 1998. Bacterial DNA and immunostimulatory CpG oligonucleotides trigger maturation and activation of murine dendritic cells. Eur. J. Immunol. 28:2045-2054. [DOI] [PubMed] [Google Scholar]
- 50.Takeshita, F., C. A. Leifer, I. Gursel, K. J. Ishii, S. Takeshita, M. Gursel, and D. M. Klinman. 2001. Cutting edge: role of Toll-like receptor 9 in CpG DNA-induced activation of human cells. J. Immunol. 167:3555-3558. [DOI] [PubMed] [Google Scholar]
- 51.Tunzi, C. R., P. A. Harper, B. Bar-Oz, E. V. Valore, J. L. Semple, J. Watson-MacDonell, T. Ganz, and S. Ito. 2000. Beta-defensin expression in human mammary gland epithelia. Pediatr. Res. 48:30-35. [DOI] [PubMed] [Google Scholar]
- 52.Underhill, D. M., A. Ozinsky, A. M. Hajjar, A. Stevens, C. B. Wilson, M. Bassetti, and A. Aderem. 1999. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 401:811-815. [DOI] [PubMed] [Google Scholar]
- 53.Wolfs, T. G., W. A. Buurman, A. van Schadewijk, B. de Vries, M. A. Daemen, P. S. Hiemstra, and C. van't Veer. 2002. In vivo expression of Toll-like receptor 2 and 4 by renal epithelial cells: IFN-gamma and TNF-alpha mediated up-regulation during inflammation. J. Immunol. 168:1286-1293. [DOI] [PubMed] [Google Scholar]
- 54.Womack, J. E., J. S. Johnson, E. K. Owens, C. E. Rexroad III, J. Schlapfer, and Y. P. Yang. 1997. A whole-genome radiation hybrid panel for bovine gene mapping. Mamm. Genome 8:854-856. [DOI] [PubMed] [Google Scholar]
- 55.Womack, J. E., and Y. D. Moll. 1986. Gene map of the cow: conservation of linkage with mouse and man. J. Hered. 77:2-7. [DOI] [PubMed] [Google Scholar]
- 56.Young, B., and M. L. M. Anderson. 1985. Quantitative analysis of solution hybridisation, p. 47-73. In B. D. Hames and S. J. Higgins (ed.), Nucleic acid hybridisation—a practical approach. IRL Press, Oxford, United Kingdom.
- 57.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, B., J. A. Smith, S. M. Tracey, B. A. Konfortov, K. Welzel, L. C. Schalkwyk, H. Lehrach, S. Kollers, J. Masabanda, J. Buitkamp, R. Fries, J. L. Williams, and J. R. Miller. 1999. A 5x genome coverage bovine BAC library: production, characterization, and distribution. Mamm. Genome 10:706-709. [DOI] [PubMed] [Google Scholar]