Abstract
Properly coordinated defense signaling networks are critical for the fitness of plants. One hub of the defense networks is centered on salicylic acid (SA), which plays a key role in activating disease resistance in plants. However, while a number of genes are known to affect SA-mediated defense, relatively little is known about how these gene interact genetically with each other. Here we exploited the unique defense-sensitized Arabidopsis mutant accelerated cell death (acd) 6-1 to dissect functional relationships among key components in the SA hub. We show that while enhanced disease susceptibility (eds) 1-2 and phytoalexin deficient (pad) 4-1 suppressed acd6-1–conferred small size, cell death, and defense phenotypes, a combination of these two mutations did not incur additive suppression. This suggests that EDS1 and PAD4 act in the same signaling pathway. To further evaluate genetic interactions among SA regulators, we constructed 10 pairwise crosses in the acd6-1 background among mutants defective in: SA INDUCTION-DEFICIENT 2 for SA biosynthesis; AGD2-LIKE DEFENSE 1, EDS5, and PAD4 for SA accumulation; and NONEXPRESSOR OF PR GENES 1 for SA signaling. Systematic analysis of the triple mutants based on their suppression of acd6-1–conferred phenotypes revealed complex and interactive genetic relationships among the tested SA genes. Our results suggest a more comprehensive view of the gene networks governing SA function and provide a framework for further interrogation of the important roles of SA and possibly other signaling molecules in regulating plant disease resistance.
IN response to pathogen infection, plants can activate different layers of defense responses and undergo global gene expression reprogramming (Maleck et al. 2000; Tao et al. 2003; Katagiri 2004). A major challenge of the postgenomic era is to identify genes that control plant innate immunity and to elucidate how they are organized into networks to orchestrate host defense responses.
One key hub in plant defense signaling networks is centered on the small phenolic molecule salicylic acid (SA). SA is important for basal defense, resistance protein-mediated defense, and systemic acquired resistance (Hammond-Kosack and Jones 1996; Ryals et al. 1996; Tsuda et al. 2008). The SA hub of Arabidopsis includes many genes, which can be further grouped into three types on the basis of how they affect SA-mediated defense (Lu 2009). Type I SA genes encode enzymes that are directly involved in SA biosynthesis. One example is SA INDUCTION-DEFICIENT 2/ENHANCED DISEASE SUSCEPTIBILITY 16 (SID2/EDS16), which encodes isochorismate synthase contributing to bulk SA biosynthesis (Wildermuth et al. 2001). Type II SA genes encode proteins that do not act directly as SA biosynthetic enzymes. Mutations in these genes lead to partially compromised SA accumulation and enhanced disease susceptibility to pathogen infection, which can be rescued by exogenous SA treatment. The precise mechanism of action for each type II SA gene, however, still remains to be resolved. Examples of type II SA genes include ACCELERATED CELL DEATH 6 (ACD6), AGD2-LIKE DEFENSE 1 (ALD1), EDS1, PHYTOALEXIN DEFICIENT 4 (PAD4), SID1/EDS5, HOPW1-1-INTERACTING 3/AVRPPHB SUSCEPTIBLE 3/GH3-LIKE DEFENSE GENE 1, and the MODIFIER OF SNC1 genes (Falk et al. 1999; Jirage et al. 1999; Nawrath et al. 2002; Lu et al. 2003; Song et al. 2004b; Palma et al. 2005, 2007; Zhang et al. 2005; Zhang and Li 2005; Goritschnig et al. 2007; Jagadeeswaran et al. 2007; Lee et al. 2007; Nobuta et al. 2007). Type III SA genes act downstream of SA accumulation. The best-characterized type III SA gene is NONEXPRESSOR OF PR GENES 1 (NPR1), which is the major SA signal transducer (Cao et al. 1997; Ryals et al. 1997; Shah et al. 1997; Dong 2004). Enhanced disease susceptibility of npr1 mutants to pathogen infection cannot be rescued by SA treatment.
Increasing evidence suggests that defense signaling networks are complex, involving crosstalk to many other signaling pathways (Feys and Parker 2000; Kunkel and Brooks 2002; Wang et al. 2007; Koornneef and Pieterse 2008; De Torres Zabala et al. 2009) and to plant development (Martinez et al. 2004; Endo et al. 2009; Wang et al. 2011a). A variety of strategies are used to interrogate the topology of defense networks. On the basis of global gene expression profiling, microarray studies have revealed some of the hierarchical structure of components in the SA hub. These studies also showed there are both positive and negative interactions between components in the SA hub and those in ethylene and/or jasmonic acid signaling pathways (Wang et al. 2008; Tsuda et al. 2009; Sato et al. 2010). Analysis of protein complexes has also added further details to defense networks. For instance, type II SA regulators EDS1 and PAD4 were shown to interact physically (Feys et al. 2001), suggesting that these two proteins function in the same pathway.
