Abstract
Allopolyploidy is an important process during plant evolution that results in the reunion of two divergent genomes into a common nucleus. Many of the immediate as well as longer-term genomic and epigenetic responses to polyploidy have become appreciated. To investigate the modifications of gene expression at the proteome level caused by allopolyploid formation, we conducted a comparative analysis of cotton seed proteomes from the allopolyploid Gossypium hirsutum (AD genome) and its model A-genome and D-genome diploid progenitors. An unexpectedly high level of divergence among the three proteomes was found, with about one-third of all protein forms being genome specific. Comparative analysis showed that there is a higher degree of proteomic similarity between the allopolyploid and its D-genome donor than its A-genome donor, reflecting a biased accumulation of seed proteins in the allopolyploid. Protein identification and genetic characterization of high-abundance proteins revealed that two classes of seed storage proteins, vicilins and legumins, compose the major component of cotton seed proteomes. Analyses further indicate differential regulation or modification of homoeologous gene products, as well as novel patterns in the polyploid proteome that may result from the interaction between homoeologous gene products. Our findings demonstrate that genomic merger and doubling have consequences that extend beyond the transcriptome into the realm of the proteome and that unequal expression of proteins from diploid parental genomes may occur in allopolyploids.
GENOME doubling, or polyploidization, is a phenomenon prevalent in eukaryotes and particularly in higher plants. Genomic studies indicate that all angiosperm species have undergone at least two rounds of polyploidization during their evolutionary history, with most lineages having experienced additional whole-genome duplications (Cui et al. 2006; Jiao et al. 2011). Allopolyploid species are particularly intriguing in that their formation entails the merger of diverged genomes, which often results in myriad dramatic and large-scale genomic and transcriptomic responses (Wendel 2000; Comai 2005), including structural and epigenetic modifications (Shaked et al. 2001; Gaeta et al. 2007; Buggs et al. 2009; Ha et al. 2009; Schnable et al. 2011), as well as changes in gene expression (Wang et al. 2006; Bottley and Koebner 2008; Flagel et al. 2008, 2009; Hovav et al. 2008; Rapp et al. 2009; Flagel and Wendel 2010; Koh et al. 2010). Compared to their progenitors, polyploids often display different physiological, morphological, and ecological phenotypes (Pires et al. 2004; Gaeta et al. 2007; Anssour et al. 2009; Ni et al. 2009; Ramsey 2011), which suggests functional and phenotypic evolution may be driven by these genomic changes.
Notwithstanding these and other recent insights into the genomic and transcriptomic consequences of genomic merger and doubling, the fate of translated gene products, i.e., the proteome, remains poorly studied in the context of polyploidization. Because protein levels are influenced by post-translational processing and inherent variation in stability, it is difficult to infer the representation and regulation of proteins and participating metabolic pathways from transcriptomic data alone, and the correlation between protein and transcript expression levels has been shown to vary extensively depending on the system being analyzed and the profiling approach used (Hajduch et al. 2010). As proteins represent the key players in cellular activities, characterizing the proteome using appropriately targeted approaches constitutes an important component of the evolutionary analysis of polyploidy and its consequences. A classical proteomic technique, two-dimensional gel electrophoresis (2-DE), has the potential to assess the expression patterns of proteins displayed by polyploid species relative to their diploid progenitors, as demonstrated in Brassica (Albertin et al. 2005, 2006, 2007). This technique allows the resolution of hundreds of protein spots within a single gel, which are accessible to identification through mass spectrometry (MS) analysis; moreover, some post-translational modifications corresponding to protein activities can be inferred via interpretation of the on-gel and MS properties. This comparative quantification of resolved spot profiles permits a proteome-scale comparison of the polyploid and its parental species.
Over the past decade, Gossypium has emerged as a model for studies of polyploidy, particularly with respect to the genomic and transcriptomic consequences of allopolyploidization (Adams et al. 2003; Senchina et al. 2003; Grover et al. 2004, 2007; Flagel et al. 2008, 2009; Hovav et al. 2008; Chaudhary et al. 2009; Rapp et al. 2009; Flagel and Wendel 2010; Salmon et al. 2010). As shown in Figure 1A, A- and D-genome Gossypium diverged for ∼5–10 million years before becoming reunited in an allopolyploid nucleus ∼1–2 million years ago (Wendel and Cronn 2003). Extensive research has identified the best models of the diploid progenitors involved in the creation of the allopolyploid lineage, the latter including the most important of the cultivated species, Gossypium hirsutum. This well-documented evolutionary framework, coupled with the substantial resources available, e.g., a comprehensive EST database (Udall et al. 2006a,b), and the prior genomic/transcriptomic research into the consequences of polyploidy (Adams et al. 2003; Senchina et al. 2003; Grover et al. 2004, 2007; Flagel et al. 2008, 2009; Hovav et al. 2008; Chaudhary et al. 2009; Rapp et al. 2009; Flagel and Wendel 2010; Salmon et al. 2010), makes Gossypium an excellent system to extend research on genomic merger and doubling to the proteomic level. In this study, we profile and analyze the proteomes of cotton seeds in the polyploid (AD genome) G. hirsutum and its two model diploid progenitors, G. herbaceum (A genome) and G. raimondii (D genome). Despite being best known for fiber production, the high-quality oil and proteins produced in the seeds of domesticated G. hirsutum have increased the agronomic and economic importance of cotton as a crop plant. The increased interest in cotton seeds (e.g., Sunilkumar et al. 2006) and the relatively simplified protein composition of mature, dormant seeds make cotton a useful model for studying protein accumulation in the context of polyploidy.
Figure 1 .
Evolutionary history of Gossypium species. (A) Phylogenetic framework of diploid and allopolyploid Gossypium, illustrating the divergence of the A- and D-genome diploids and their polyploidization leading to the evolution of the AD-genome allopolyploid cottons. (B) Morphology of mature cottonseeds. Key to species: AD, G. hirsutum var. Acala Maxxa; A, G. herbaceum (A1-73); D, G. raimondii. Shown are seeds with (top) and without (bottom) lint.
Materials and Methods
Plant materials
Three Gossypium species were used in the present study: one polyploid species G. hirsutum var. Acala Maxxa (AD genome) and two diploid species that represent the model diploid progenitors of allopolyploid cotton, namely G. herbaceum (A1-73; A genome) and G. raimondii (D genome). For each species, seeds were collected and pooled from three to four plants that were grown in the Pohl Conservatory at Iowa State University, Ames, Iowa. After boll opening, mature seeds were hand harvested and air dried at room temperature for at least 1 month. Prior to protein extraction, the fiber-containing seeds were delinted with concentrated sulphuric acid. The weight of the seeds collected was measured both before and after delinting.
Protein extraction
Total protein was isolated from mature desiccated seeds on the basis of a phenol extraction procedure successfully applied to other oilseeds (Hajduch et al. 2005, 2006, 2007; Houston et al. 2009). Briefly, for each sample, 250 mg of delinted seeds was ground to a fine powder in liquid nitrogen with a mortar and pestle and homogenized with 10 ml of a 1:1 mixture of extraction buffer [100 mM Tris-HCl, pH 8.8, 900 mM sucrose, 10 mM EDTA, and 0.4% (v/v) 2-mercaptoethanol] and Tris-saturated phenol. The homogenate was agitated for 30 min and centrifuged at 4000 rpm for 30 min at 4°. The upper phenol phase was extracted and combined with 5 vol of 0.1 M ammonium acetate in methanol and placed at −20° for overnight protein precipitation. The protein pellet was subsequently collected via centrifugation for 30 min at 4°. The recovered pellet was thoroughly washed over four washing steps: once with 0.1 M ammonium acetate in methanol, twice with 80% acetone, and once with 70% ethanol. The final pellet was dried at room temperature and solubilized in isoelectric focusing (IEF) buffer [8 M urea, 2 M thiourea, 2% (w/v) 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate, 2% (v/v) Triton X-100, 50 mM DTT]. Protein concentration was determined using the modified Bradford total protein assay (Bradford 1976) from Bio-Rad (Hercules, CA), using bovine gamma globulin as standard.
