Abstract
Analysis of genomic data requires an efficient way to calculate likelihoods across very large numbers of loci. We describe a general method for finding the distribution of genealogies: we allow migration between demes, splitting of demes [as in the isolation-with-migration (IM) model], and recombination between linked loci. These processes are described by a set of linear recursions for the generating function of branch lengths. Under the infinite-sites model, the probability of any configuration of mutations can be found by differentiating this generating function. Such calculations are feasible for small numbers of sampled genomes: as an example, we show how the generating function can be derived explicitly for three genes under the two-deme IM model. This derivation is done automatically, using Mathematica. Given data from a large number of unlinked and nonrecombining blocks of sequence, these results can be used to find maximum-likelihood estimates of model parameters by tabulating the probabilities of all relevant mutational configurations and then multiplying across loci. The feasibility of the method is demonstrated by applying it to simulated data and to a data set previously analyzed by Wang and Hey (2010) consisting of 26,141 loci sampled from Drosophila simulans and D. melanogaster. Our results suggest that such likelihood calculations are scalable to genomic data as long as the numbers of sampled individuals and mutations per sequence block are small.
THE coalescent process is highly variable: samples from even a single well-mixed population rapidly coalesce down to a few ancestral lineages, so that their deeper ancestry is determined by just a few random coalescence events (Felsenstein 1992). Thus, small samples taken from a large number of loci give much more information than large samples from a few loci. For example, the distribution of coalescence times, and hence the history of effective population size, has been inferred from single diploid genomes (Li and Durbin 2011). Although it is now feasible to sample very large numbers of markers, or indeed whole genomes, we urgently need methods for analyzing such data. In principle, we can calculate likelihoods from very large data sets, if we have loosely linked blocks of sequence within which recombination is negligible. Provided that only a few genomes are sampled, we can tabulate the probability that any particular configuration of mutations will be seen at each locus and then multiply across large numbers of loci to find the likelihood of our model (Takahata et al. 1995).
Wilkinson-Herbots (2008) and Wang and Hey (2010) derive the distribution of coalescence times for a pair of genes sampled from two populations that separated at some time in the past and subsequently exchanged migrants. This “isolation-with-migration” (IM) model is of particular interest in evaluating the role of gene flow during speciation. Hobolth et al. (2011) show how this and similar calculations can be done more efficiently using matrix exponentials.
Here, we present an alternative method, based on generating functions, which provides direct information about the pattern of mutational variation and can be automated using symbolic algebra packages such as Mathematica. We give the IM model as an example and show how the method extends to linked loci.
The Generating Function of a Genealogy
The ancestry of a sample of genes, Ω, is described by the lengths of the branches that are ancestral to every possible subset. For example, suppose that we have three genes at a locus, labeled Ω = {a, b, c}. We label lineages by the set of genes to which they are ancestral. Thus, if lineages ancestral to genes b and c coalesced most recently, then the branches {b} and {c} have the same length; i.e., t{b} = t{c}, and t{a} = t{b} + t{b,c}. With this topology, there are no lineages ancestral to {a, b} or {a, c} and t{a,b} = t{a,c} = 0. Thus, both the topology and the branch lengths are encoded by the vector of all possible branches t, which has elements tS for S ⊆ Ω.
The generating function (GF) for the branch lengths t depends on a set of corresponding dummy variables, ω and is defined as the expectation . It is more convenient to use this form—a Laplace transform—rather than the alternative . Generating functions are widely used, primarily because the distribution of the sum of two independent variables is given by the product of the corresponding GF. In particular, Latter (1973) used a GF approach to find the solution for the expected frequency of heterozygotes under the symmetric IM model and Griffiths (1981b) used the GF for the numbers of types to calculate sampling distributions for the infinite-alleles model. Griffiths (1991) applied this to the two-locus problem (see also Jenkins 2008). In the context of the coalescent, the GF has a concrete interpretation: under the infinite-sites model, it is the probability of seeing no mutations, given mutation rate ωS along branch S.
Information about the branch lengths themselves can be recovered from the GF. The mean lengths, E[tS], are found by differentiating with respect to ωS and setting ω to zero; higher moments are found by differentiating more than once. The actual distribution can be found by taking the inverse Laplace transform, which may be done either algebraically (if the GF has a certain form) or by numerical integration.
In practical applications, we wish to know the probability that there are kS mutations on branch S. Under the infinite-sites model, with mutation rate μ, this is given by taking the expectation of a Poisson distribution with mean μt over the distribution of coalescence times,
| (1) |
which is proportional to the th differential of the GF with respect to ωS, taken at ωS = μ, and setting all other ω’s to zero. We see that Equation 1 defines a term in a Taylor series, so that the probability of a particular configuration of mutations is given by the coefficient in the expansion of ψ. In other words, if we set ωS = μ − xS and expand around the point xS = 0, then the probability of seeing kS mutations on branch S is the coefficient of , multiplied by . Similarly, the joint probability of seeing a configuration of mutations on branches S1, S2, … is the coefficient of , multiplied by . In the following, we scale time relative to twice the effective population size, 2N; i.e., the scaled mutation rate is 2Nμ = θ/2.