An alternative approach to study defense networks is through analyzing mutants with two or more signaling components having been knocked out. This is a classical genetic way to assign genes to specific functional groups and has been widely used in model organisms, such as yeast and Escherichia coli, to understand gene functions and the architecture of signaling networks (Collins et al. 2006, 2007; Roguev et al. 2007; Breslow et al. 2008; Typas et al. 2008). Arabidopsis is also a premier system for this type of genetic analysis. However, when a mutation in one defense gene already leads to enhanced disease susceptibility, additional susceptible phenotypes caused by mutations in two or more defense genes can be difficult to detect on the basis of standard disease assays. Therefore, more sensitive methods should be developed to assess the functional relationships between certain components on defense networks.
A small Arabidopsis mutant acd6-1 is being used to develop one such method and has already revealed new insights into the SA signaling networks (Song et al. 2004b; Lu et al. 2009). ACD6 is a type II SA regulator that was shown to be a major determinant of fitness in Arabidopsis (Lu et al. 2003; Todesco et al. 2010). acd6-1 is a gain-of-function mutant that demonstrates constitutive defense, severe cell death, and extreme dwarfism (Rate et al. 1999; Vanacker et al. 2001; Lu et al. 2003). The cell death and dwarf phenotypes are sensitized to the change of defense levels in acd6-1. We have taken advantage of this unique feature of acd6-1 in a genetic analysis to understand functional relationships between several SA regulators and in a suppressor screen to identify novel defense genes (Song et al. 2004b; Lu et al. 2009; Wang et al. 2011a,b).
In this article, we exploited acd6-1 in a systematic dissection of functional relationships among components in the SA hub. We showed that while eds1-2 and pad4-1 individually suppressed acd6-1–conferred phenotypes, a combination of these two mutations did not incur additional suppression. Thus, we provided direct genetic evidence to show that EDS1 and PAD4 act in the same signaling pathway. We further conducted a comprehensive evaluation in acd6-1 of pairwise genetic interactions among five SA components: type I SA gene SID2; type II SA genes ALD1, EDS5, and PAD4; and type III SA gene NPR1. Systematic analysis of a total of 10 triple mutants for their defense and cell death phenotypes have revealed complex genetic interactions among the SA genes and suggest interconnected defense signaling networks.
Materials and Methods
Plant materials
All plants used in this report are in Columbia-0 background. Plants were grown in growth chambers with light intensity at 200 µmol m−2 s−1, 60% humidity, and 22°. The single mutants (acd6-1, ald1-1, npr1-1, pad4-1, eds5-1, and sid2-1), the double mutants (acd6-1ald1-1, acd6-1eds5-1, acd6-1npr1-1, acd6-1pad4-1, and acd6-1sid2-1), and the triple mutants (acd6-1ald1-1pad4-1 and acd6-1npr1-1pad4-1) were previously described (Song et al. 2004b; Lu et al. 2009). eds1-2, which was introgressed to the Col-0 background, was kindly provided by Jane Parker at Max Planck Institute for Plant Breeding Research. eds1-2 was crossed to acd6-1 to make acd6-1eds1-2. Additional triple mutants were constructed by crossing respective double mutants in the acd6-1 background and were screened for homozygotes in the F2 population using specific derived cleaved amplified polymorphic sequence (dCAPS) markers and/or other polymerase chain reaction (PCR) markers. Genotyping primers used in this study were listed in supporting information, Table S2.
Pseudomonas infection and bacterial growth assay
Pseudomonas infection was performed with 25-day-old plants grown in a chamber with a 12-hr light/12-hr dark cycle. Freshly cultured Pseudomonas syringae pv. maculicola (Pma) ES4326 strain DG3 (OD600 = 0.6–0.8) was diluted to a final concentration 1 × 105 cfu ml−1 and infiltrated into the fourth to sixth leaves of each genotype. Three days postinfection, discs of 7 mm diameter from the infected leaves were excised with a core borer and ground in 10 mM MgSO4. Serial bacterial dilutions were made and spread on KB plates containing kanamycin (50 μg/ml). Six independent leaf samples were used for each data point. Statistical analysis was performed with Student's t-test (StatView 5.0.1).
Cell death staining
For trypan blue staining, the fourth to sixth leaves of the plants were boiled in lactophenol [phenol:glycerol:lactic acid:water = 1:1:1:1 (v/v)] containing 0.01% trypan blue for 2 min, cleared off with boiling alcoholic lactophenol (95% ethanol:lactophenol = 2:1), and rinsed with 50% ethanol. The stained leaves were examined with a Stemi SV 1.1 stereomicroscope (Zeiss) and pictures were taken with an AxioCam MRc5 camera (Zeiss) connected to the microscope.
RNA analysis
Twenty-five-day-old plants were harvested for total RNA extraction with TRIzol reagent (Invitrogen) according to the protocol provided by the manufacturer. Northern blotting was performed as previously described (Lu et al. 2003). Radioactive probe for PR1 was made by PCR with a specific antisense primer for a PR1 fragment in the presence of [32P] dCTP.
SA measurement
Twenty-five-day-old plants were harvested for SA extraction followed by HPLC analysis as previously described (Wang et al. 2011a).
