Abstract
Understanding the genetic basis of reproductive isolation between recently diverged species is a central problem in evolutionary genetics. Here, I present analyses of the genetic architecture underlying hybrid male sterility and segregation distortion between the Bogota and USA subspecies of Drosophila pseudoobscura. Previously, a single gene, Overdrive (Ovd), was shown to be necessary but not sufficient for both male sterility and segregation distortion in F1 hybrids between these subspecies, requiring several interacting partner loci for full manifestation of hybrid phenomena. I map these partner loci separately on the Bogota X chromosome and USA autosomes using a combination of different mapping strategies. I find that hybrid sterility involves a single hybrid incompatibility of at least seven interacting partner genes that includes three large-effect loci. Segregation distortion involves three loci on the Bogota X chromosome and one locus on the autosomes. The genetic bases of hybrid sterility and segregation distortion are at least partially—but not completely—overlapping. My results lay the foundation for fine-mapping experiments to identify the complete set of genes that interact with Overdrive. While individual genes that cause hybrid sterility or inviability have been identified in a few cases, my analysis provides a comprehensive look at the genetic architecture of all components of a hybrid incompatibility underlying F1 hybrid sterility. Such an analysis would likely be unfeasible for most species pairs due to their divergence time and emphasizes the importance of young species pairs such as the D. pseudoobscura subspecies studied here.
STUDYING the genetics and evolution of reproductive isolating barriers between populations is key to understanding speciation. Over the past two decades, we have gained a good understanding of various aspects of the evolution of reproductive isolation and made particularly rapid progress in our understanding of the genetics of intrinsic postzygotic isolation, i.e., hybrid sterility and inviability (Coyne and Orr 2004). We have advanced from understanding several key comparative patterns characterizing intrinsic isolation—e.g., Haldane’s rule and the preferential sterility and inviability of hybrids of the heterogametic sex (Haldane 1922)—to identifying and characterizing the genes that cause hybrid sterility and hybrid inviability.
Bateson, Dobzhansky, and Muller independently described a model for the evolution of genetic hybrid incompatibilities that underlie intrinsic postzygotic isolation (Bateson 1909; Dobzhansky 1937; Muller 1942; Orr 1996). A hybrid incompatibility involves a negative epistatic interaction among genes from two species that causes hybrid sterility or hybrid inviability. Under the simplest version of their model, functional divergence at only two loci is sufficient to cause a hybrid incompatibility. Theory and empirical data suggest, however, that hybrid incompatibilities—particularly those involved in hybrid male sterility—may often be complex; i.e., hybrids may have to carry the “correct” alleles at three or more loci for sterility or inviability to arise (Orr 1995; Davis and Wu 1996). This pattern is known as complex epistasis, as several alleles must come from the appropriate species for the manifestation of full hybrid sterility.
Despite the identification of several genes that cause hybrid sterility or inviability (Presgraves 2010), our understanding of the complex genetic architecture of hybrid incompatibilities is still lagging. Although a single incompatibility consisting of two genes can, in theory, cause postzygotic isolation, we have little understanding of how many incompatibilities typically separate species. Furthermore, we do not know how many partner genes typically interact in single incompatibility. While mapping and identifying individual genes that cause hybrid sterility or inviability in species hybrids is extremely informative for many important questions in speciation, it provides few clues about the number and effect sizes of genes that take part in incompatibilities and the number of distinct incompatibilities that cause postzygotic reproductive isolation. Indeed, of all the hybrid incompatibility genes that have been identified so far, we do not have a single case where we know how many partner loci these genes interact with to complete the hybrid incompatibility.
According to theory and recent genetic evidence, hybrid incompatibilities accumulate between taxa faster than linearly with divergence, a pattern known as the “snowball effect” (Orr 1995; Matute et al. 2010; Moyle and Nakazato 2010). This rapid accumulation of genic incompatibilities continues even after reproductive isolation is complete. This process can, obviously, confound the interpretation of the number of genes that cause intrinsic reproductive isolation. In particular, it is difficult to say which incompatibilities were important during speciation vs. which accumulated after the completion of reproductive isolation. It is clear that the genetic dissection of interacting partner genes that form Dobzhansky–Muller incompatibilities in young species is crucial to gaining an understanding of the genetic architecture and evolution of hybrid sterility.