While we assume an infinite-sites mutation model for simplicity throughout, the GF can also be used to obtain the probabilities of mutational configurations for more complex mutation models. For example, under the Jukes-Cantor (Jukes and Cantor 1969) model mutations to a different state happen at rate (3/4)μ and the chance of a back mutation is (1/4)μ. The probabilities that two sequences differ or are the same at any particular site are and respectively. Given a pair of sequences of length n the probability of seeing j sites in a different and n − j in the same state is given by taking the expectation of a Binomial distribution over the distribution of coalescence times:
| (2) |
This can be written as a sum of the GFs of pairwise coalescence times:
| (3) |
Thus, in principle, we can obtain results under a finite-sites mutation model directly from the GF without the need to take derivatives.
The generating function is a sum of terms, each corresponding to a particular topology. For a given topology, many branches will have zero length by definition, and so the GF will be independent of the corresponding ωS; some branches will have the same lengths (e.g., t{b} = t{c}) and so the corresponding terms will be a function of the sum of the respective dummy variables (e.g., ω{b} + ω{c}). Under the infinite-sites model, this brings a substantial simplification if we see mutations on internal branches, because any terms that do not depend on the corresponding dummy variables can be dropped from the GF: they represent topologies inconsistent with the data. The joint likelihood for a given mutational configuration can then be calculated by multiple differentiation of the remaining terms, which involves a sum over only the possible topologies.
The General Recursion
The recursion for the generating function of genealogical branch lengths can be derived by tracing back from the present to the most recent event, which might be a coalescence, a recombination, a movement between demes, a change in population structure, or whatever. Events i occur at rate λi and (tracing back in time) change the configuration of genes from the sampling configuration Ω to Ωi. Configurations include the number of lineages and—depending on the model—their locations and/or genetic backgrounds. For example, suppose that we start with three lineages {a}, {b}, and {c}. A coalescence between lineages {b} and {c} generates a new configuration {{b, c}, {a}}, in which there are now two lineages—one ancestral to {b, c} and the other to {a}. We derive a recursion that expresses the GF ψ[Ω] as a sum over the possible configurations before the previous event. The time back to that event is exponentially distributed with rate , and so the distribution of the lengths of the terminal branches is just the convolution of this with their previous distribution. Taking Laplace transforms, this corresponds simply to multiplication by the factor , since a convolution of distributions transforms to a product of the previous GF and the GF of an exponential distribution with rate . Summing over all possible events we have
| (4) |
The denominator gives the total rate of events, in the interval from the present to the first event, plus the sum of the ωS that correspond to terminal branches (the “leaves” of the tree). The numerator is the sum over all possible generating functions at the previous event; Ωi denotes the configuration prior to event i. This recursion yields a set of linear equations for the ψ[Ω] that is readily solved; the limit is set by the number of possible sample configurations of genes that have to be tracked. To see how this works, we give a series of examples.
A Single Population
In the simplest case of a single well-mixed population, we need to track only coalescence events. Scaling time relative to twice the effective population size, 2N, the rate of coalescence is given by the number of pairs of lineages in a given sample configuration where there are |Ω| lineages. Thus
| (5) |
where the sum is over all the possible pairwise coalescences, between genes x and y. denotes the sample configuration after coalescence, i.e., Ω with lineages {x}, {y} replaced by the new lineage {x, y}. Since we define the GF for a single gene as 1, we have for two genes
| (6) |
This is equivalent to the probability of identity in state with ωa + ωb = θ. Note that for brevity, we have condensed the notation so that ψ[a, b] represents the GF for two lineages ancestral to genes a and b, respectively; and ψ[ab, c] represents two lineages, one ancestral to a and b, and the other to c. For automated recursions (File S1), the full (and unambiguous) notation ψ[ω, {{a}, {b}}], ψ[ω, {{a, b}, {c}}] would be used. For three genes
| (7) |
Each of the three terms corresponds to one of the three possible topologies. For example, the last term depends on ωbc and corresponds to coalescence between {b} and {c}, so that the interior branch tbc > 0. To find the probability of each topology, we set all the ωS to zero, and see that each term contributes . To find the probability that there are k mutations ancestral to b and c, we differentiate k times with respect to ωbc, set ωbc to equal the scaled mutation rate θ/2 and all other ωS to zero, and multiply by (−θ/2)k/k! (Equation 1). This gives the geometric distribution for k > 0, the factor 3 arising because there is a 1/3 probability that b and c coalesce first, allowing mutations of this class to exist. Alternatively, we could set all the ωS to 0, except for , and then expand around xbc = 0; the coefficients of are proportional to the chance of seeing kbc mutations that are ancestral to b and to c. The joint probabilities of other mutational configurations can be found in a similar way.
Migration
Suppose that two populations exchange migrants at a scaled rate 2Nm. For simplicity, we assume that migration is symmetric and both demes are of the same size (the generalization to more demes, different population sizes, and asymmetric migration is obvious) and that a set Ω1 of genes is sampled from one deme and Ω2 from the other. Now, there can be coalescence, which reduces the size of one or the other set, or migration, which transfers a lineage x from one deme to the other creating, for example, new sample configurations Ω1,+x and Ω2,−x. Thus
| (8) |
This leads to a set of linear equations that can readily be solved. We need to distinguish only sample configurations where the genes are in different demes, ψ[a\b], or in the same demes, ψ[a, b\Ø], say (again, we have condensed the notation; Ø represents the empty set, and \ the separation between the two demes). From Equation 8 and using the symmetry of the model,
| (9) |
This has the solution
| (10) |
where M = 4Nm. Note that the GF is a function only of ωa + ωb, given the constraint ta = tb. Equation 10 has been previously derived as the probability of identity in state with ωa + ωb = θ (Griffiths 1981a, equation 10). Taking the inverse Laplace transform gives the probability of pairwise coalescent times,
| (11) |
where and . This result was derived directly by Herbots (1997), using a partial fraction expansion (see Griffiths 1981a; Wilkinson-Herbots 2008, equation 18), but can also be found from the discrete time transition matrix (Wakeley 1996). In fact, λ0 and λ1 are the eigenvalues of the symmetric transition matrix Q given by Hobolth et al. (2011) with S1 = S2, S11 = S22, and m1 = m2.