Results
acd6-1 is a sensitive tool to detect both additive and nonadditive interactions between SA regulators
We previously used the defense-sensitized mutant acd6-1 to elucidate functional relationships between several SA genes (Song et al. 2004b; Lu et al. 2009; Wang et al. 2011a). In this genetic analysis, we introduced two mutations that affect SA-mediated defense, each of which is known to cause suppression of acd6-1–conferred phenotypes, into the acd6-1 background and assessed whether the two mutations together result in additive or nonadditive suppression of acd6-1–conferred phenotypes. On the basis of the phenotypes of the triple mutant, we made inferences on the interaction between the respective genes. A nonadditive suppression would indicate that the two genes act in the same pathway, whereas an additive suppression would suggest that the genes function in different pathways. Using this analysis, we demonstrated additive interactions between several SA genes (Song et al. 2004b; Lu et al. 2009; Wang et al. 2011).
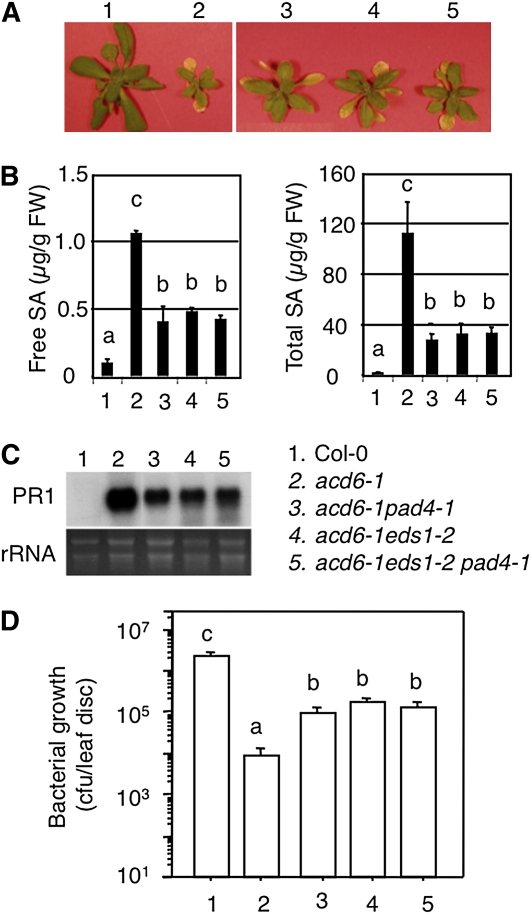
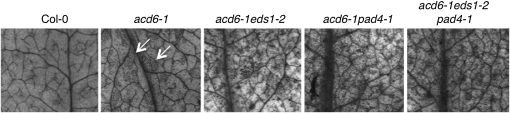
To further test the validity of acd6-1 as an effective tool to detect interactions between defense genes, we set out to analyze in the acd6-1 background the functional relationship between two previously characterized SA genes, EDS1 and PAD4, that are presumed to act in the same pathway on the basis of biochemical and microarray studies (Feys et al. 2001; Bartsch et al. 2006; Sato et al. 2007). We crossed acd6-1 with eds1-2, which was introgressed to Col-0 by five times of crosses (J. Parker, personal communication). Both pad4-1 and eds1-2 partially suppressed acd6-1–conferred phenotypes, namely small size, SA accumulation, expression of the defense marker gene PATHOGENESIS-RELATED GENE 1 (PR1), and constitutive disease resistance to the pathogen P. syringae (Figure 1, A–D and Lu et al. 2003). In addition, large patches of cell death in acd6-1 were also greatly reduced by eds1-2 (Figure 2). Compared to the two parental double mutants, the triple mutant acd6-1eds1-2pad4-1 showed a similar level of suppression of these phenotypes, suggesting a nonadditive interaction between eds1-2 and pad4-1. These results provide direct genetic evidence to demonstrate that EDS1 and PAD4 act in the same signaling pathway to regulate acd6-1–conferred phenotypes, consistent with evidence from previous studies based on global gene expression profiling and protein–protein interactions (Feys et al. 2001; Bartsch et al. 2006; Sato et al. 2007). Our data also suggest that the acd6-1 background can be used to unravel both additive and nonadditive genetic interactions among defense genes.
Figure 1 .
eds1-2 acts nonadditively with pad4-1 in suppressing acd6-1–conferred phenotypes. (A) Picture of 25-day-old plants. (B) SA quantitation. Free and total SA were extracted from plants shown in A and analyzed by HPLC. (C) Expression of PR1. Total RNA was extracted from uninfected 25-day-old plants for Northern blot analysis. rRNA was used as a loading control. (D) Bacterial growth assay. Plants of 25 days old were infected with PmaDG3 (OD600 = 0.0001) and bacterial growth was assessed 3 days after infection. Data represent the average of bacterial numbers in six samples (n = 6) ± SE. In B and D, statistical analysis was performed with Student’s t-test (StatView 5.0.1). Letters indicate significant difference among the samples (P < 0.05). The key for the genotypes used in these experiments is shown in C, right.