2-DE
As described in Hajduch et al. (2005), 1 mg of seed proteins was dissolved in 450 μl IEF buffer and separated by IEF in the first dimension and by SDS–PAGE in the second dimension. IEF was performed in a Bio-Rad PROTEAN IEF System using 24-cm linear immobilized pH gradient (IPG) strips with pH ranges of 4–7 and 3–10 (GE Healthcare), using the following conditions: active rehydration at 50 V for 10 hr, 100 V for 100 V hr, 500 V for 500 V hr, and 8000 V for 99 kV hr. After completion of IEF, the strips were prepared for SDS–PAGE as follows: the strips were reduced for 15 min with 2.0% (w/v) DTT in equilibration buffer [6 M urea, 50 mM Tris-HCl, pH 8.8, 30% (v/v) glycerol, 2% (w/v) SDS], alkalized for 15 min with 2.5% (w/v) iodoacetamide in equilibration buffer, rinsed with SDS running buffer [1.5 M Tris-HCl, 6 M urea, 30% (v/v) glycerol, 5% (w/v) SDS], and then transferred onto 12% self-cast polyacrylamide gels. The second-dimension SDS–PAGE was performed in an Ettan DALT 12 System (GE Healthcare), using 1 W/strip for 1 hr and 2 W/strip for 15 hr. Finished gels were washed twice in deionized water for 10 min and stained overnight with colloidal Coomassie [20% (v/v) ethanol, 1.6% (v/v) phosphoric acid, 8% (w/v) ammonium sulfate, 0.08% (w/v) Coomassie Brilliant Blue G-250]. Stained gels were stored in 250 ml storage solution [10% (v/v) Colloidal Coomassie solution, 0.02% (w/v) sodium azide] per gel at 4°.
Image and statistical analysis
Gels were imaged with a ScanMaker 9800XL (Microtek, Carson, CA), using a resolution of 300 dpi and 16-bit grayscale pixel depth. Image analysis was conducted with ImageMaster 2D platinum software version 6.0 (Amersham Biosciences, Uppsala, Sweden), which allows spot detection, quantification, and cross-image spot matching. Using the built-in normalization method implemented in the ImageMaster 2D platinum software, spot expression was represented by relative spot abundance, dividing each absolute spot volume by the total volume of all spots selected for analysis. For spots shared by all three genomes, differential protein expression was tested using a one-way ANOVA model with a fixed effect: Yij = µ + Gi + eij, where µ represents the overall mean, Gi denotes a genome fixed effect, and eij is the random error term used for significance test. When spots were shared by all three genomes, the hypothesis of additive parental expression in the allopolyploid was tested; for this, a spot was considered additive with respect to expression if the spot abundance in the polyploid AD genome was equal to the average abundance found in the parental A and D genomes. Any deviation from the average parental value was considered nonadditive expression, which then was further categorized by comparing the AD-genome value to both diploid values. Possible deviations from additivity include parental genomic dominance (where the expression found in the AD genome is statistically equal to one parental value for spots differentially expressed between parents) and transgressive expression (where the expression in the polyploid AD genome statistically falls either below or above that found in both parental genomes). The P-values of these analyses were adjusted for multiple testing (Benjamini and Hochberg 1995), and the false discovery rate (FDR) was controlled at 5%.
MS analyses
Selected spots for protein identification were excised from gels and subjected to in-gel trypsin digestion followed by liquid chromatography–tandem mass spectrometry (LC-MS/MS), using an LTQ XL ProteomeX ion trap mass spectrometer (Thermo-Fisher, San Jose, CA). Tandem mass spectral data were searched against the Arabidopsis protein database and an in-house Gossypium EST/contig translated database (provided by J. Udall, Department of Plant and Animal Sciences, Brigham Young University, Provo, Utah), using SEQUEST, which is part of the BioWorks 3.1SR1 software suite (Thermo-Fisher). The instrument and searching parameters were applied according to Hajduch et al. (2006).
Genetic analysis
Primer sequences (Supporting Information, Table S1) designed to amplify the suite of seed storage protein (SSP) genes were derived from the conserved regions of each SSP gene in Gossypium, identified from the alignment of publicly available G. hirsutum sequences (GenBank accession nos. M19378.1, M16891.1, M69188.1, and M16936.1) and SSP ESTs (identified by blast hits against an in-house cotton EST database). Amplified PCR products were excised from 1.0% agarose gels, purified using a Qiaquick gel purification kit (Qiagen, Valencia, CA), and cloned with the pGEM-T Easy Vector System (Promega, Madison, WI). Cloned products were sequenced using the Applied Biosystems 3730xl DNA Analyzer at the Iowa State University DNA facility. Because the PCR products are ∼2000 bp in length, internal primers were also designed for sequencing. The resulting sequences were aligned with those derived from GenBank and the cotton EST database using MUSCLE (Edgar 2004) and inspected manually. The obtained Gossypium SSP gene sequences were deposited in GenBank under accession nos. JN602029–JN602047. Neighbor-joining analysis was conducted on the aligned sequences using PAUP* (Swofford 2001). Uncorrected (“p”) DNA/RNA distances were set for distance analysis and missing data were ignored for affected pairwise comparisons.
Results
The proteomic profiles of mature cotton seeds
Allopolyploid G. hirsutum var. Acala Maxxa (AD genome) and representatives of its diploid progenitors (A genome G. herbaceum and D genome G. raimondii) were used to profile the cotton seed proteome. Mature seed mass and size of intact and delinted seeds were recorded before protein extraction (Figure 1B). The protein yields from phenol extraction ranged from 6.3 to 8.9% (dry weight) without significant variation among three genomes, which is in agreement with the seed protein contents previously reported for Gossypium (Frampton et al. 1958; Pandey and Thejappa 1975).
Mature seed proteins from each genome were isolated in biological quadruplicate for 2-DE separation (Figure S1). To construct a high-quality proteomic map, two overlapping ranges of IPG strips were used for the first-dimensional IEF: a broader-range pH 3–10 strip and a narrower-range pH 4–7 strip. As shown in Figure 2A, the use of pH range 4–7 largely enhanced spot resolution in the signal-dense area of pH 3–10; therefore, the proteomic profiles were constructed using spots detected in the pH 4–7 gels and the subsection of the pH 3–10 gels containing the pH 7–10 region. For each gel of each pH range, detected spots were matched within biological replicates and then between genomes. Spot detection was considered only for spots reproducibly represented by at least three biological replicates; spots meeting this criterion were selected for profiling and subjected to qualitative and quantitative analyses. According to this criterion, 646 spots were confidently detected from pH 4–7 gels, and 208 spots were resolved from pH 3–10 gels (pH 7–10 region only). Of the 854 total spots, 315 were present in the polyploid (AD-genome) seed proteome, and fewer were recovered from the A- and D-genome seed proteomes (250 and 289 spots, respectively).
Figure 2 .