Drosophila pseudoobscura pseudoobscura (hereafter USA) and Drosophila pseudoobscura bogotana (hereafter Bogota) provide a powerful system for addressing these questions. USA and Bogota are young subspecies estimated to have diverged between 150,000 and 230,000 years (Schaeffer and Miller 1991; Wang et al. 1997). The USA and Bogota subspecies are geographically separated by >2000 km: the USA subspecies is distributed across western North America, whereas the Bogota subspecies is found in regions of high elevation near Bogota, Colombia (Dobzhansky et al. 1963). These taxa are incompletely reproductively isolated: hybrid F1 males from Bogota mothers are sterile, whereas reciprocal hybrid males and hybrid females from both directions of the cross are fertile (Prakash 1972). There is no detectable prezygotic isolation between these subspecies (Noor 1995).
The genetic basis of hybrid male sterility between these subspecies involves a small number of chromosomal regions that interact in a complex pattern (Orr and Irving 2001). Regions on the XL and XR from Bogota (Muller elements A and D, respectively) were found to interact with dominant factors on the second and third autosomes from USA (Muller elements E and C, respectively) to cause hybrid sterility (Orr and Irving 2001). The fourth autosome and fifth dot chromosome (Muller elements B and F, respectively) have no detectable effect on postzygotic isolation. None of the important regions shows much effect singly on hybrid sterility: all interacting partners must be simultaneously present for the full expression of hybrid sterility.
More recently, Bogota–USA hybrid F1 males were discovered to be slightly fertile (Orr and Irving 2005). Orr and Irving (2005) found that “sterile” F1 hybrid males become very weakly fertile when aged. Surprisingly, these F1 hybrid males produce almost all daughters. Several lines of evidence show that this sex-ratio distortion is caused not by hybrid male inviability, but by an overrepresentation of X-bearing sperm among the functional gametes of hybrid males (Orr and Irving 2005). The precise stage at which this segregation distortion arises is unclear. The genetic basis of segregation distortion appears similar to that of hybrid male sterility (Orr and Irving 2001, 2005; Phadnis and Orr 2009). Hybrid segregation distortion involves the same approximate regions on the Bogota XL and XR, but the effects of particular autosomes have not been characterized (Orr and Irving 2005). Pure Bogota individuals show normal segregation because they are homozygous for recessive autosomal suppressors.
Phadnis and Orr (2009) performed fine-scale mapping of the sepia (se) region on Bogota XR—a region that has a large effect on both male sterility and segregation distortion in the hybrid F1 males. Through a series of recombination mapping and transgenic experiments in Drosophila pseudoobscura, they showed that a single gene, Overdrive (Ovd), plays a causal role in both hybrid sterility and hybrid segregation distortion (Phadnis and Orr 2009). These results indicate that hybrid male sterility and segregation distortion in Bogota–USA hybrids share a common genetic basis, raising the possibility that genetic conflict involving segregation distorters and their suppressors may drive the evolution of hybrid sterility.
Ovd is necessary but not sufficient to cause hybrid male sterility or segregation distortion; it requires the “correct” alleles at several interacting loci to yield a strong hybrid incompatibility. The number and locations of these interacting partners of Ovd that are essential for both hybrid phenotypes remain unknown. Indeed, we cannot even be sure about the number of relevant loci on the Bogota X—the best-studied chromosome in this hybridization—that are involved in hybrid sterility. Even less is known about the genetic basis of segregation distortion in hybrids and its suppression in pure Bogota individuals.
Here, I perform mapping of hybrid sterility and hybrid segregation distortion genes on the X, second, and third chromosomes, chromosomes previously shown to be important for hybrid sterility and segregation distortion (Orr and Irving 2001, 2005). I perform two independent crosses and analyses for the X and the autosomes, using different mapping strategies that are optimal for each chromosome.