Figure 2 .
eds1-2 acts nonadditively with pad4-1 in suppressing cell death in acd6-1. The fourth to sixth leaves of the indicated genotypes were stained with trypan blue. Photographs were taken with a dissecting microscope connected to an AxioCam MRc5 camera (Zeiss). eds1-2 and pad4-1 showed no detectable cell death (data not shown). Note the large patches of cell death shown in acd6-1 (arrows) were reduced in the double and triple mutants.
Multiple SA regulators contribute to both SID2-dependent and -independent defense pathways and/or cell death control
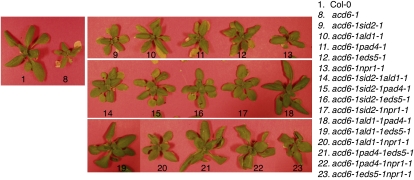
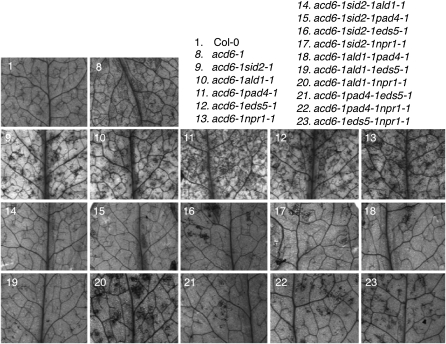
Next we set out to dissect the genetic interactions among several known SA genes, SID2 (type I SA gene), ALD1, EDS5, and PAD4 (type II SA genes), and NPR1 (type III SA gene). Loss-of-function mutations in each of these genes partially suppressed acd6-1–conferred phenotypes (Song et al. 2004b; Lu et al. 2009). We made combinatorial pairwise crosses of these SA mutants in the acd6-1 background and obtained a total of 10 triple mutants, two of which were reported previously (Song et al. 2004b; Lu et al. 2009). Here we performed a systematic analysis of all 10 triple mutants for their defense and cell death phenotypes.
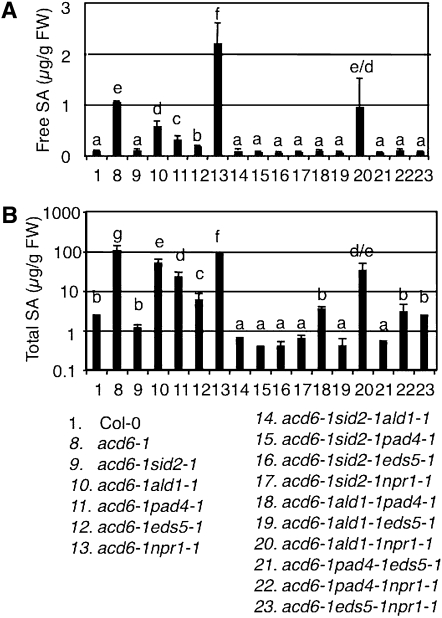
We first examined how type I gene SID2 interacts with type II genes. Consistent with SID2’s major role in SA biosynthesis, we detected only residual SA levels but no expression of the SA marker gene PR1 in acd6-1sid2-1. Compared with acd6-1, acd6-1sid2-1 was larger and had partially reduced resistance to P. syringae infection and cell death (Figures 3–7 and Lu et al. 2009). Since the double mutants acd6-1ald1-1, acd6-1eds5-1, and acd6-1pad4-1 accumulated more SA than acd6-1sid2-1 but less SA than acd6-1, we conclude that these type II SA genes only partially affect SID2-mediated SA biosynthesis (Lu et al. 2009). When each of these type II SA mutations was introduced into acd6-1sid2-1, we detected a small further reduction of glucosyl-conjugated SA (total SA) in the respective triple mutants (Figure 4 and Table S1). These results suggest that type II SA genes regulate both SID2-dependent and SID2-independent SA accumulation and the SID2-independent pathway only plays a minor role in affecting SA accumulation.
Figure 3 .
Genetic interactions among SA mutants lead to altered acd6-1 morphology. Plants were photographed 25 days postplanting. The single mutants largely resemble Col-0 (data not shown).
Figure 7 .
Genetic interactions among SA mutants lead to altered cell death in acd6-1. The fourth to sixth leaves of the indicated genotypes were stained with trypan blue. Photographs were taken with a dissecting microscope connected to an AxioCam MRc5 camera (Zeiss). The single mutants showed no detectable cell death (data not shown).
Figure 4 .
Genetic interactions among SA mutants lead to altered SA accumulation in acd6-1. SA was extracted from 25-day-old plants and analyzed by HPLC for free (A) and total SA (B). Note B has a log scale. The single mutants have similar SA levels as Col-0 (data not shown). Letters indicate significant difference among the samples (P < 0.05).