Proteomic profile of G. hirsutum seeds. (A) Experimental design for two-dimensional gel electrophoresis using two pH ranges, 3–10 and 4–7. (B) Synthetic proteome map of G. hirsutum constructed with images from pH 4–7 (left) and pH 3–10 (pH 7–10 region only; right). Identified protein spots are indicated as follows: vicilin A, circle; vicilin B, rectangle; vicilin-like, parallelogram; legumin A, diamond; legumin B, triangle; others, cross.
Comparative proteomics of allopolyploid G. hirsutum and models of its diploid progenitors
Proteomic profiles of the allopolyploid and two progenitor diploid species were first compared qualitatively through spot matching between genomes (Table 1 and File S1). The pairwise comparison between genomes revealed that only 92 spots were observed in all cotton seed proteomes, corresponding to 29.0%, 36.8%, and 32.2% of the total spots detected from the AD, A, and D genomes, respectively. Surprisingly, given the high degree of genetic similarity between A- and D-genome orthologs (Senchina et al. 2003), about one-third of the spots from each genome were found to be genome specific. This result shows that considerable proteomic variation exists not only between diploid species but also among polyploid and diploid genomes in Gossypium. Spots represented by only two genomes were also noted, and interestingly, 26.2% of AD-genome spots were found in the D genome, whereas only 10.7% of the polyploid proteome was represented by the A genome, suggesting a higher similarity between AD- and D-genome seed proteomes than between the AD- and A-genome proteomes. This compositional bias toward the D genome suggests an unequal contribution of the diploid genomes to global protein expression in cotton seeds, a potential response to hybridization and genome doubling similar to that experienced by the transcriptome (Flagel et al. 2008; Rapp et al. 2009).
Table 1 . Qualitative comparison of seed proteomes of allopolyploid G. hirsutum (AD genome) and its parental diploid A and D genomes.
| Pattern of qualitative expression | No. | A genome | D genome | AD genome |
|---|---|---|---|---|
| Shared spots in all three genomes | 92 | 36.8% | 32.2% | 29.0% |
| Genome-specific spots | ||||
| A-specific | 78 | 31.2% | ||
| D-specific | 65 | 22.7% | ||
| AD-specific | 108 | 34.1% | ||
| Spots found in two genomes | ||||
| A and D | 46 | 18.4% | 16.1% | |
| A and AD | 34 | 13.6% | 10.7% | |
| D and AD | 83 | 29.0% | 26.2% | |
| Total no. of spots | 250 | 286 | 317 |
In addition to the qualitative variation observed in the 2-DE spot patterns, differential quantitative expression of shared spots (illustrated in Figure 3) was analyzed. Quantitative changes attributed to polyploidy were inferred by testing the additivity of parental contributions to the allopolyploid proteome and by classifying the expression patterns (see examples in Figure 4) using a two-step procedure. This procedure first tested for nonadditivity, i.e., spots that deviated significantly in abundance from the average of the parental diploids. These nonadditive spots were further categorized by comparison to their homologous expression levels in both parents (Table 2). Of the 92 common spots analyzed, 33 spots (35.9%) were detected as nonadditive, among which 8 spots were expressed transgressively; that is, their expression was either greater or less than that of both parental diploids. Twenty-two nonadditive spots displayed statistically equivalent expression as one of the two diploid parents, and 13 and 9 spots were sorted into the D-dominant and A-dominant patterns, respectively. The remaining 3 spots displayed an intermediate level of expression between the parental values and were considered codominant.
Figure 3 .
Differential expression patterns of cotton seed proteomes. Partial 2-DE gels are shown (pH 5–6, molecular weight 26–17 kDa) of AD-genome and diploid A- and D-genome seed proteomes. Spots shared by all species and having consistent expression levels are shown as black circles. Variation in either expression level or presence is indicated by color, where red denotes expression in the allopolyploid and blue and gold represent expression levels in the A- and D-genome diploids, respectively. Circle sizes correspond to spot volumes.
Figure 4 .
Representative 2-DE gels illustrating additive and nonadditive quantitative expression patterns. Proteins 621, 616, 627, and 1437 display additive patterns. Proteins deviating from statistical additivity (see text for details) were further categorized as follows: 626 and 636, D dominant; 1345, A dominant; 657, transgressive expression above that of both diploids; 1321, transgressive expression below that of both diploids. These proteins were identified by mass spectrometry as vicilin A (657 and 1321), vicilin B (1437), and legumin A (621, 616, 627, 636, and 1345).
Table 2 . Quantitative analysis of protein additivity of shared spots in allopolyploid Gossypium seed proteomes.
| Pattern of shared spots | No. | Percentage |
|---|---|---|
| Additive | 59 | 64.1 |
| Nonadditive | 33 | 35.9 |
| Higher than both diploids | 6 | 6.5 |
| A-genome dominant | 9 | 9.8 |
| Codominant | 3 | 3.3 |
| D-genome dominant | 13 | 14.1 |
| Lower than both diploids | 2 | 2.2 |
| Total no. of spots | 92 |
Identification of major components of cotton seed proteomes
To characterize and compare the major components of seed proteomes in the three Gossypium species, high-abundance spots (>1% mean relative volume of each genome at each pH range) were targeted for mass spectrometry (MS)-based protein identification. Because it is also of interest to determine the proteins or functional categories that contribute to the expression patterns observed in the polyploid, representative spots from the expression categories defined in Table 2 were also included for protein identification. According to these nonexclusive criteria, a total of 199 spots corresponding to ∼80% of the total spot abundance for the three species were subjected to tandem MS analysis. Searching against a custom Gossypium and Arabidopsis protein database successfully identified 155 spots (62 from the AD genome, 55 from the A genome, and 53 from the D genome; see File S2 for spot selection and identification). The majority of the identified spots (140 spots) belonged to the category of SSPs, including vicilin A (19 spots), vicilin B (5 spots), legumin A (83 spots), and legumin B (27 spots), and one vicilin-like gene (6 spots) not previously reported in Gossypium. The remaining spots (9.7%, 15/155) identified were classified to the functional categories of cellular organization (4 spots), molecular function (4 spots), and stress response (3 spots).
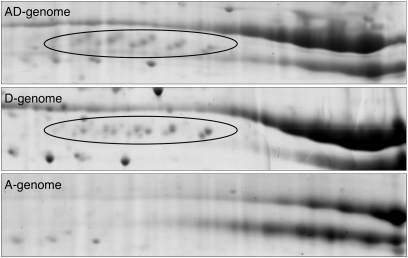
Due to the experimental design of using two IEF pH ranges, the relative expression of identified spots was independently profiled from pH ranges 4–7 (pH 4–7 gels) and 7–10 (subsection of the pH 3–10 gels). To estimate the overall protein composition of each cotton seed proteome, the percentages of spot abundances for pH 4–7 or pH 7–10 subproteomes were normalized by their composition relative to the full pH 4–10 range (0.4 to pH 4–7, 0.6 to pH 7–10; calculated using the ratio of total spot abundances of pH 4–7 and 7–10 subsections on the pH 3–10 gels) and summed for the identified proteins, as shown in Table 3. Two principal SSP families, vicilin and legumin, constituted a major fraction of the cottonseed proteomes, representing 71%, 68%, and 72% of the total protein in the AD-, A-, and D-genome species, respectively. Although the overall accumulation of SSPs appears constant among the three species, variation in relative expression of each individual SSP was observed among the diploid parents and the polyploid. For example, vicilin A was more highly expressed in the AD genome (19% of the total protein abundance) than in its diploid progenitors (13% and 10% in the A and D parents, respectively); in contrast, the allopolyploid species utilized less legumin A (18%) as a nutrient reservoir than either of the two diploid species (26% and 33% in the A and D parents, respectively). Moreover, the composition of both legumin B and the vicilin-like protein in the AD-genome seeds displayed a similar expression pattern to the D-genome progenitor, which could clearly be distinguished from the pattern of the A-genome progenitor (Table 3). This previously unreported vicilin-like protein was identified via BLAST homology to a Pistacia vera vicilin protein (GenBank accession no. ABO36677.1) and was detected only in the AD- and D-genome species, while no corresponding spots or peptides were detected in the A-genome species (Figure 5); this pattern was also evident by surveying the Gossypium EST database (data not shown). This indicates that expression of this vicilin-like gene may be specific to the diploid D-genome species and was subsequently recruited into AD-genome species during or postallopolyploidization.