My study departs from and builds on previous analyses in several ways. First, whereas previous studies were limited by the small number of visible markers available in D. pseudoobscura, I use molecular markers to increase marker density. Second, I employ distinct crossing designs that capitalize on information about gene locations from previous studies, allowing me to focus on mapping genes that cause F1 hybrid phenomena and to avoid the confounding effects of any recessively acting genes that appear in F2 or backcross-like genotypes. Complex epistatic interactions, such as those that may underlie hybrid sterility, can dramatically reduce mapping power. I therefore use two separate crossing designs to exert greater control over the mapping population and thus provide an increase in mapping power—despite the lack of good balancer chromosomes in D. pseudoobscura. Finally, while previous studies used sperm motility to measure hybrid fertility, I count progeny. This allows for the simultaneous mapping of hybrid sterility and hybrid segregation distortions in single crosses and allows comparison with earlier findings. It also allows me, for the first time, to address the relationship between hybrid sterility and hybrid segregation distortion on the autosomes. From these studies, a coherent picture emerges that implicates three major-effect loci (and four minor-effect loci) on Bogota X chromosome and USA autosomes as the genetic basis of hybrid sterility. I also find a strong, but not perfect, overlap in regions implicated in segregation distortion, confirming previous findings for a shared genetic basis of both hybrid phenotypes.
Materials and Methods
Fly stocks
All flies were maintained and crosses performed at 22° using standard cornmeal–sugar–yeast–agar food. Most strains used are as in Orr and Irving (2001, 2005). Mapping of X-linked factors was performed between the Bogota-ER strain and a USA strain with a multiply marked X chromosome cut (ct; 1–22.5), scalloped (sd; 1–43.0), yellow (y; 1–74.5), and se (1–156.5). Mapping of autosomal genes was performed between Bogota-ER and Treeline Lobe (L; 3–13.3) strain from USA. These strains were chosen as they carry collinear third chromosomes (Treeline arrangement), allowing free recombination along the entire third chromosome between Bogota and USA.
Autosomal mapping also involved a se introgression line, INT104, produced in previous work (Phadnis and Orr 2009), and a newly constructed YUSA stock. Briefly, INT104 is a se introgression line that derives its genetic material almost entirely from Bogota except for a small region near se, which is derived from USA (Phadnis and Orr 2009). Hybrid males produced by crossing INT104 females to USA males are completely fertile and show normal segregation as they carry the USA allele at Ovd, near se. The YUSA stock was constructed by first crossing males from a USA strain with individually marked chromosomes y (1–74.5), glass (gl; 2–83.3), orange (or; 3–0.0), and incomplete (inc; 4–0.0) males to INT104 females. Multiple lines were established by repeatedly backcrossing the resulting males (carrying unrecombined autosomes) to Bogota females for 10 generations. The YUSA stock has a genome entirely derived from Bogota, except the Y chromosome, which derives from USA. This was confirmed in two ways. First, I crossed virgin females from the YUSA strain to y, gl, or, and inc males and confirmed that all major chromosomes were homozygous for Bogota (the fifth dot chromosome was unmarked). Second, I sequenced a Y-linked region and confirmed that it was derived from USA. In particular, I scored five diagnostic SNPs in the Y-linked gene CG12218Y using the forward primer 5′-GCAGTCGAACCAGTGCAAT-3′ and the reverse primer 5′-GTGCGGGCAATGGATAAT-3′ (Carvalho and Clark 2005). YUSA males are fertile and show normal sex-chromosome segregation.
Crosses
Male sterility in Bogota–USA hybrids was previously shown to involve complex epistasis between 5 and 15 loci (Orr and Irving 2001). Because highly epistatic interactions can dramatically reduce mapping power, separate crosses that yielded mapping populations enriched for individuals of appropriate genotypes were used to map the X-linked and autosomal loci. The mapping crosses were designed to take advantage of information about the genetics of hybrid sterility and hybrid segregation distortion from previous studies (Orr and Irving 2001, 2005; Phadnis and Orr 2009).