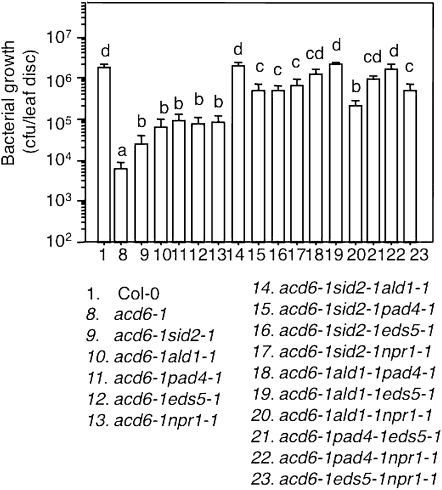
Although the effect of SID2-independent pathway(s) on SA accumulation is minor, we observed a strong influence of this pathway on other acd6-1–conferred phenotypes. Compared to the respective double mutants in the acd6-1 background, the triple mutants acd6-1sid2-1ald1-1, acd6-1sid2-1eds5-1, and acd6-1sid2-1pad4-1 exhibited more reduced cell death visible to the naked eye, which was further confirmed by trypan blue staining (Figures 3 and 7). They also had more reduced disease resistance (Figure 6). These results suggest that type II SA genes, ALD1, EDS5, and PAD4, act additively with type I SA gene SID2 in regulating disease resistance and cell death in acd6-1.
Figure 6 .
Genetic interactions among SA mutants lead to altered disease resistance in acd6-1. Bacterial growth was assessed 3 days after infection with PmaDG3 (OD600 = 0.0001). Statistical analysis was performed with Student's t-test (StatView 5.0.1). Different letters indicate significant difference among the samples (P < 0.05; n = 6).
Multiple SA regulators contribute to both NPR1-dependent and -independent defense pathways and cell death control
To study how type I and type II SA genes interact with type III SA gene NPR1, we analyzed mutants defective for these genes in the acd6-1npr1-1 background. NPR1, an ankyrin-repeat–containing protein, acts as an SA signal transducer and can also positively or negatively influence SA accumulation (Cao et al. 1997; Ryals et al. 1997; Shah et al. 1997; Dong 2004; Lu et al. 2009). Compared to acd6-1, acd6-1npr1-1 was slightly larger, had reduced PR1 expression, disease resistance, and cell death, but accumulated higher levels of free SA (Figures 3–7 and Lu et al. 2009).
First we examined the interaction between type I SA gene SID2 and type III SA gene NPR1. The triple mutant acd6-1sid2-1npr1-1 only accumulated residual SA, suggesting that the high SA levels observed in acd6-1npr1-1 are largely produced via the SID2-dependent SA biosynthesis. Compared with the two double mutants, acd6-1sid2-1npr1-1 also displayed further reduced cell death and disease resistance (Figures 3, 6, and 7). While it is possible that one or more SID2-independent pathways are responsible for further suppressed disease resistance and cell death in acd6-1npr1-1sid2-1, the additive suppression could also occur if SID2 acts through both NPR1-dependent and -independent pathways.
Next we examined the interaction between type II genes (ALD1, EDS5, and PAD4) and NPR1. The pad4-1 mutant was previously shown to greatly suppress the high SA levels in acd6-1npr1-1. On the basis of this result, we proposed that the negative role of NPR1 in regulating SA levels requires the function of PAD4 (Lu et al. 2009). We confirmed this result in this study. We further observed a similar suppression effect of eds5-1 on SA accumulation in acd6-1npr1-1 (Figure 4), suggesting that NPR1’s negative regulation of SA accumulation also involve EDS5 besides PAD4. Since the SA levels in the triple mutants, acd6-1eds5-1npr1-1 and acd6-1pad4-1npr1-1, were even lower than those in the double mutants, acd6-1eds5-1 and acd6-1pad4-1, we speculate that EDS5 and PAD4 act through both NPR1-dependent and -independent pathways to regulate SA levels. Alternatively, these results can be explained that besides its roles as a positive SA signal transducer and as a negative regulator of SA accumulation, NPR1 plays a positive role in regulating SA accumulation in a separate pathway from those mediated by EDS5 and PAD4. Consistent with reduced SA levels, acd6-1eds5-1npr1-1 and acd6-1pad4-1npr1-1 had much reduced PR1 expression, disease resistance, and cell death compared with the respective double mutants (Figures 5–7).
Figure 5 .
Genetic interactions among SA mutants lead to altered PR1 expression in acd6-1. Total RNA was extracted from uninfected 25-day-old plants for northern blot analysis. rRNA was used as a loading control. No PR1 expression was detected in the single mutants, sid2-1, ald1-1, pad4-1, eds5-1, and npr1-1 (data not shown).
In contrast to acd6-1eds5-1npr1-1 and acd6-1pad4-1npr1-1, the acd6-1ald1-1npr1-1 mutant expressed a high level of PR1 transcripts, accumulated similar levels of SA, and displayed similar degrees of disease resistance and cell death as acd6-1ald1-1 or acd6-1npr1-1 (Figures 4–7). These data indicate a nonadditive interaction between ALD1 and NPR1. Given that ALD1 is a type II SA gene and NPR1 is a type III SA gene, we propose that ALD1 acts upstream of NPR1 in the same pathway to regulate plant defense and cell death.