Table 3 . Composition of cotton seed proteomes (percentage of abundance ± SE).
| AD genome: G. hirsutum | A genome: G. herbaceum | D genome: G. raimondii | |
|---|---|---|---|
| Vicilin A | 19.14 ± 1.53 | 13.06 ± 2.48 | 10.41 ± 1.95 |
| Vicilin B | 23.20 ± 3.89 | 24.66 ± 1.64 | 18.03 ± 2.04 |
| Legumin A | 18.20 ± 2.27 | 25.78 ± 4.90 | 32.94 ± 2.75 |
| Legumin B | 10.34 ± 1.14 | 5.95 ± 0.75 | 10.06 ± 0.87 |
| Vicilin-like | 0.20 ± 0.09 | 0 | 0.13 ± 0.04 |
| Non-SSP | 0.63 ± 0.19 | 0.36 ± 0.04 | 0.79 ± 0.21 |
| Unknowna | 7.26 ± 1.54 | 4.73 ± 1.13 | 6.48 ± 0.86 |
| Total | 78.96 ± 2.00 | 74.54 ± 2.43 | 78.85 ± 1.51 |
Spots were selected for protein identification, but no matched peptides were retrieved from databases.
Figure 5 .
2-DE gels of Vicilin-like isoforms. Vicilin-like isoforms are indicated by ovals.
Genetic analysis of Gossypium SSP genes
The major Gossypium SSPs have previously been characterized (Dure and Chlan 1981; Galau et al. 1983; Chlan et al. 1986, 1987; Dure 1989; Galau et al. 1991), although complete gene sequences were limited to G. hirsutum. In surveying the Gossypium EST databases, a considerable level of nucleotide diversity became evident, not only for the orthologous genes obtained from the diploid species, but also for copies found in the G. hirsutum EST database. These data indicated that some of the major SSPs in Gossypium are encoded by multigene families (data not shown). Thus, to understand the genetic basis of the proteomic profiles and their compositional diversity, gene family structures were characterized using conserved primers designed to amplify each SSP from four Gossypium species: AD-genome G. hirsutum var. Acala Maxxa, D-genome G. raimondii, and two A-genome species G. herbaceum (which was used in the proteomic analysis, noted as A1 here) and G. arboreum (another putative A-genome progenitor of allopolyploid cotton, noted as A2, included for additional perspective on the genetic diversity of Gossypium SSP genes).
Vicilin A and vicilin B, which share 72% amino acid similarity, belong to the vicilin (7S globulin or α-globulin) gene family (Chlan et al. 1987) and represent the first discovered cotton SSPs. To test the single-copy status for both vicilins, >10 sequences per gene were generated from each of the four Gossypium species mentioned above. After removing sequencing errors and redundancy, both vicilins were determined to exist as single-copy genes in the diploid genomes (A1, A2, and D), corresponding to two homoeologous copies (AT deriving from the A-genome progenitor, modeled by A1 or A2; DT deriving from the D-genome progenitor) that were retained in the allopolyploid AD genome. Gene trees for both vicilin genes were generated using the neighbor-joining method (Figure 6), and the same tree topologies were resolved using maximum-likelihood and maximum-parsimony methods (data not shown). These trees also were congruent with the phylogeny of Gossypium (Cronn et al. 2002; Wendel and Cronn 2003) shown in Figure 1A. Nucleotide variation, including indels and nonsynonymous and synonymous substitutions, were identified among the orthologous and homoeologous gene copies for each gene (see File S3 for gene sequences and alignments). As noted on the branches of gene trees, the majority of substitutions were inferred to have occurred since the divergence of the A and D genomes from their common ancestor, with a few lineage-specific substitutions having arisen after polyploidization 1–2 million years ago. The AT copy of both vicilin genes exhibited less lineage-specific nucleotide substitution than did the DT copies in the polyploid AD genome, which likely is explained by the fact that the A-genome diploid species used are better models of the actual A-genome progenitor than the D-genome diploid is of the actual D-genome progenitor of allopolyploid cotton (Senchina et al. 2003).
Figure 6 .
Neighbor-joining trees of SSP genes in Gossypium. Numbers of total and nonsynonymous (in parentheses) nucleotide substitutions are indicated on branches. Those distinguishing the (A, AT) from (D, DT) clades are unpolarized and hence are shown at the root of each tree. AT and DT refer to homoeologous copies in the allopolyploid genome. The symbol * indicates a sequence with a stop codon.
The legumin (11-12S globulin or β-globulin) gene family is the other major SSP group found in Gossypium, and its members, legumin A and legumin B, are more diverged compared to the vicilin gene family members, sharing only 58.5% similarity in amino acid sequences. Multiple sequences of legumin A and legumin B were also generated and characterized from allopolyploid and diploid Gossypium. As with the vicilins, both legumins were also found as single-copy genes in the diploid genomes; however, only the D-genome–derived copy was detected for legumin B in the AD genome. This observation was further supported by the absence of an A-genome–derived copy in the cotton EST database. Two possible explanations exist for the loss of the original A-genome–derived copy in the polyploid: gene deletion and concerted evolution that resulted in the homogenization of the homoeologous pair toward the D-genome–derived copy, a phenomenon previously demonstrated for ribosomal genes in allotetraploid Gossypium (Wendel et al. 1995) and more recently for numerous protein-coding genes (Salmon et al. 2010). Interestingly, despite the fact that both A- and D-genome–derived copies were recovered for legumin A, a nonsynonymous substitution in the A-genome–derived copy of legumin A caused a premature stop codon. Additionally, an accelerated rate of nucleotide substitution in the legumin A A-genome–derived copy was also observed (Figure 6), which suggests that this copy is nonfunctional. Together, these observations suggest that different regulatory mechanisms and uneven selection pressures exist on vicilin and legumin genes, even though they both function as storage proteins in cotton seeds.