Population Splits: The IM Model
Now, suppose that the two populations derive from a single ancestral population T generations ago. Dealing with finite times explicitly leads to complicated expressions (Wang and Hey 2010). However, we can retain the simple form of the GF by taking the Laplace transform with respect to the divergence time, with dummy variable Λ. This has a concrete interpretation, as the expectation over a model in which the divergence time is exponentially distributed with rate Λ, times a normalizing factor Λ. We can either fit this model directly or take the inverse Laplace transform with respect to Λ, to find the GF of the genealogy for a given divergence time T, which we denote P. (More precisely, we take the inverse Laplace transform of Λ−1ψ, since .)
The recursion is now
| (12) |
The additional term Λψ[Ω ∪ Ω] represents the replacement of the GF for two separate demes by the GF for a single population, which follows the standard coalescent (see Equation 5). Expression (12) is otherwise identical to Equation 8.
As a simple example, consider two genes,
| (13) |
which have a solution similar to Equation 10:
| (14) |
With complete isolation (i.e., M = 0), differentiation of these expressions yields the explicit formula for the numbers of pairwise differences in the complete isolation model given by Takahata et al. (1995).
For three genes we have
| (15) |
Although there are only two types of configuration with three genes, there are three permutations of the first. Thus, in our symmetric model, we have four coupled linear equations, which can be written in matrix form,
where and j is the number of pairs that can coalesce given a particular sample configuration.
This has an explicit solution, which we derive in detail in File S1 using a simple symbolic algorithm. If the demes were not equivalent because of asymmetric migration and/or differences in effective population size, then we would need to distinguish configurations such as ψ[a\b, c] and ψ[b, c\a] and would have eight coupled equations.
With coalescence or population splits alone, the recursions can be solved directly: every event leads back to a simpler configuration, with either fewer lineages or fewer demes. However, with migration, we must solve a set of coupled equations. This is easily done numerically, for specific ω, but beyond the simplest cases leads to cumbersome algebraic expressions that cannot readily be differentiated. One way around this problem (which we employ in File S1) is to condition on the topology. Another simplification is to expand the GF in M = 4Nm, writing . Then, each migration event leads back to a lower-order expression, and we can again find the solution directly. This procedure is equivalent to separating out the GF into a sum of terms, each corresponding to 0, 1, 2, … migration events.
In comparison, it is straightforward to obtain results for summaries of the genealogy from the GF. For instance, the distribution of the total number of mutations X can be found by setting all ω to be the same and taking the inverse Laplace transform (see File S1). Similarly, the probability of a particular topology can be found by taking the limit of the ωS corresponding to internal branches that are incompatible with this topology at infinity with all other ωS evaluated at zero. For a triplet with sampling configuration {a\b, c} this gives
| (16) |
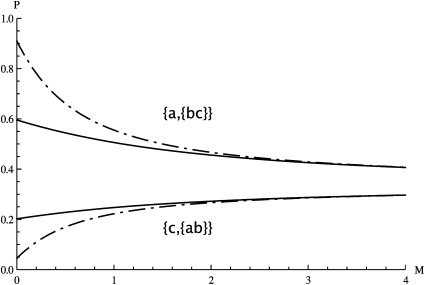
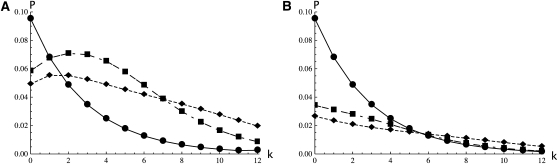
For the case of three genes in the IM model Equation 16 yields Figure 1. Furthermore, for a given topology, {a, {b, c}} say, one can find the distribution of the number of mutations on the internal branch, P[kbc|{a, {b, c}}], by differentiating the limit in Equation 16 with respect to ωbc and setting all other ωS to zero as before. Plotting these distributions (Figure 2) reveals that genealogies congruent with the sampling, i.e., with topology {a, {b, c}}, tend to have a longer internal branch than those with incongruent topologies {b, {a, c}} or {c, {a, b}} (Figure 2A vs. 2B). This is to be expected, given that coalescence events between lineages sampled from the same population, in this case {b, c}, occur relatively faster, leaving a long time tbc during which mutations can occur on the internal branch. In contrast, coalescence events between lineages sampled from different populations are likely to occur deeper in the past, within the ancestral population. These new results extend previous theory on pairwise coalescence times in the IM model (Wilkinson-Herbots 2008; Wang and Hey 2010) to topologically informative samples. Likewise, it is straightforward to use the GF to extend pairwise results for the IM model beyond the two-deme case. Larger numbers of populations (d) would be incorporated into Equation 9 by an additional term (d − 1); e.g., the rate at which pairs of lineages in different demes are brought together in the same population becomes M/(d − 1).