Genetic analysis reveals additive interactions among type II SA regulators, ALD1, EDS5, and PAD4
We further analyzed genetic interactions among the three type II SA mutants, ald1-1, eds5-1, and pad4-1 in the acd6-1 background, to learn more about the pathway(s) in which these genes act. Compared with ald1-1 and pad4-1, eds5-1 had a greater suppression of SA levels in acd6-1, suggesting that among these type II SA genes, EDS5 plays a greater role in regulating SA accumulation (Figure 4). This notion is consistent with previous studies (Nawrath and Metraux 1999; Nawrath et al. 2002). When any two of these mutants were genetically combined, we observed further suppression of acd6-1–conferred small size, SA accumulation, PR1 expression, disease resistance, and cell death (Figures 3–7 and Song et al. 2004b). These results suggest that type II SA genes do not act in one linear pathway but rather in separate pathways to regulate SA-mediated defense and cell death in acd6-1.
Discussion
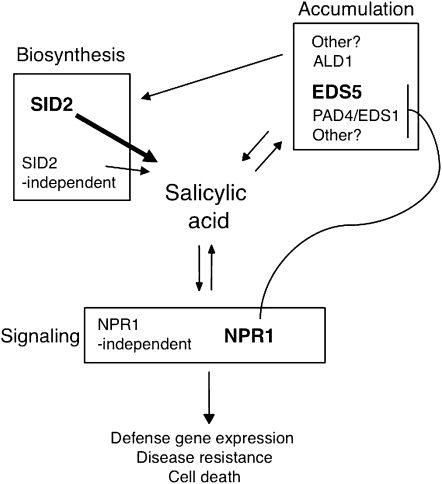
Genetic analysis directly associates gene functions with phenotypes; thus a genetic approach to study relationships between genes can reveal functional information invisible to other approaches, such as protein–protein interaction and microarray analysis. Here we exploited a sensitive Arabidopsis mutant acd6-1, whose phenotypes (small size and cell death) are easily perturbed by the changes of defense levels, in a genetic interpretation of relationships among several key components in the SA signaling networks. Our data have revealed both additive and nonadditive relationships among these SA genes and suggest highly interactive SA signaling networks (Figure 8).
Figure 8 .
SA-mediated defense signaling networks. SA represents a key hub on the defense signaling networks. Genes regulate SA-mediated defense can be viewed in three types. Type I genes are responsible for SA biosynthesis, with SID2 contributing to the major SA production and SID2-independent pathway playing a minor role. Type II genes encode protein products that do not act directly as SA biosynthetic enzymes. It is possible that these SA regulators might influence SA biosynthetic processes by either modifying the activities of the SA biosynthetic enzymes or regulating precursor availability for SA biosynthesis. Alternatively, they can affect SA stability, sequestration, transport, and/or conjugation. Among the known type II SA genes, EDS5 plays a major role in regulating SA accumulation. Other components also partially affect SA levels. Type III genes include NPR1 as the main SA signal transducer and other signal transducers independent of NPR1. Expression of some SA regulatory genes is known to be regulated by SA, suggesting the existence of multiple signal amplification loops involving SA and SA regulatory genes. In this report, we described a genetic analysis based on the defense-sensitized mutant acd6-1 to elucidate the functional relationships among the SA genes. We showed that the same type of SA genes can additively interact with each other and they can also additively interact with other types of SA genes to affect SA accumulation, defense gene expression, disease resistance, and/or cell death phenotypes in the acd6-1 background. Nonadditive interactions were observed between EDS1 and PAD4 and between ALD1 and NPR1.
EDS1 and PAD4 are type II SA regulators that share similarities in their protein sequences. Although the two proteins might have distinct roles in regulating plant defense and other processes (Feys et al. 2005; Rietz et al. 2011), evidence also suggests that they can act together in the same pathway under certain conditions (Feys et al. 2005; Bartsch et al. 2006; Sato et al. 2007). Our data that eds1-2 and pad4-1 act nonadditively to suppress acd6-1–conferred phenotypes support the latter notion, suggesting that the two genes function in the same pathway to regulate plant defense and cell death in the acd6-1 background. ALD1 encodes an aminotransferase and was proposed to generate an amino acid-derived signal to activate plant defense (Song et al. 2004a). Like npr1 mutants, ald1-1 is defective in both local defense and systemic acquired resistance (Song et al. 2004b). A nonadditive interaction between the ald1-1 and npr1-1 mutations was observed in acd6-1 and in the syntaxin double mutant syp121-1syp122-1 (this study and Zhang et al. 2007), suggesting that ALD1 and NPR1 act in the same branch of a defense pathway.