Detailed proteomic characterization of Gossypium SSPs
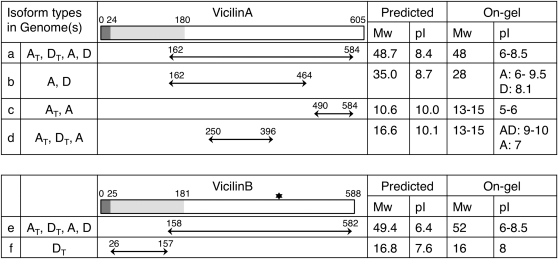
An observation common to 2-DE gels, and pertinent to the analysis of the SSP proteome in Gossypium, is that numerous spots often correspond to isoforms of the same protein accession, as previously demonstrated with 2-DE analyses of SSPs in pea, soybean, rapeseed, and Arabidopsis (Hajduch et al. 2005, 2006; Higashi et al. 2006; Bourgeois et al. 2009). By contrasting the isoform peptide sequences obtained through MS analysis to the full-length proteins, the on-gel spot location and the computationally predicted location can be examined to determine the formation and features of SSP isoforms, used here to characterize the isoforms of the vicilins (Figure 7). In the three Gossypium species studied, the most abundant vicilin A and vicilin B spots were identified to have molecular weights of 48 kDa (Figure 7, isoform “a”) and 52 kDa (Figure 7, isoform “e”), respectively, and both were composed of a horizontal isoform series spanning from pH 6 to pH 8.5. These spots were previously reported as the mature forms of vicilin that were processed through a series of post-translational modifications (Dure and Chlan 1981; Dure and Galau 1981). By mapping the peptides derived from MS analysis to the full-length protein sequences, these two isoform series were characterized and determined to derive from the ∼70 kDa vicilin A and vicilin B prepropolypeptides through the cleavage of signal peptides together with the N-terminal fragments, respectively. Similarly, less abundant vicilin isoforms (Figure 7, isoforms “b”, “c”, “d”, and “f”) observed at a lower molecular weight were also evaluated and characterized as the products of proteolytic cleavage or peptide degradation. Protein modifications (e.g., glycosylation, phosphorylation, acetylation, and methylation) also likely contributed to the formation of these vicilin isoforms, which can be inferred by slight shifts in spot pI and/or molecular weight and thus were evident when comparing the isoform on-gel and predicted locations.
Figure 7 .
Distribution and polypeptides of vicilin isoforms in Gossypium. The symbol ✶ indicates a glycosylation site.
It is worth noting that these isoform patterns varied not only among polyploid and diploid species, but also between the AT and DT homoeolog-derived isoforms within the allopolyploid AD genome. Using the analyzed SSP gene sequences as the reference, amino acid variations between the homoeologous peptides were identified, to enable diagnosis of whether the AD-genome polypeptides and their corresponding modifications were inherited from the A- or the D-parental species or whether they exhibited a novel pattern after polyploidization. As shown in Figure 7, the 48 kDa (Figure 7, isoform a) and 52 kDa (Figure 7, isoform e) polypeptides common to all species analyzed were expressed by both homoeologous genes in the AD genome. Alternatively, only AT polypeptides (Figure 7, isoform c) were recovered in the acidic 12–15 kDa region, consistent with the A-genome–specific pattern in diploids. A novel modification in the allopolyploid was observed for the 17-kDa vicilin B polypeptides (Figure 7, isoform f), which appears to be the result of retention of the N-terminal fragment of the vicilin B precursor in the allopolyploid only (whereas the parental diploids experience cleavage and degradation of this fragment). The polypeptides of this fragment were further determined to originate from expression of the DT homoeolog of vicilin B; however, not enough peptide information was recovered to completely rule out the presence of AT homoeolog products. Altogether, these findings suggest differential regulation or modification of homoeologous gene products, as well as novel patterns in the polyploid proteome that may result from the interaction between homoeologous gene products.
More than 30 spots were identified corresponding to legumin isoforms in each Gossypium species, commonly distributed at molecular weights of 30 kDa, 17–20 kDa, and 11–12 kDa as legumin A and at a molecular weight of 11–13 kDa as legumin B. As with vicilins, these legumin isoforms are also processed through a series of modifications, including proteolytic cleavage and peptide degradation. Isoform analysis through peptide mapping indicated that the 30-kDa polypeptides of legumin A derived from the C-terminal fragment of the 58 kDa prepropolypeptide (see File S4 for peptide mapping analysis). Other isoform peptide sequences obtained through MS analysis failed to be clustered and mapped to continuous polypeptide regions, perhaps reflecting a lower peptide coverage recovered from MS analysis compared to that of vicilins. The contribution of homoeologous polypeptides within the allopolyploid was also evaluated, showing that all peptides detected for legumin A and legumin B were encoded by the DT gene copy. This result is consistent with the gene family structure of legumins: in the allopolyploid AD genome, only the D-genome–derived copy of legumin B exists, and the A-genome–derived copy of legumin A appears to be nonfunctional, due to a premature stop codon (as noted above). Considering this strict DT homoeologous expression of legumin isoforms in the AD genome, the legumin SSPs are possibly the key components that contribute to the biased accumulation of cotton seed proteins in allopolyploid cotton.
Discussion
Vicilin and legumin are the major proteins in mature cotton seeds
Seed storage proteins, which accumulate during seed filling and store nutrients for seed germination and seedling growth, compose one of the most important protein categories in plant seeds. Due to their high abundance in nature and their economic importance as a major source of dietary protein, detailed studies of SSPs date to the early part of the 20th century (Osborne 1924), when Osborne classified them according to their solubility in water (albumin), neutral saline (globulin), alcohol/water mixtures (prolamin), and acids or alkalis (glutelin). The most widely distributed and prevalent SSP group is globulin, which can be divided on the basis of the sedimentation rate of its aggregated forms into the 7S vicilins and 11/12S legumins (Shewry et al. 1995). In our survey of the most abundant cotton seed proteins, nearly all of the proteins identified belong to the vicilin and legumin families, comprising 60–70% of the total seed proteins in abundance and suggesting that vicilins and legumins are the major component of mature cotton seeds, as well as the major cotton SSPs.
Quantification of the SSPs, made possible by 2-DE technology, permitted the precise estimation of each SSP category in cotton seeds. In agreement with prior research, which characterized the two principal forms of vicilin as occurring at 48 kDa and 52 kDa (Dure and Chlan 1981; Dure and Galau 1981; Dure et al. 1981; Chlan et al. 1986), these vicilin isoforms were also observed as the most abundant proteins on our proteomic maps. Their relative abundances (37% in the AD genome, 36% in the A genome, and 28% in the D genome), however, were a little higher than the previous estimate of 27% by cylindrical SDS–PAGE (Dure and Chlan 1981). In addition to these vicilin isoforms, the overall composition of vicilins and legumins was also estimated, together with a water-soluble fraction of SSPs termed as albumin in prior research, which suggested that each of these three SSP categories may account for up to one-third of the total protein amount in cotton seeds (Youle and Huang 1979, 1981). Subsequent research, which characterized the albumin mRNA, noted that not only does albumin encode a low molecular weight protein of only 139 amino acids, but also the albumin mRNA makes up a much smaller proportion of the total mRNA pool (2%) in developing seeds, when compared to vicilins (15%) and legumins (30%) (Hughes and Galau 1989; Galau et al. 1992). It is not surprising, then, that this protein was not detected in our proteomic analyses, which encompass ∼80% of total seed protein abundance. The discrepancy between previous protein quantifications and the current analysis is likely due to the more ambiguous classification of globulin and albumin in early studies, which were based on protein solubility and sedimentation rates instead of actual sequences. Although intact globulins are mostly insoluble in water, their degraded or cleaved forms can gain higher water solubility and display a molecular weight similar to that of albumin; therefore, these albumin-like globulin forms could contribute to overestimation of the amount of albumin in cotton seeds. Because mature albumins are typically cleaved into smaller polypeptides that fall outside of the effective separation range of SDS–PAGE, we cannot rule out the possibility that the poor representation of albumin in the present protein profiles may be due to a technical limitation of 2-DE profiling; however, other estimates of protein abundances, which rely on amino acid composition, concur with our assessment. That is, cotton seeds have been characterized as deficient in sulfur-containing amino acids, indicating that sulfur-rich proteins (such as albumin) constitute a low fraction of the total seed proteins (Bressani et al. 1966; Chlan et al. 1986; Galau et al. 1991, 1992).