Figure 1 .
Topological probabilities (Equation 16) for a sample of three genes in the IM model, plotted against the scaled migration rate M for two splitting times, T = 0.5 (solid lines) and T = 2 (dashed lines). The chance of observing an incongruent genealogy with topology {c, {a, b}} or {b, {a, c}} (bottom) increases with M, as congruent topologies {a, {b, c}} (top) become less likely.
Figure 2 .
The distribution of the number of mutations (k) on the internal branches for a sample of three genes {a, {b, c}} in the IM model with symmetric migration θ = 5, M = 0.8 plotted for three different splitting times T = 0 (circles, solid line), T = 2 (squares, long-dashed line), and T = 4 (diamonds, short-dashed line). Congruent genealogies with topology {a, {b, c}} (A) tend have longer internal branches than those with incongruent topologies {c, {a, b}} or {b, {a, c}} (B). Note that for T = 0 the distributions for the two topologies are identical as expected in a panmictic population.
Recombination Between Linked Loci
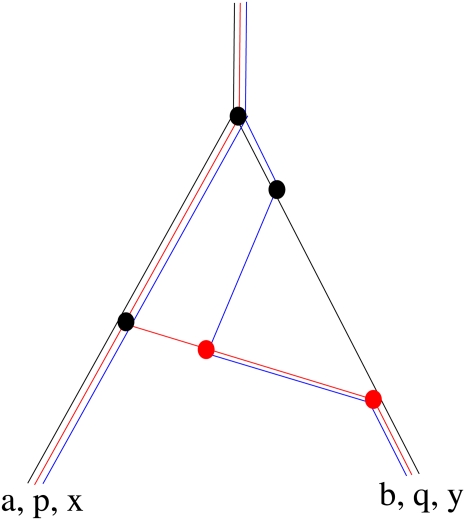
The GF method readily extends to multiple linked loci. Each individual is represented as a list, which for each locus gives the set of genes to which it is ancestral; Figure 3 gives an example with three loci. Suppose that we have k individuals, carrying lineages Ω = Ω1, … , Ωk,
| (17) |
where , i.e., we need to sum the ωS leaves over both loci and individuals, ℛR is the set of all possible recombination events and rα is the rate of a recombination of type α ε∈ ℛR. The first sum on the right in Eq. (4) is over all possible coalescences between the k individuals. At each coalescence, the lists of genes at each locus are merged. For example, a coalescence between {a, p, x} and {∅, q, ∅} gives an ancestral lineage {a, pq, x}, in which the second locus is now ancestral to genes p and q. The second sum on the right is over all possible recombination events α ε∈ ℛR, each resulting in a new set of lineages Ωα; these increase the number of lineages to k + 1. For example, a recombination in the parent of an individual {b, q, y}, between the first locus and the other two, gives two ancestral lineages {b, ∅, ∅} and {∅, q, y} (Figure 3). Note that this recursion does capture the non-Markovian nature of recombination: the distribution of coalescence times at a locus depends on the genealogies at all the other loci, not just the adjacent locus. The GF gives the joint distribution of genealogies rather than the full ancestral recombination graph (which includes additional information about which loci were carried by the ancestors).
Figure 3 .
An example of coalescence and recombination between three loci. At the present generation (bottom), there are two individuals: one carries genes a, p, x and the other carries b, q, y. Lineages ancestral to the three loci are colored black, red, and blue, respectively. This is denoted as {{a, p, x}, {b, q, y}}. Tracing back, the most recent event is a recombination (red dot) giving three individuals {{a, p, x}, {b, ∅, ∅}, {∅, q, y}}, where ∅ is the empty set. There is then another recombination event, preceded by three coalescence events (black dots); these produce the configurations {{a, p, x}, {b, ∅, ∅}, {∅, q, ∅}, {∅, ∅, y}}; {{a, pq, x}, {b, ∅, ∅}, {∅, ∅, y}}; {{a, pq, x}, {b, ∅, y}}; and {{ab, pq, xy}}. Recombination and coalescence events prior to this single common ancestor do not affect the observed genealogy.
Consider the simplest case, of two genes at two loci; when these are in two individuals, the configuration is denoted {a, x}, {b, y} and ωL = ωa + ωb + ωx + ωy:
| (18) |
By symmetry, we need only these three recursions, for the cases where the four genes are distributed over two, three, or four individuals. Note that ψ[{ab, xy}] = 1, ψ[{a, ∅}, {b, xy}] = ψ[{a}, {b}], and so on, connecting these two-locus recursions to the one-locus GF.
This has the solution
| (19) |
where , and R = 2Nr. These formulas correspond to those previously obtained by Simonsen and Churchill (1997), using a Markov chain method. For example, the covariance of coalescence times between two loci is
| (20) |
which can be found straightforwardly from the GF by taking derivatives with respect to ωa and ωx and evaluating at ω = 0, noting that E[Tab] = E[Txy] = 1:
| (21) |
This agrees with Simonsen and Churchill (1997, equation 52).