While our results revealed two cases of nonadditive interactions between SA regulators, the additive interactions appear to be more prevalent. Each SA gene of one type was found to act additively with genes of the other two types. Such additive effects are less likely due to the leakiness of the mutations, since the SA mutants used in this report are generally considered as null mutants (Cao et al. 1997; Falk et al. 1999; Jirage et al. 1999; Wildermuth et al. 2001; Nawrath et al. 2002; Song et al. 2004b). Instead, our results indicate that there are multiple regulatory pathways feeding into the regulation of SA biosynthesis, accumulation, and signaling. Consistent with our results, Tsuda et al. (2009) reported an additive interaction between type I mutant sid2 and type II mutant pad4 in response to bacterial infection and elicitor treatments in the absence of acd6-1.
Interestingly, we found that the degree of suppression of acd6-1–conferred phenotypes by two mutations together is often smaller than the added value from two single mutations. For instance, on the basis of total SA quantification, the degree of suppression of SA accumulation in acd6-1 by ald1-1, eds5-1, and pad4-1 is 52, 94, and 78%, respectively (Table S1). However, the corresponding triple mutants showed 99% reduction in SA levels, a value smaller than any two combined values from above. These observations suggest a negative interaction between most SA genes. The negative interaction can be explained that while most SA genes act in different pathways, they can also functionally compensate each other, possibly due to some genes sharing redundant function and/or they can regulate each other’s function. Indeed, many prior studies showed that expression of some SA genes is dependent on other genes in the SA networks. For instance, expression of ALD1 and EDS5 is known to be PAD4 dependent (Nawrath et al. 2002; Song et al. 2004b) and PAD4 to be NPR1 dependent (Jirage et al. 1999). In addition, expression of some SA genes can also be regulated by SA treatment (Zhou et al. 1998; Falk et al. 1999; Nawrath et al. 2002; Lu et al. 2003; Song et al. 2004b; Jagadeeswaran et al. 2007; Lee et al. 2007). Thus, there are likely interlocked signal amplification loops that involve SA and multiple SA regulators. Consistent with a picture of interactive SA signaling networks, a previous microarray study placed NPR1 both downstream and upstream of type II SA regulators and type I SA gene SID2 (Wang et al. 2008). The highly interactive SA networks suggest that plant innate immunity is robust, involving multiple key components acting in concert to regulate disease resistance to broad-spectrum pathogens.
Understanding how genes function and their interactions with each other to form complex signaling networks governing cellular processes and behavior of organisms has become increasingly important in the postgenomic era. In this report, we have demonstrated the utility of a unique Arabidopsis mutant acd6-1 in elucidating the functional relationships among key components in the SA signaling networks. Together with those results obtained from complementary approaches related to biochemistry and global gene expression profiling, the results from this study have revealed a picture of a complex and interactive genetic map for the SA signaling networks. Therefore, this study provides a framework for further systematic interrogation of the important role of SA and other signaling molecules in plant disease resistance, leading to a better understanding of mechanisms of plant disease resistance.
Acknowledgments
We thank members in the Lu laboratory for assistance in this work and Stephen Miller at University of Maryland Baltimore County for critical comments on the manuscript. We thank William LaCourse for sharing the usage of his HPLC instrument, Charles Bieberich for sharing the use of his dissecting microscope, and Tim Ford for taking pictures for this publication. This work was supported by startup funds from University of Maryland Baltimore County and a grant from the National Science Foundation (RIG-0818651) to H.L. and a scholarship from the China Scholarship Council to C.Z.
Literature Cited
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J. L., et al. , 2006. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D. K., Cameron D. M., Collins S. R., Schuldiner M., Stewart-Ornstein J., et al. , 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5: 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J. D., Volko S., Dong X., 1997. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Collins S. R., Schuldiner M., Krogan N. J., Weissman J. S., 2006. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 7: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., et al. , 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M., Bennett M. H., Truman W. H., Grant M. R., 2009. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 59: 375–386 [DOI] [PubMed] [Google Scholar]
- Dong X., 2004. NPR1, all things considered. Curr. Opin. Plant Biol. 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Endo J., Takahashi W., Ikegami T., Beppu T., Tanaka O., 2009. Induction of flowering by inducers of systemic acquired resistance in the Lemna plant. Biosci. Biotechnol. Biochem. 73: 183–185 [DOI] [PubMed] [Google Scholar]
- Falk A., Feys B. J., Frost L. N., Jones J. D., Daniels M. J., et al. , 1999. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B. J., Moisan L. J., Newman M. A., Parker J. E., 2001. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B. J., Parker J. E., 2000. Interplay of signaling pathways in plant disease resistance. Trends Genet. 16: 449–455 [DOI] [PubMed] [Google Scholar]
- Feys B. J., Wiermer M., Bhat R. A., Moisan L. J., Medina-Escobar N., et al. , 2005. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goritschnig S., Zhang Y., Li X., 2007. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 49: 540–551 [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack K. E., Jones J. D., 1996. Resistance gene-dependent plant defense responses. Plant Cell 8: 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran G., Raina S., Acharya B. R., Maqbool S. B., Mosher S. L., et al. , 2007. Arabidopsis GH3-LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 51: 234–246 [DOI] [PubMed] [Google Scholar]