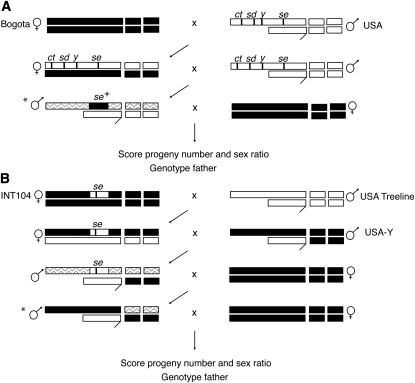
For X-linked mapping, Bogota-ER females were crossed to ct sd y se males (Figure 1A). The resulting F1 hybrid females were backcrossed to ct sd y se males. Because se hybrid males carry USA material at Ovd, they are almost always fertile and show normal segregation regardless of genotype at other hybrid sterility loci (Phadnis and Orr 2009). Such males are therefore uninformative for mapping partner hybrid sterility and segregation distortion genes. Because se hybrid males are uninformative and constitute 50% of a backcross population, inclusion of these males in the mapping population can severely reduce mapping power. Therefore, only se+ backcross hybrid males, which carry the Bogota material at Ovd, were scored for progeny number and sex ratio.
Figure 1 .
Crossing designs for mapping loci that cause male sterility and segregation distortion in F1 hybrids between D. pseudoobscura Bogota and USA. Bogota material is solid, USA material is open, and recombinant chromosomal regions are shaded. The large metacentric X chromosome of D. pseudoobscura is denoted as a long bar, the Y chromosome as a hooked bar, and the autosomes as small bars. Only the second and third autosomes are shown for simplicity; the fourth autosome and the fifth dot chromosome play no role in postzygotic isolation between these subspecies (Orr and Irving 2001). (A) Crosses for mapping loci on the Bogota X chromosome. The focal males (indicated by *) bear one set of unrecombined USA autosomes that carry dominant sterility factors, Bogota material at se, and a recombinant X chromosome. (B) Crosses for mapping loci on the autosomes. The focal males (indicated by *) bear an unrecombined Bogota X chromosome, a USA Y chromosome, an entire set of unrecombined Bogota autosomes, and a set of recombined autosomes. INT104 is an introgression line that carries USA material near se in an otherwise Bogota genetic background. This se introgression, which includes Ovd, is used to make sure that males in the intermediate crossing generations are fertile. The YUSA stock has a USA Y chromosome in an otherwise Bogota genetic background.
Mapping of dominant autosomal genes for hybrid sterility and segregation distortion requires a mapping population that has one set of recombinant autosomes along with a nonrecombinant Bogota X chromosome, a USA Y chromosome, and at least one copy of nonrecombinant Bogota autosomes. In most hybridizations, generating individuals of such a genotype is unfeasible because of the sterility of individuals in the intermediate generations of crosses. I circumvent this problem in producing the autosomal mapping population by utilizing the introgression line INT104 analogous to a hybrid fertility rescue mutation. More precisely, introgression line INT104, which carries a USA allele of Ovd in an otherwise Bogota genetic background, ensures that recombinant hybrid males in the penultimate generation are fertile (Figure 1B). The crossing scheme used here also produces males that are homozygous for the nonrecombinant Bogota autosomes, similar to pure Bogota individuals. Because these males are uninformative for mapping, they were excluded from the mapping population.
Fertility and segregation distortion assay
Five-day-old focal virgin hybrid males were mated singly to two three-day-old Bogota-ER virgin females. Males were frozen at −80° and females were discarded after 7 days. Progeny number and sex ratio were scored 21 days after the parents were removed.
Markers and genotyping
DNA extraction was performed using the protocol described by Gloor et al. (1993). Primers for amplification of microsatellite loci were based on previously published marker locations (Ortiz-Barrientos et al. 2006; Chang and Noor 2007) or designed from the D. pseudoobscura whole-genome sequence (Richards et al. 2005) using the web-based program Tandem Repeats Finder (Benson 1999) and Primer 3 (Rozen and Skaletsky 2000) (Table S1). An attempt was made to distribute the markers evenly across linkage groups using previously published local recombination rate data (Ortiz-Barrientos et al. 2006). Fluorescently labeled primers were used to amplify microsatellite loci from the extracted DNA following a touchdown PCR protocol: 95° for 2 min; 5 cycles of 95° for 30 sec, 55° for 30 sec, 72° for 30 sec; 10 cycles of 95° for 30 sec, 52° for 30 sec, 72° for 30 sec; and 15 cycles of 95° for 30 sec, 49° for 30 sec, 72° for 30 sec. Up to five markers were multiplexed in single PCR reactions when possible. The PCR products were analyzed for fragment size using the Applied Biosystems 3730 Genetic Analyzer.