Including recombination leads to sets of coupled linear equations, whose solution involves an unwelcome matrix inversion. As with migration, this problem can be avoided by expanding in powers of R, which is equivalent to summing over histories that involve 0, 1, … recombination events. Moreover, these recombination events are uniformly distributed across the genetic map, and so we have a description of the ancestry of the whole genome and not just of two linked loci. The recursions give us the probability that there are no recombination events, that there is one event producing two blocks with different genealogies, that there are three events producing three blocks of genome, and so on. This may allow likelihoods to be calculated for short sequence blocks, provided that R is small.
Slatkin and Pollack (2006) calculate the probabilities of alternative topologies for genes at two loci in three completely isolated species; their recursion is essentially the same as ours, but tracks just the distribution of topologies rather than the full distribution of coalescence times. Since no coalescence can occur until two of the genes are brought together in the same ancestral population prior to the most recent speciation event, this reduces to the case of three linked pairs of genes in two completely isolated species. This case can be solved by the above method, by including a rate of population splits, Λ, which corresponds to the time, T, between the two speciation events.
Drosophila melanogaster–D. simulans Divergence
To illustrate the feasibility of the GF method for inference in practice, we applied it to both real and simulated data. We first reanalyzed the genomic data set of Drosophila melanogaster–D. simulans compiled and analyzed by Wang and Hey (2010), using a likelihood method for pairwise samples. The data (kindly provided by Y. Wang) consist of alignments of 30,247 blocks of intergenic sequence of 500 bp each sampled from two inbred lines of D. simulans and one inbred line each of D. melanogaster and D. yakuba (the latter used as an outgroup to account for mutational heterogeneity and, in the triplet analysis, to polarize mutations). Following Wang and Hey (2010), low-quality sequences, indels, and positions next to indels were removed. Rather than using the divergence to the outgroup to scale the mutation rate at each locus (Yang 2002; Wang and Hey 2010), each locus was trimmed after a fixed number of mutational differences between D. yakuba and D. melanogaster. We chose a cutoff of 16 divergent sites, which corresponds roughly to a third of the observed mean divergence across all loci in the full data set. A total of 2,090 loci that were below this cutoff were excluded from the analysis. Since our method assumes infinite-sites mutations, sites with more than two segregating states (12.9% of all polymorphic sites) were excluded. We also filtered out shared derived mutations that were topologically incongruent with the majority class of shared derived mutations in each block (2.5% of all polymorphic sites). A total of 2,016 loci, which contained equal numbers of topologically conflicting shared derived mutations, were excluded. The final, trimmed data set consisted of 26,141 loci. To convert scaled parameter estimates into absolute values (Ne = θ/4μ, t = g2NeT), we followed Wang and Hey (2010) and assumed that D. yakuba and D. melanogaster split 10 MYA and with a generation time per year of g = 0.1, which gives a mutation rate per block of 8 × 10−8.
Given that Wang and Hey (2010) detected a signal of gene flow from D. simulans to D. melanogaster but not in the reverse direction, we fitted an IM model with asymmetric migration. The GF for this case can be obtained using Equation 4 and, given that each genealogy can be affected by only one migration event at most, is considerably simpler than the analogous expression with symmetric migration given by solving Equation 15 (details are provided in File S1). To investigate the effect (in terms of bias and power) of including a third sample and thus topology information on parameter estimation, we performed analogous likelihood analyses on pairwise (one sample from each of D. melanogaster and D. simulans) and triplet data. To assess the effect of removing positions that violate the infinite-sites mutation model, we also ran a pairwise analysis on the full, untrimmed data set. Mutational heterogeneity in this analysis was incorporated by binning loci according to their outgroup divergence and specifying mutation rate scalars for each bin (we used 10 bins).
To speed up calculations in the triplet analysis the GF was conditioned on the topology (by taking limits as shown in Equation 16). Probabilities of all observed mutational configurations were tabulated separately for each topology class (congruent, incongruent, and topologically uninformative loci) (see File S1). Using the FindMaximum function in Mathematica, the joint likelihood of M, T, and θ can be maximized very efficiently (a few seconds or minutes for pairs or triplets, respectively). A Mathematica notebook for this calculation is provided in File S1; scripts for preprocessing input data are available from the authors on request.