Biased accumulation of D-genome proteins in polyploid cotton seeds
Allopolyploidization involves the merger of two different, and often divergent, genomes, whose reconciliation in a common nucleus often leads to myriad changes, including unequal integration and expression of the two merging genomes. Recent studies into the consequences of allopolyploidization have underscored this possibility of nonequivalence by demonstrating biased expression among homoeologs and a phenomenon termed transcriptional genome dominance (Rapp et al. 2009). In F1 hybrids between the allotetraploids Arabidopsis thaliana and A. arenosa, an analysis of nonadditively expressed genes revealed that, for those genes more highly expressed in A. thaliana, the F1 allotetraploid hybrid preferentially exhibited repressed expression, much like that in A. arenosa (Wang et al. 2006). In the recently formed natural allotetraploid Tragopogon miscellus, higher levels of expression have been reported for homoeologs originating in T. dubius vs. those originating in T. pratensis (Buggs et al. 2010). Similar studies in allopolyploid wheat have also demonstrated nonequivalent expression patterns among homoeologs (Bottley et al. 2006, 2008; Bottley and Koebner 2008; Pumphrey et al. 2009). In cotton, biased expression of D-genome homoeologs has been reported for petal and fiber tissues in G. hirsutum (Flagel et al. 2008; Hovav et al. 2008). These data were later extended to a synthetic F1 hybrid and the other four natural allotetraploid species that originated from the same genomic merger and doubling, an analysis that suggested that the D-genome homoeolog bias was established during genome merger and was subsequently retained during the divergence of all five extant allopolyploid species (Flagel and Wendel 2010). This observation was later augmented by the discovery of the phenomenon of transcriptional genomic dominance, whereby gene expression levels in a nascent (synthetic) AD-genome allopolyploid mimicked those in the parental D genome more often than those in the A genome (Rapp et al. 2009). Because this was true both for genes that were up- and downregulated in D relative to A genome, we termed this phenomenon genomic dominance (in this case biased toward D genome).
A natural extension of these transcriptional characterizations regarding the nonequivalence accompanying polyploidy is to ask whether similar patterns are exhibited at the protein level and whether any observed nonequivalencies are linked to phenotypic and functional variations. Attempts to address these questions were first made in allopolyploid Brassica using the neosynthesized tetraploid (Albertin et al. 2006, 2007), where little qualitative variation (<1% deviation in spot presence/absence) was observed between the neosynthesized B. napus allotetraploid and its diploid progenitors, B. oleracea and B. rapa. For the 25–38% of spots displaying quantitative difference (i.e., those expressed nonadditively), expression patterns were slightly closer to that of the B. rapa parent rather than B. oleracea, in accordance with a previous study that suggested bias toward the B. rapa genome in the transcriptional expression of rRNA genes (Chen and Pikaard 1997). The lack of genomic and transcriptomic data, however, makes it difficult to infer the structural and functional significance of these observations in Brassica. In the present work, we profiled total mature seed proteins in a naturally formed allotetraploid whose genomic and transcriptomic reactions to genomic merger and doubling have been extensively studied (Adams et al. 2003; Senchina et al. 2003; Grover et al. 2004, 2007; Flagel et al. 2008, 2009; Hovav et al. 2008; Chaudhary et al. 2009; Rapp et al. 2009; Flagel and Wendel 2010; Salmon et al. 2010) and address the question whether the D-genome bias and dominance previously observed in the transcriptome of polyploid cotton is reflected at the protein level.
Consistent with transcriptomic data suggesting D-genome dominance (Rapp et al. 2009), the proteome of the allopolyploid was more similar to the D-genome parent, with 26.2% of the 2-DE spots detected in the allopolyploid being present only in the progenitor D genome vs. 10.7% that were present only in the A-genome diploid (Table 1). This observation was extended by quantitative profiling of shared spots, which displayed a higher level of nonadditive expression equivalent to that of the progenitor D genome than to that of the A genome (14.1% “D-genome dominant” vs. 9.8% “A-genome dominant”; Table 2). Hence, the cotton seed proteome displays an overall dominance reflecting its D-genome component; however, by parsing the qualitative and quantitative expressions for each individual SSP, additional patterns of diversity in dominance become evident, including dominant expression by both progenitor genomes, particularly with respect to multiple isoforms corresponding to each SSP (Table 4). Similar patterns of differential and uncoordinated expression of protein isoforms were also demonstrated in the synthetic allopolyploid B. napus (Albertin et al. 2007).
Table 4 . Multiplicity of expression patterns displayed by SSP isoforms.
| Genome specific | Found in two genomes | Shared by all three genomes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SSP | AD | A | D | AD-A | AD-D | A-D | Additive | Aa | Da | Coa | Transgressive |
| Vicilin A | 3 | 1 | 2 | 2 | |||||||
| Vicilin B | 1 | 1 | |||||||||
| Vicilin-like | 3 | ||||||||||
| Legumin A | 6 | 8 | 1 | 1 | 5 | 13 | 3 | 3 | 1 | 2 | |
| Legumin B | 1 | 3 | 1 | 1 | 1 | ||||||
Expression dominance. Co, codominant.
Biased expression of D-genome–derived homoeologs, another remarkable transcriptional feature of allopolyploid Gossypium (Flagel et al. 2008, 2009; Hovav et al. 2008; Flagel and Wendel 2010), was also observed in the biased accumulation of cottonseed proteins expressed by D-genome–derived homoeologs (e.g., the vicilin-like protein as shown in Figure 5). Biased expression is generally more difficult to uncover in complex protein data than in transcriptomic sequence data and is most easily recognized by the absence of A- or D-genome–derived homoeologous copies (e.g., for legumins). Sequence changes at the protein level accumulate slowly relative to the scale of genetic diversification among cotton lineages, such that exons differ by only ∼1%, mostly at synonymous sites, among A- and D-genome orthologs (Senchina et al. 2003), making it difficult to identify and distinguish homoeologous protein isoforms. Thus, the biased expression reported here is likely a significant underestimate of biased protein expression in the polyploid, as it relies on the limited cases of homoeolog loss that are most readily detected.
Unexpectedly high variation among Gossypium seed proteomes
Comparative proteomics, which permits the characterization of protein-level variation among related species, is in its infancy as an evolutionary approach, particularly with respect to polyploid species. The few studies that have applied modern proteomic techniques in a comparative fashion have found little variation among species (Albertin et al. 2005, 2006, 2007). In contrast to the expectations implied by this work as well as the high level of coding sequence conservation among the species studied, the cotton seed proteomes analyzed here display extraordinary variation. This variation occurs not only between diploid species, but also between the allopolyploid species and its model diploid progenitors; only one-third of protein features were common to the three Gossypium species profiled. Although amino acid sequence variation of SSPs can in principle account for some of this variation, the fact that there is only ∼1% synonymous nucleotide differences among orthologous Gossypium exons, both in previous studies (Senchina et al. 2003) and for the SSP genes analyzed here, indicates that amino acid substitutions account for only a very small part of the variation detected.