Recombination mapping
Genotype data were extracted using GeneMapper (Applied Biosystems) and confirmed by visual inspection. Linkage maps for the markers were constructed using JoinMap (Kyazma). Composite interval mapping (CIM) (Zeng 1994) was performed using Windows QTL Cartographer V 2.5 (Wang et al. 2011). Fertility was analyzed in two ways: as a continuous trait and as a binary trait (males producing no offspring vs. some offspring). The latter method produced data similar to sperm motility assays that are typically used in such experiments. Sex-ratio distortion is measured as the ratio of the number of females to total progeny number, and mapping was performed using data from males that produced five or more progeny. Because distortion is measured as a ratio of the number of females to the total number of progeny, data from crosses that produced very few progeny are obviously meaningless. Significance thresholds were calculated using permutations of data in QTL Cartographer (P = 0.05 and n = 500). Because only individuals that produced more than five progeny were used in the analysis for segregation distortion, the sample sizes for these analyses are smaller than those used for analysis of hybrid sterility. One consequence is reduced mapping power for segregation distortion relative to that for hybrid sterility. A total of 576 males were used for mapping the X chromosome, and 480 males were used for the autosomal mapping.
Results
Mapping X-linked Bogota factors
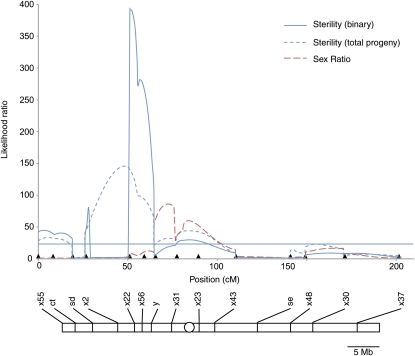
The D. pseudoobscura X is a large metacentric chromosome that constitutes nearly 40% of the entire genome. I mapped loci affecting hybrid sterility and segregation distortion on the Bogota X chromosome using 14 markers, 4 of which were visible, including se. The first striking observation of the mapping population prior to genotyping was that hybrid sterility is a nearly Mendelian trait. Nearly half of the mapping population (which is controlled for the large effect of Ovd on XR) is completely sterile, indicating the possibility of a single large-effect locus affecting hybrid sterility. This large effect manifests in the CIM as a large peak tightly linked to the marker X22 on XL (Figure 2). Indeed, when Bogota Ovd is present, X22 is a strong predictor of whether a recombinant male is fertile or sterile; nearly all males that carry the Bogota allele at X22 are sterile and vice versa. Previous studies showed a small, but nonsignificant, effect of the ct region (Orr and Irving 2001). Consistent with this previous report, I find this small but now statistically significant effect of the ct region in my analysis. I also detect two other small-effect loci near X2 and X23, respectively. These results are consistent with previous studies (Orr and Irving 2001, 2005), where single-marker analysis revealed a large effect of XL on hybrid sterility. To simplify analysis and to render my results comparable with those from previous studies, I rely on CIM of X-linked loci that treat hybrid sterility as a binary trait. I conclude from these mapping studies that, in addition to Ovd on XR, there is one major-effect locus contributing to hybrid sterility on Bogota XL at X22 and three loci of modest effect at ct, X2, and X23 (Figure 2).
Figure 2 .
Mapping of loci on the Bogota X chromosome that affect fertility and segregation distortion in hybrids. The X chromosome of D. pseudoobscura is a large metacentric chromosome that represents 40% of the entire genome. Likelihood ratio (LR) test statistic profiles from CIM of male fertility and segregation distortion in the X-chrosomosome mapping population. All genotyped males are Bogota at the sepia locus to control for the effects of Overdrive (Ovd), and therefore this prominent locus does not appear in this analysis. Solid blue line represents mapping of hybrid sterility as a binary trait, dashed blue line represents mapping of hybrid sterility as a continuous trait based on total progeny numbers, and red dashed line represents mapping of segregation distortion loci. LR significance thresholds from permutation tests for all phenotypes were close to 25.00 and are shown as a single horizontal line for clarity. Marker locations are shown as solid triangles. Markers names are presented below their respective solid triangles, and their positions on the physical map are shown to scale. The centromere is represented as an open circle.