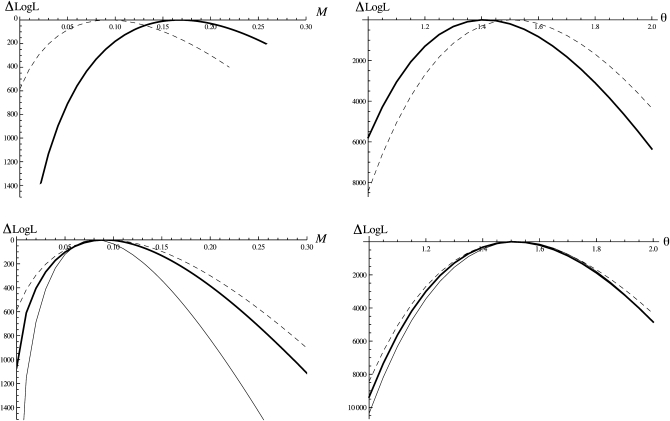
Despite the fact that we are assuming an infinite-sites mutation model [Wang and Hey (2010) used a Jukes–Cantor (Jukes and Cantor 1969) model], the results from the pairwise analysis on the full data (Table 1) agree well with those obtained by Wang and Hey (2010). As expected, our maximum-likelihood estimate (MLE) of Ne (5.5 × 106) falls in between the Ne estimates obtained by Wang and Hey (2010, Table 7) for the ancestral population (3.1 × 106) and D. simulans (5.9 × 106) (note that Wang and Hey 2010 fit a slightly more complex history with separate Ne parameters for each species). Likewise, estimates of M and T agree well with the results of Wang and Hey (2010). The trimming of back mutations and topologically incongruent mutations led to a slight decrease in Ne and increased estimates of M in the pairwise analysis. This effect was more pronounced in the triplet analysis; in particular, the MLE for M was threefold higher than the estimate of Wang and Hey (2010) (Table 1). Furthermore (and perhaps unexpectedly) we found no increase in power in the triplet analysis (Figure 4). To investigate this further, we repeated these analyses on simulated data generated using ms (Hudson 2002) under the IM history estimated for the two Drosophila species, i.e., using the MLE obtained from the pairwise analysis on the trimmed data (Table 1). In contrast to the Drosophila analyses, we found no bias in parameter estimates and higher power to estimate M and T in triplet compared to pairwise analyses of these simulated data (Figure 4). This suggests that the differences between pairwise and triplet analyses seen in the Drosophila example result from violations of the infinite-sites mutation model rather than from an inherent bias of our method. An obvious interpretation is that the use of shared derived mutations to infer the topology at each locus in the triplet analysis makes our method sensitive to misinference of ancestral states resulting from backmutations on the outgroup branch. In other words, mispolarized mutations artificially inflate the proportion of loci with incongruent topologies and hence the estimate of M. As a simple check, we can ask what the expected frequencies of congruent, incongruent, and topologically uninformative loci are (these can be derived from the GF analogous to Equation 16; see File S1). Given the MLE for trimmed pairwise and triplet analysis (Table 1), we expect 2.1% incongruent and 15.7% topologically uninformative loci on the basis of the pairwise results and 2.6% incongruent and 19.3% uninformative loci on the basis of the triplet results. However, the observed frequencies in the data set are 6.2% and 18.8% for topologically incongruent and uninformative loci, respectively. This confirms that there is an apparent (and likely artificial) excess of incongruent topologies in the data that explains the bias seen the triplet MLEs. While this illustrates the problems of assuming infinite-sites mutations when dealing with old divergence events, it is actually surprising how little effect ignoring back mutations had in this case, considering the large distance between in- and outgroup.
Table 1 . Population parameters estimated for D. melanogaster–D. simulans using 26,141 loci (data from (Wang and Hey 2010).
| Data set | θ (Ne) | M = 4Nm | T (t) | logL |
|---|---|---|---|---|
| Pair, full data | 1.85 (5.52 × 106) | 0.051 | 2.70 (2.98 × 106) | −93,466 |
| Pair, trimmeda | 1.51 (4.72 × 106) | 0.093 | 3.34 (3.15 × 106) | −65,717 |
| Triplet, trimmeda | 1.40 (4.37 × 106) | 0.174 | 3.34 (2.92 × 106) | −149,556 |
| Pair, simulated | 1.53 (4.79 × 106) | 0.098 | 3.24 (3.10 × 106) | −65,619 |
| Triplet, simulated | 1.51 (4.72 × 106) | 0.092 | 3.29 (3.11 × 106) | −151,483 |
Absolute values are in parentheses. MLEs for M and t in the pairwise analysis agree well with the results of Wang and Hey (2010) who estimated t = 3.04 and M = 0.059 (after correction for differences in scaling M). The filtering necessary to satisfy the infinite-sites model leads to a decrease in the estimate of Ne and an increase in M. The last two rows show parameters estimated from data simulated using the MLE from the pairwise analysis (boldface type).
Trimmed refers to shortening each locus to a fixed outgroup divergence and removing back mutations and topologically incongruent mutations.
Figure 4 .
Profile log-likelihood curves for M (left plots) and θ (right plots) for pairwise (dashed lines) and triplet analyses (thick solid lines) calculated from 26,141 loci for D. melanogaster and D. simulans (Wang and Hey 2010) (top row) and simulated data under an IM model with migration from D. simulans to D. melanogaster (bottom row). Analysis of the Drosophila data suggests an apparent bias of the triplet MLE of M and no improvement in power. Comparison with data simulated under the same history (using the MLE obtained in the pairwise analysis, see Table 1) shows no bias and tighter log-likelihood for the triplet analyses as expected. The improvement in power when adding a third individual is greater if this is sampled from the species receiving migrants [i.e., the reverse sampling as in the Drosophila example (thin solid lines)].
We also analyzed triplet data simulated under the reverse sampling scheme (two individuals from the species/population receiving migrants). The GF for this is slightly more complicated and is derived in File S1. The power to estimate M in this case increases substantially when analyzing triplets (Figure 4). This is expected given that most migration events will result in incongruent genealogies with relatively long internal branches.
Discussion
The GF framework provides a general method to derive likelihoods under a variety of models that include migration, changes in population structure, and recombination and applies to arbitrary sample sizes. Here our aim is to set out the method and show that it can be implemented for indefinitely large numbers of loci. So, we have focused on small samples for simplicity. Assuming that populations are exchangeable in size and rate of migration reduces both the number of parameters to be estimated and the number of configurations to track. In the case of the symmetric IM model, we do not need to distinguish the two demes, which halves the number of sample configurations. At the opposite extreme, under a highly asymmetric model with unidirectional migration (as in the Drosophila example above), each lineage in the receiving population can be affected by only a single migration event at most, which also greatly simplifies the problem. More generally, although it is possible to calculate the GF for fairly complex problems (up to six genes in the IM model, say), it is harder to extract useful information from it. Thus, while we can readily find the properties of chosen summary statistics (for example, the number of segregating sites), tabulating the probability of all observed mutational configurations is limited by their sheer number, rather than by the difficulty of finding the GF itself. These computational issues are explored in File S1, using automated recursions for the IM model with three genes.