This exaggerated interspecific expression variation observed in the Gossypium seed proteomes, when compared to similar research in Brassica, which revealed 15% divergence between diploid species and only 1% between the synthesized allotetraploid B. napus and its diploid progenitors (Albertin et al. 2006), may reflect a gradual accumulation pattern of differential protein expressions in allopolyploid cotton naturally formed 1–2 million years ago vs. newly synthesized B. napus, as well as, at least in part, differences in the tissues examined. That is, the stem and root proteomes studied in Brassica are likely more complex with respect to their proteomes than are the seed proteomes studied here, which tend to be composed of fewer protein types that are extensively modified into many isoforms (Hajduch et al. 2005; Higashi et al. 2006; Bourgeois et al. 2009; Larre et al. 2010). Thus, relatively few underlying differences in post-translational modification programs among cotton species may propagate to affect multiple isoforms, in the process generating a relatively large impact on inferences of similarity, at least in comparisons of seed vs. stem or root proteomes. In addition to this speculation, it may be that the magnitude of proteomic variation is dependent not only on tissue type, but also on ploidy level (Feldman et al. 1986). Classic isozyme analyses, which are able to detect variable protein expression in the form of inferred gene losses or silencing, were previously applied to analyze homoeologous expression patterns in polyploids (Wendel 2000) and suggested higher variability in expression of seed storage proteins vs. other classes of genes in allopolyploid wheat (Galili and Feldman 1983; Feldman et al. 1986). Furthermore, some of the differential expression patterns observed between diploid and polyploid wheat were inferred to result from proteomic interactions between the contributing genomes (Islam et al. 2003). As the very nature of allopolyploid species involves the coexistence of homoeologous genomes, which itself often involves conflict or competition between regulatory machineries that independently evolved in progenitor species, one can readily envision that the merger of diverged regulatory and post-translational machineries will lead to vastly enhanced combinatorial complexity, which in turn is detected in studies such as ours as “novel” spot presence/absences and transgressive expression levels.
Conclusions
This work presents the first high-quality proteomic map for mature seeds in cotton, a vital oil and meal seed crop. In total, 155 SSP spots and 5 nonstorage protein spots were identified. In addition to this comprehensive characterization of protein composition, proteomic profiles were generated, revealing a pattern of interspecific complexity and nonadditive protein accumulation in cotton allopolyploids. The biased accumulation of seed proteins toward the D-genome progenitor, combined with the genetic analyses presented here, provides a novel perspective on the proteomic consequences of polyploidization. One caveat to our study is that we included only one accession of allopolyploid Gossypium and its diploid progenitors; therefore, some of the proteomic variation observed might reflect choice of accession rather than between species. The accessions studied were selected as the most widely used models for exploring the genomic and transcriptomic consequences of polyploidy, thereby providing additional among-study perspective.
Further exploration into comparative proteomics, including the analysis of additional accessions, will be necessary to identify and characterize the regulatory mechanisms involved in generating the proteomic complexity and novelty observed in these and other species. It also will be interesting to explore the relationships among tissue choice, ploidy level, and multiple experimental variables in developing an enhanced understanding of the effects of hybridization and genome doubling on the proteome of higher plants.
Acknowledgments
We thank Kara Grupp and Anna Krush for their help in tissue collection and Corrinne Grover for helpful discussions and comments on the manuscript. We also thank Cotton Incorporated and the National Science Foundation Plant Genome Program for financial support. Dharminder Pathak was supported by a training grant from Punjab Agricultural University, Ludhiana, India.
Literature Cited
- Adams K. L., Cronn R., Percifield R., Wendel J. F., 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin W., Brabant P., Catrice O., Eber F., Jenczewski E., et al. , 2005. Autopolyploidy in cabbage (Brassica oleracea L.) does not alter significantly the proteomes of green tissues. Proteomics 5: 2131–2139 [DOI] [PubMed] [Google Scholar]
- Albertin W., Balliau T., Brabant P., Chevre A. M., Eber F., et al. , 2006. Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics 173: 1101–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin W., Alix K., Balliau T., Brabant P., Davanture M., et al. , 2007. Differential regulation of gene products in newly synthesized Brassica napus allotetraploids is not related to protein function nor subcellular localization. BMC Genomics 8: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anssour S., Krugel T., Sharbel T. F., Saluz H. P., Bonaventure G., et al. , 2009. Phenotypic, genetic and genomic consequences of natural and synthetic polyploidization of Nicotiana attenuata and Nicotiana obtusifolia. Ann. Bot. (Lond.) 103: 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Bottley A., Koebner R. M., 2008. Variation for homoeologous gene silencing in hexaploid wheat. Plant J. 56: 297–302 [DOI] [PubMed] [Google Scholar]
- Bottley A., Xia G. M., Koebner R. M., 2006. Homoeologous gene silencing in hexaploid wheat. Plant J. 47: 897–906 [DOI] [PubMed] [Google Scholar]
- Bottley A., Chapman N. H., Koebner R. M., 2008. Homoeologous gene silencing in tissue cultured wheat callus. BMC Genet. 9: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois M., Jacquin F., Savois V., Sommerer N., Labas V., et al. , 2009. Dissecting the proteome of pea mature seeds reveals the phenotypic plasticity of seed protein composition. Proteomics 9: 254–271 [DOI] [PubMed] [Google Scholar]
- Bradford M. M., 1976. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bressani R., Elias L. G., Baham E., 1966. Cottonseed protein in human foods, pp. 75–100 World Protein Resources, edited by Altschul A. M. American Chemical Society, Washington, DC [Google Scholar]
- Buggs R. J., Doust A. N., Tate J. A., Koh J., Soltis K., et al. , 2009. Gene loss and silencing in Tragopogon miscellus (Asteraceae): comparison of natural and synthetic allotetraploids. Heredity 103: 73–81 [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Chamala S., Wu W., Gao L., May G. D., et al. , 2010. Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol. Ecol. 19: 132–146 [DOI] [PubMed] [Google Scholar]
- Chaudhary B., Flagel L., Stupar R. M., Udall J. A., Verma N., et al. , 2009. Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., Pikaard C. S., 1997. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl. Acad. Sci. USA 94: 3442–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlan C. A., Pyle J. B., Legocki A. B., Dure L., 1986. Developmental biochemistry of cottonseed embryogenesis and germination. XVIII. cDNA and amino acid sequences of members of the storage protein families. Plant Mol. Biol. 7: 475–489 [DOI] [PubMed] [Google Scholar]
- Chlan C. A., Borroto K., Kamalay J. A., Dure L., 1987. Developmental biochemistry of cottonseed embryogenesis and germination. XIX. Sequences and genomic organization of the α globulin (vicilin) genes of cottonseed. Plant Mol. Biol. 9: 533–546 [DOI] [PubMed] [Google Scholar]
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Cronn R. C., Small R. L., Haselkorn T., Wendel J. F., 2002. Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am. J. Bot. 89: 707–725 [DOI] [PubMed] [Google Scholar]
- Cui L., Wall P. K., Leebens-Mack J. H., Lindsay B. G., Soltis D. E., et al. , 2006. Widespread genome duplications throughout the history of flowering plants. Genome Res. 16: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L., 1989. Characteristics of the storage proteins of cotton. J. Am. Oil Chem. Soc. 66: 356–359 [Google Scholar]