Analysis of segregation distortion reveals peaks near X31 and X23, a region that flanks the centromere of the X chromosome and that also affects hybrid sterility. No peak for segregation distortion is detected near X22, the region that has the largest effect on hybrid sterility. This suggests that the genetic bases of hybrid sterility and segregation distortion may be separable at X22. However, it is important to note that mapping power to detect the segregation distortion effect of a locus that has a large effect on hybrid sterility is—essentially by definition—reduced in this experimental design. Put differently, if X22 has a large effect on both hybrid sterility and segregation distortion, then the progeny count data necessary to detect the effect of this locus on segregation distortion will be reduced as nearly all males carrying the Bogota allele at X22 are sterile. My mapping results indicate that the genetic bases of hybrid sterility and segregation distortion are at least partially overlapping, but the extent of this overlap cannot be addressed further with this experimental design.
Mapping dominant USA autosomal factors
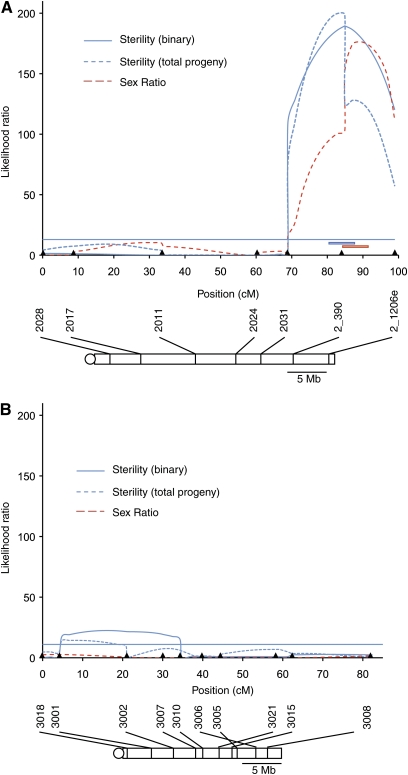
The second chromosome is the largest autosome in D. pseudoobscura. I mapped loci for hybrid sterility and segregation distortion on the second chromosome using seven markers. CIM reveals a single large-effect QTL for both hybrid sterility and segregation distortion at marker 2_390 (Figure 3A).
Figure 3 .
Mapping of dominantly acting USA autosomal loci. Likelihood ratio (LR) test statistic profiles from CIM of male fertility and segregation distortion in the autosomal mapping population. Solid blue line represents mapping of hybrid sterility as a binary trait, dashed blue line represents mapping of hybrid sterility as a continuous trait based on total progeny numbers, and red dashed line represents mapping of segregation distortion loci. LR significance thresholds from permutation tests for all phenotypes were around 10.00 and are shown as a single horizontal line for clarity. Conventions for marker and centromere locations are as in Figure 2. (A) Mapping of factors on the second chromosome. The blue and red bars show one standard deviation around the QTL for hybrid sterility and segregation distortion, respectively. (B) Mapping of factors on the third chromosome. A scrambling of marker order between 3006 and 3021 is observed because the linkage map is based on the Treeline arrangement in the mapping population and the physical map is based on the sequenced strain of D. pseudoobscura, which harbors the Arrowhead arrangement. Treeline and Arrowhead differ by multiple overlapping inversions on the distal part of the third chromosome.
I also mapped loci for hybrid sterility and segregation on the third chromosome using 10 markers to genotype the autosomal mapping population. I detected one locus with a small effect on hybrid sterility between the markers 3001 and 3002 (Figure 3B). This region lies in the collinear part of the third chromosome that is not tied up in a chromosomal inversion difference. CIM does not detect regions with an effect on segregation distortion on chromosome 3.