Our GF approach is more flexible than those of Wang and Hey (2010) and Hobolth et al. (2011) in two ways. First, the recursions for a given data set can be simplified by dropping terms that are incompatible with the observed mutational pattern. This strategy is closely related to importance sampling schemes (e.g., Griffiths and Tavaré 1994). Thus, instead of summing over all possible topologies, the calculation is reduced to histories that are possible, given the data. For a sample with a fully resolved topology, the total number of terms is given by the number of configurations due to migration, so that for n = 4 and 6 there are only 28 and 124 configurations, respectively. Thus, solutions at least for symmetric cases are feasible. Second, other processes, such as recombination or changes in population size, can easily be incorporated into the GF framework. Since, under the IM model, genealogies involving migration events tend to be shorter and thus more likely to be shared between linked loci, incorporating recombination should improve inference.
Given that species may diverge gradually in space and/or ecology, it makes sense to model population separation as an explicit process, rather than an instantaneous event, followed by constant gene flow. We must distinguish here between our GF method, which calculates an average over exponentially distributed split times, and more general models that allow varying rates of gene flow. We follow the IM model in assuming that populations split abruptly and that subsequently, genes flow at a constant rate. Our initial assumption of an exponential distribution of separation times (with rate Λ) can be viewed either as a technical ruse to allow us to recover the distribution at a specific time, T, by taking an inverse Laplace transform or, in Bayesian terms, as expressing our prior beliefs about T. In reality, gene flow is likely to decrease gradually as populations diverge, and we can imagine a variety of models for the way rates of gene flow vary through time. However, even with large data sets there may be little power to detect changes in the rate of gene flow (Becquet and Przeworski 2009); the question of whether rates of gene flow vary across loci as a result of selection is yet more challenging, but crucial to identifying genes responsible for reproductive isolation (e.g., Machado 2002).
Yang (2010) recently introduced a model that is related to both approaches just described. This assumes that populations separate suddenly, with no subsequent gene flow, but that the split time varies across loci, following a beta distribution—which can be regarded as an approximation to a biologically feasible model in which migration causes variation in coalescence time across loci. This is related to, but different from, our assumption of an exponential rate, Λ, of separation times. If, following Yang (2010), we assumed exponentially distributed split times across loci, we would fix Λ to find the probability of mutational configurations. On the other hand, if we assumed a definite separation time T, we would take the inverse Laplace transform at T and calculate the probabilities from that. If we then averaged the multilocus likelihood over a prior distribution of T, we would get a quite different result from that yielded by Yang’s (2010) procedure.
As our application to the Drosophila data demonstrates, the GF method outlined here provides an efficient way to calculate and maximize the joint likelihood of divergence parameters from very many nonrecombining blocks of sequence for topologically informative samples. Not only do triplet samples (as opposed to pairs) give better information about branch lengths but also, more importantly, the joint distribution of topologies and branch lengths provides qualitatively new information about historical parameters. As our simulation example demonstrates, dependent on the sampling scheme, this substantially increases power. Our analytic solutions have three key advantages over previous methods. First, the probabilities of mutational configurations need to be tabulated only once, so in contrast to simulation-based methods computation time does not increase with the number of loci and an indefinite number of loci can be analyzed. Second, derivatives can be used to maximize the joint log-likelihood, which greatly speeds up calculations. Thus our computation takes a fraction of the time of, for example, an IMa analysis (Hey and Nielsen 2004) on a handful of loci and is also more efficient than the numerical method of Wang and Hey (2010) (Y. Wang, personal communication). Finally, the GF method allows us to separate topology and branch length information, which provides a way to incorporate additional sources of information. For example, topology information contained in the patterns of shared derived indels could be included without the need to model indel evolution explicitly.
In practice, however, our method is currently limited to the infinite-sites mutation model and thus can deal with only relatively recent divergence events for which close outgroups are available. However, it is encouraging how small the bias resulting from assuming infinite-sites mutations is in the Drosophila example, despite the considerable divergence of the outgroup. Fortunately, researchers are commonly interested in fitting IM histories to sister taxa or populations that have diverged much more recently than the Drosophila species analyzed here (and for which more closely related outgroups are available). The use of multiple outgroups to correct for misinferred ancestral states should also help to overcome this problem. Another limitation is that the GF can be used to find exact solutions only if the number of mutations per genealogical branch is relatively small (e.g., the most diverse locus in the trimmed Drosophila data set contained 26 mutations). For much larger numbers of mutations per block, numerical calculations, which involve finding the coefficients in a series expansion, become unfeasible. Although it may be possible to use a Gaussian approximation in this case, the assumption of no recombination within blocks restricts our and related methods (Hey and Nielsen 2004; Wang and Hey 2010) to short blocks of sequence anyway, so this may not be relevant in practice.