- Dure L., Chlan C., 1981. Developmental biochemistry of cottonseed embryogenesis and germination: XII. Purification and properties of principal storage proteins. Plant Physiol. 68: 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L., Galau G. A., 1981. Developmental biochemistry of cottonseed embryogenesis and germination: XIII. Regulation of biosynthesis of principle storage proteins. Plant Physiol. 68: 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dure L., 3rd, Greenway S. C., Galau G. A., 1981. Developmental biochemistry of cottonseed embryogenesis and germination: changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 20: 4162–4168 [DOI] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Galili G., Levy A., 1986. Genetic and evolutionary aspects of allopolyploidy in wheat, pp. 88–100 The Origin and Domestication of Cultivated Plants, edited by Barigozzi C. Elsevier, Amsterdam [Google Scholar]
- Flagel L. E., Wendel J. F., 2010. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 186: 184–193 [DOI] [PubMed] [Google Scholar]
- Flagel L., Udall J., Nettleton D., Wendel J., 2008. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L. E., Chen L., Chaudhary B., Wendel J. F., 2009. Coordinated and fine-scale control of homoelogous gene expression in allotetraploid cotton. J. Hered. 100: 487–490 [DOI] [PubMed] [Google Scholar]
- Frampton V., Pons W., Kerr T., 1958. A comparison of chemical properties of seeds of Gossypium species. Econ. Bot. 14: 197–199 [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E., Osborn T. C., 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galau G. A., Chlan C. A., Dure L., 1983. Developmental biochemistry of cottonseed embryogenesis and germination. Plant Mol. Biol. 2: 189–198 [DOI] [PubMed] [Google Scholar]
- Galau G. A., Wang H. Y., Hughes D. W., 1991. Sequence of the Gossypium hirsutum D-genome alloallele of legumin A and its mRNA. Plant Physiol. 97: 1268–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galau G. A., Wang H. Y., Hughes D. W., 1992. Cotton Mat5-A (C164) gene and Mat5-D cDNAs encoding methionine-rich 2S albumin storage proteins. Plant Physiol. 99: 779–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G., Feldman M., 1983. Diploidization of endosperm protein genes in polyploid wheats, pp. 1119–1123 6th International Wheat Genetics Symposium, Kyoto, Japan [Google Scholar]
- Grover C. E., Kim H., Wing R. A., Paterson A. H., Wendel J. F., 2004. Incongruent patterns of local and global genome size evolution in cotton. Genome Res. 14: 1474–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C. E., Kim H., Wing R. A., Paterson A. H., Wendel J. F., 2007. Microcolinearity and genome evolution in the AdhA region of diploid and polyploid cotton (Gossypium). Plant J. 50: 995–1006 [DOI] [PubMed] [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandran V., Kasschau K. D., et al. , 2009. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. USA 106: 17835–17840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch M., Ganapathy A., Stein J. W., Thelen J. J., 2005. A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 137: 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch M., Casteel J. E., Hurrelmeyer K. E., Song Z., Agrawal G. K., et al. , 2006. Proteomic analysis of seed filling in Brassica napus. Developmental characterization of metabolic isozymes using high-resolution two-dimensional gel electrophoresis. Plant Physiol. 141: 32–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch M., Casteel J. E., Tang S., Hearne L. B., Knapp S., et al. , 2007. Proteomic analysis of near-isogenic sunflower varieties differing in seed oil traits. J. Proteome Res. 6: 3232–3241 [DOI] [PubMed] [Google Scholar]
- Hajduch M., Hearne L. B., Miernyk J. A., Casteel J. E., Joshi T., et al. , 2010. Systems analysis of seed filling in Arabidopsis: using general linear modeling to assess concordance of transcript and protein expression. Plant Physiol. 152: 2078–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y., Hirai M. Y., Fujiwara T., Naito S., Noji M., et al. , 2006. Proteomic and transcriptomic analysis of Arabidopsis seeds: molecular evidence for successive processing of seed proteins and its implication in the stress response to sulfur nutrition. Plant J. 48: 557–571 [DOI] [PubMed] [Google Scholar]
- Houston N. L., Hajduch M., Thelen J. J., 2009. Quantitative proteomics of seed filling in castor: comparison with soybean and rapeseed reveals differences between photosynthetic and nonphotosynthetic seed metabolism. Plant Physiol. 151: 857–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovav R., Udall J. A., Chaudhary B., Rapp R., Flagel L., et al. , 2008. Partitioned expression of duplicated genes during development and evolution of a single cell in a polyploid plant. Proc. Natl. Acad. Sci. USA 105: 6191–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. W., Galau G. A., 1989. Temporally modular gene expression during cotyledon development. Genes Dev. 3: 358–369 [DOI] [PubMed] [Google Scholar]
- Islam N., Tsujimoto H., Hirano H., 2003. Proteome analysis of diploid, tetraploid and hexaploid wheat: Towards understanding genome interaction in protein expression. Proteomics 3: 549–557 [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., et al. , 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Koh J., Soltis P. S., Soltis D. E., 2010. Homeolog loss and expression changes in natural populations of the recently and repeatedly formed allotetraploid Tragopogon mirus (Asteraceae). BMC Genomics 11: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larre C., Penninck S., Bouchet B., Lollier V., Tranquet O., et al. , 2010. Brachypodium distachyon grain: identification and subcellular localization of storage proteins. J. Exp. Bot. 61: 1771–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Kim E. D., Ha M., Lackey E., Liu J., et al. , 2009. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne T. B., 1924. The Vegetable Proteins. Longmans Green & Co., London [Google Scholar]
- Pandey S., Thejappa N., 1975. Study on relationship between oil, protein, and gossypol in cottonseed kernels. J. Am. Oil Chem. Soc. 52: 312–315 [DOI] [PubMed] [Google Scholar]
- Pires J. C., Zhao J., Schranz M. E., Leon E. J., Quijada P. A., et al. , 2004. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. Lond. 82: 675–688 [Google Scholar]
- Pumphrey M., Bai J., Laudencia-Chingcuanco D., Anderson O., Gill B. S., 2009. Nonadditive expression of homoeologous genes is established upon polyploidization in hexaploid wheat. Genetics 181: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J., 2011. Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. USA 108: 7096–7101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp R. A., Udall J. A., Wendel J. F., 2009. Genomic expression dominance in allopolyploids. BMC Biol. 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon A., Flagel L., Ying B., Udall J. A., Wendel J. F., 2010. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 186: 123–134 [DOI] [PubMed] [Google Scholar]
- Schnable J. C., Springer N. M., Freeling M., 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senchina D. S., Alvarez I., Cronn R. C., Liu B., Rong J., et al. , 2003. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20: 633–643 [DOI] [PubMed] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A. A., 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry P. R., Napier J. A., Tatham A. S., 1995. Seed storage proteins: structures and biosynthesis. Plant Cell 7: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunilkumar G., Campbell L. M., Puckhaber L., Stipanovic R. D., Rathore K. S., 2006. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. USA 103: 18054–18059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L., 2001. PAUP* Phylogenetic Analysis Using Parsimony and Other Methods, Version 4 Sinauer Associates, Sunderland, MA [Google Scholar]
- Udall J. A., Swanson J. M., Haller K., Rapp R. A., Sparks M. E., et al. , 2006a. A global assembly of cotton ESTs. Genome Res. 16: 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall J. A., Swanson J. M., Nettleton D., Percifield R. J., Wendel J. F., 2006b. A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics 173: 1823–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H. S., Wei N. E., Jiang H., et al. , 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249 [PubMed] [Google Scholar]
- Wendel J. F., Cronn R. C., 2003. Polyploidy and the evolutionary history of cotton, pp. 139–186 Advances in Agronomy. Academic Press, New York/London/San Diego [Google Scholar]
- Wendel J. F., Schnabel A., Seelanan T., 1995. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl. Acad. Sci. USA 92: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R. J., Huang A. H., 1979. Albumin storage protein and allergens in cottonseeds. J. Agric. Food Chem. 27: 500–503 [DOI] [PubMed] [Google Scholar]
- Youle R. J., Huang A. H. C., 1981. Occurrence of low molecular weight and high cysteine containing albumin storage proteins in oilseeds of diverse species. Am. J. Bot. 68: 5 [Google Scholar]