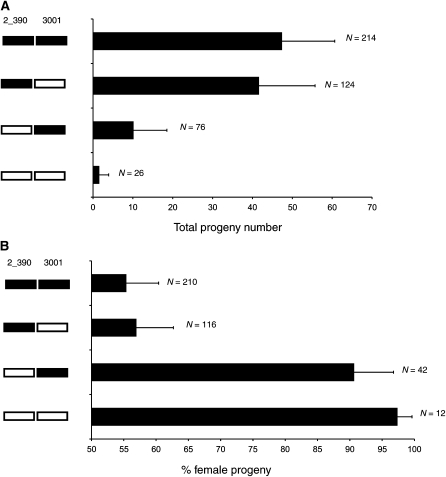
I tested the patterns of epistasis between the loci on the second and third chromosomes (Figure 4). In presence of a Bogota X chromosome, the 2_390 region on the second chromosome alone has a large effect on hybrid sterility and segregation distortion. The 3001 region on the third chromosome has a small effect of hybrid sterility. Full expression of hybrid sterility and segregation distortion is seen when both second and third chromosome loci carry material from USA, highlighting the epistatic interactions between these loci.
Figure 4 .
Pattern of conspecific epistasis between the autosomes. Bogota material is solid and USA material is open. 2_390 is a QTL on the second chromosome and 3001 is a QTL on the third chromosome. While the 2_390 region on the second has a large effect and the 3001 region on the third chromosome has a small effect on hybrid sterility and segregation distortion, the full expression of both phenotypes requires both regions from USA to be present simultaneously. (A) Pattern of epistasis for hybrid sterility. Fertility for each genotype is presented as the mean and standard deviation of the total progeny count. (B) Pattern of epistasis for hybrid segregation distortion. Segregation distortion for each genotype is presented as the mean and standard deviation of the percentage of female progeny.
Discussion
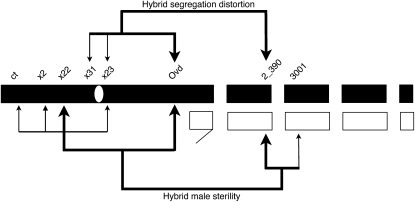
The Dobzhansky–Muller model elegantly describes the evolution of intrinsic reproductive barriers between species. Much effort has been devoted to identifying single genes involved in such incompatibilities, and several are now known. However, several outstanding questions remain about the structure of Dobzhansky–Muller interactions that separate species. In particular, it is difficult to say how many genetically independent incompatibilities typically cause postzygotic isolation between taxa and how many loci typically interact in a single incompatibility. Taken together with past studies (Orr and Irving 2001, 2005; Phadnis and Orr 2009), my results provide a more complete picture of the complex genetic architecture that underlies hybrid F1 male sterility between the Bogota and USA subspecies of D. pseudoobscura. Bogota and USA are young taxa in the earliest stages of speciation, and the genes that cause sterility of hybrid males are essential for reproductive isolation between these taxa. We can map this set of genes that are essential for reproductive isolation and can say with some certainty how many incompatibilities underlie hybrid sterility, how many individual loci are involved in Dobzhansky–Muller interactions, and the extent to which hybrid sterility may share a genetic basis with segregation distortion in these young species (Figure 5).
Figure 5 .
The genetic architecture of male sterility and segregation distortion in hybrid F1 males between Bogota and USA. Hybrid male sterility is caused by loci on the Bogota X chromosome that interact with dominantly acting loci on the USA autosomes. Segregation distortion is caused by loci on the Bogota X chromosome; Bogota material at 2_390 acts as a recessive suppressor of this segregation distortion. Hybrid male sterility and segregation distortion show a partial overlap in their genetic bases. Bogota material is solid, and USA material is open. The large metacentric X chromosome of D. pseudoobscura is denoted as a long bar and the centromere as an open oval, the Y chromosome as a hooked bar, and the autosomes as small bars. Thick lines denote large-effect loci, and thin lines denote small-effect loci.
A single Dobzhansky–Muller incompatibility causes hybrid F1 male sterility between Bogota and USA: replacing an incompatible allele with a compatible allele at any single hybrid incompatibility locus results in restored fertility. For example, replacing the incompatible Bogota allele at X22 in an F1 hybrid male with a compatible USA allele results in nearly completely restored fertility. Similarly, replacing the incompatible USA allele at 2_390 in an F1 hybrid male with a compatible Bogota allele results in nearly completely restored fertility. This single incompatibility involves a relatively modest number of loci that interact in a complex pattern. A total of seven loci—two large-effect loci (X22 and Ovd) and three small-effect loci (ct, X2, and X23) on the Bogota X chromosome along with two dominantly acting loci on the USA autosomes (2_390 and 3001)—interact to cause hybrid F1 male sterility (Figure 5). Interactions between these Bogota X chromosomal alleles and USA autosomal alleles form the basis of this hybrid incompatibility. The Bogota–USA hybridization is one of a few cases in which the Y plays little or no role in hybrid male sterility.