Implementing efficient inference schemes for biologically realistic histories clearly requires further work. For instance, it would be worthwhile to extend our inference scheme to the general IM model (i.e., allowing for asymmetric migration in both directions and different population sizes) and more realistic mutation models and incorporate recombination explicitly. In contrast, the catastrophic increase of possible sample and mutational configurations with the number of individuals frustrates full results for large numbers of individuals. Nevertheless, full results for small but topologically informative samples under a range of models of structure and history should be of considerable interest for at least three reasons: first, although thorough investigations of the trade-offs of various sampling schemes are lacking, it is clear that in general replication across loci is far more profitable than analyzing a few loci sampled from a large number of individuals (Felsenstein 1992; Li and Durbin 2011). Second, minimal sampling in terms of individuals reflects the practical limitations of current sequencing technologies. While massively paralleled sequencing has made it affordable to sequence small numbers of genomes in any organism, obtaining multilocus sequence data for many individuals remains challenging in nonmodel organisms. Finally, under a wide range of models of population structure, large samples quickly coalesce down to a few lineages that dominate their genealogical history, allowing a separation of timescales to be applied (Wakeley 2009). Thus, we envisage that new analytic solutions of simple cases, such as those derived here for the total number of mutations and topological probabilities of triplets under the IM model, will provide a guide to the development of approximate methods (involving importance sampling and summary statistics) with wide applicability.
Acknowledgments
We thank Yong Wang and Jody Hey for sharing the Drosophila data set. We also thank two anonymous reviewers and Jerome Kelleher for thoughtful comments on earlier versions of this manuscript. This work was supported by a grant from the European Research Council (250152) (to N.B.) and a grant from the United Kingdom Natural Environment Research Council (NE/I020288/1) (to K.L.).
Literature Cited
- Becquet C., Przeworski M., 2009. Learning about modes of speciation from computational approaches. Evolution 63(10): 2547–2562 [DOI] [PubMed] [Google Scholar]
- Felsenstein J., 1992. Estimating effective population size from samples of sequences: inefficiency of pairwise and segregating sites as compared to phylogenetic estimates. Genet. Res. 59: 139–147 [DOI] [PubMed] [Google Scholar]
- Griffiths R. C., 1981a. The number of heterozygous loci between two randomly chosen completely linked sequences of loci in two subdivided population models. J. Math. Biol. 12: 251–261 [Google Scholar]
- Griffiths R. C., 1981b. Transient distribution of the number of segrating sites in a neutral infinite-sites model with no recombination. J. Appl. Probab. 18: 42–51 [Google Scholar]
- Griffiths R. C., 1991. The two-locus ancestral graph, pp. 100–117 in, editors, Selected Proceedings of the Symposium of Applied Probability, edited by I. V. Basawa and R. I. Taylor. Institute of Mathematical Statistics, Haywards, CA [Google Scholar]
- Griffiths R. C., Tavaré S., 1994. Sampling theory for neutral alleles in a varying environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 344(1310): 403–410 [DOI] [PubMed] [Google Scholar]
- Herbots H., 1997. The structured coalescent, pp. 231–255 in Progress in Population Genetics and Human Evolution (IMA Volumes in Mathematics and Its Applications, No. 87), edited by P. Donelly and S. Tavare. Springer-Verlag, Berlin/Heidelberg, Germany/New York [Google Scholar]
- Hey J., Nielsen R., 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167: 747–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobolth A., Andersen L. N., Mailund T., 2011. On computing the coalescent time density in an isolation-with-migration model with few samples. Genetics 187: 1241–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338 [DOI] [PubMed] [Google Scholar]
- Jenkins P. A., 2008. Importance sampling on the coalescent with recombination. Ph.D. Thesis, Oxford University, Oxford [Google Scholar]
- Jukes T. H., Cantor C. R., 1969. Evolution of protein molecules, pp. 21–123 Mammalian Protein Metabolism, edited by Munro H. N. Academic Press, New York [Google Scholar]
- Latter B. D. H., 1973. The island model of population differentiation: a general solution. Genetics 73: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357): 493–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado C. A., 2002. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Mol. Biol. Evol. 19: 472–488 [DOI] [PubMed] [Google Scholar]
- Simonsen K. L., Churchill G. A., 1997. A Markov chain model of coalescence with recombination. Theor. Popul. Biol. 52: 43–59 [DOI] [PubMed] [Google Scholar]
- Slatkin M., Pollack J. L., 2006. The concordance of gene trees and species trees at two linked loci. Genetics 172: 1979–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Satta Y., Klein J., 1995. Divergence time and population size in the lineage leading to modern humans. Theor. Popul. Biol. 48: 198–221 [DOI] [PubMed] [Google Scholar]
- Wakeley J., 1996. Pairwise differences under a general model of subdivision. J. Genet. 75(1): 81–89 [Google Scholar]
- Wakeley J., 2009. Coalescent Theory. Roberts & Co., Greenwood Village, CO [Google Scholar]
- Wang Y., Hey J., 2010. Estimating divergence parameters with small samples from a large number of loci. Genetics 184: 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson-Herbots H. M., 2008. The distribution of the coalescence time and the number of pairwise nucleotide differences in the “isolation with migration” model. Theor. Popul. Biol. 73(2): 277–288 [DOI] [PubMed] [Google Scholar]
- Yang Z., 2002. Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci. Genetics 162: 1811–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2010. A likelihood ratio test of speciation with gene flow using genomic data. Genome Biol. Evol. 2: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]