- Jirage D., Tootle T. L., Reuber T. L., Frost L. N., Feys B. J., et al. , 1999. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F., 2004. A global view of defense gene expression regulation–a highly interconnected signaling network. Curr. Opin. Plant Biol. 7: 506–511 [DOI] [PubMed] [Google Scholar]
- Koornneef A., Pieterse C. M., 2008. Cross talk in defense signaling. Plant Physiol. 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel B. N., Brooks D. M., 2002. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Lee M. W., Lu H., Jung H. W., Greenberg J. T., 2007. A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. Mol. Plant Microbe Interact. 20: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Lu H., 2009. Dissection of salicylic acid-mediated defense signaling networks. Plant Signal. Behav. 4: 713–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Rate D. N., Song J. T., Greenberg J. T., 2003. ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Salimian S., Gamelin E., Wang G., Fedorowski J., et al. , 2009. Genetic analysis of acd6–1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J. 58: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K., Levine A., Eulgem T., Morgan A., Schmid J., et al. , 2000. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Martinez C., Pons E., Prats G., Leon J., 2004. Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 37: 209–217 [DOI] [PubMed] [Google Scholar]
- Nawrath C., Metraux J. P., 1999. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Heck S., Parinthawong N., Metraux J. P., 2002. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K., Okrent R. A., Stoutemyer M., Rodibaugh N., Kempema L., et al. , 2007. The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 144: 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K., Zhang Y., Li X., 2005. An importin alpha homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Palma K., Zhao Q., Cheng Y. T., Bi D., Monaghan J., et al. , 2007. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 21: 1484–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate D. N., Cuenca J. V., Bowman G. R., Guttman D. S., Greenberg J. T., 1999. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz S., Stamm A., Malonek S., Wagner S., Becker D., et al. , 2011. Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 191: 107–119 [DOI] [PubMed] [Google Scholar]
- Roguev A., Wiren M., Weissman J. S., Krogan N. J., 2007. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat. Methods 4: 861–866 [DOI] [PubMed] [Google Scholar]
- Ryals J., Weymann K., Lawton K., Friedrich L., Ellis D., et al. , 1997. The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J. A., Neuenschwander U. H., Willits M. G., Molina A., Steiner H. Y., et al. , 1996. Systemic acquired resistance. Plant Cell 8: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Mitra R. M., Coller J., Wang D., Spivey N. W., et al. , 2007. A high-performance, small-scale microarray for expression profiling of many samples in Arabidopsis-pathogen studies. Plant J. 49: 565–577 [DOI] [PubMed] [Google Scholar]
- Sato M., Tsuda K., Wang L., Coller J., Watanabe Y., et al. , 2010. Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog. 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J., Tsui F., Klessig D. F., 1997. Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA- inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10: 69–78 [DOI] [PubMed] [Google Scholar]
- Song J. T., Lu H., Greenberg J. T., 2004a Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant growth and death2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. T., Lu H., McDowell J. M., Greenberg J. T., 2004b A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 40: 200–212 [DOI] [PubMed] [Google Scholar]
- Tao Y., Xie Z., Chen W., Glazebrook J., Chang H. S., et al. , 2003. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Balasubramanian S., Hu T. T., Traw M. B., Horton M., et al. , 2010. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Glazebrook J., Cohen J. D., Katagiri F., 2008. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53: 763–775 [DOI] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F., 2009. Network properties of robust immunity in plants. PLoS Genet. 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A., Nichols R. J., Siegele D. A., Shales M., Collins S. R., et al. , 2008. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat. Methods 5: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H., Lu H., Rate D. N., Greenberg J. T., 2001. A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J. 28: 209–216 [DOI] [PubMed] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A. H., Dong X., 2007. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17: 1784–1790 [DOI] [PubMed] [Google Scholar]
- Wang G. F., Seabolt S., Hamdoun S., Ng G., Park J., et al. , 2011a Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol. 156: 1508–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.-Y., Shi J.-L., Ng G., Battle S. L., Zhang C., et al. , 2011b Circadian clock-regulated phosphate transporter PHT4;1 plays an important role in Arabidopsis defense. Mol. Plant 4: 516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Mitra R. M., Hasselmann K. D., Sato M., Lenarz-Wyatt L., et al. , 2008. The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: roles of major regulators and the phytotoxin coronatine. Mol. Plant Microbe Interact. 21: 1408–1420 [DOI] [PubMed] [Google Scholar]
- Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M., 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li X., 2005. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1–1, constitutive 1. Plant Cell 17: 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cheng Y. T., Bi D., Palma K., Li X., 2005. MOS2, a protein containing G-patch and KOW motifs, is essential for innate immunity in Arabidopsis thaliana. Curr. Biol. 15: 1936–1942 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Feechan A., Pedersen C., Newman M. A., Qiu J. L., et al. , 2007. A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49: 302–312 [DOI] [PubMed] [Google Scholar]
- Zhou N., Tootle T. L., Tsui F., Klessig D. F., Glazebrook J., 1998. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]