Two different explanations have been offered to explain the pattern of complex epistasis that often characterizes hybrid sterility. According to the first explanation, sterility is caused by the cumulative effects of a large number of factors, each of small effect (Naveira and Fontdevila 1986, 1991 Davis and Wu 1996). An extreme version of this hypothesis maintains that even the identities of the genes may not be important: sterility is caused when the amount of foreign genetic material crosses a threshold (Naveira and Fontdevila 1986, 1991). According to the second explanation, hybrid sterility may be caused by the interactions of a few genes of large effect (Orr 1995). Here, I found a modest number of loci involved in hybrid sterility, with only three large-effect loci accounting for most of the effect (X22, Ovd, and 2_390). These results support the idea that few genes of large effect may underlie hybrid male sterility. The number of incompatibilities is predicted to increase faster than linearly with time [the “snowball effect” (Orr 1995; Matute et al. 2010; Moyle and Nakazato 2010)]. It may, therefore, be difficult to tease apart the effects of individual hybrid sterility loci in older taxa, where many distinct incompatibilities may underlie postzygotic isolation. This further highlights the importance of genetic analysis of postzygotic isolation in very young taxa that are in the early stages of speciation.
In addition, my analyses find that the genetic basis of hybrid segregation distortion and its suppression also appears to be fairly simple. Sex-chromosome segregation distortion requires three loci on the X chromosome: loci near X31 on XL and X23 and Ovd on XR cause sex chromosome distortion. This distortion is almost completely suppressed by a recessive suppressor(s) near 2_390 on the Bogota second chromosome.
A comparison of QTL locations for hybrid sterility and segregation distortion suggests that the two phenotypes at least partially share their genetic bases. Although some loci, such as ct and X22 on XL, appear exclusive to hybrid sterility, other loci such as X23 on XR and 2_390 have an effect on both phenotypes. As noted earlier, however, mapping power for genes causing segregation distortion decreases in regions that have a strong effect on hybrid sterility in this experimental design. These results are consistent with the previously observed correlation between hybrid sterility and segregation distortion (Orr and Irving 2005; Phadnis and Orr 2009). It is important to note that, although loci that cause hybrid sterility and segregation distortion reside near each other, they need not always share exactly the same genetic basis. Instead, the genetic bases of the two hybrid phenomena may only partially overlap. Determining whether the same genes cause both hybrid sterility and segregation distortion will require fine mapping of individual loci and characterization of the effect of the genes on both phenotypes (e.g., Phadnis and Orr 2009).
It is important to emphasize that the hybrid incompatibility loci studied here play an essential role in the sterility of hybrid F1 males. Studies of genes that cause hybrid F1 sterility or inviability—as opposed to those of recessive loci that cause problems in specific F2-like genotypes—are rare (Barbash et al. 2003; Slotman et al. 2004; Brideau et al. 2006; Chang and Noor 2007; Phadnis and Orr 2009). Genome-wide introgression studies that rely on introducing single chromosomal regions from one species into another detect abundant recessive hybrid sterility loci, but miss dominant loci that underlie hybrid F1 sterility (True et al. 1996; Tao et al. 2003; Masly and Presgraves 2007). A likely reason for this discrepancy is that complex conspecific epistasis between several genes may underlie F1 hybrid sterility. More precisely, because multiple loci from the same species may be required together to complete a hybrid incompatibility, moving single chromosomal regions alone from one species into another may cause no F1 hybrid sterility.
This problem can be seen more clearly in the light of the D. pseudoobscura Bogota–USA hybridization. For example, the Bogota alleles at X22 and Ovd together have large effects on hybrid sterility, but have little or no effect when considered singly. Introgressing either region singly from Bogota into USA is not expected to show any hybrid phenotype. On the other hand, when only a single incompatibility is involved in postzygotic isolation, removing any single essential partner breaks the incompatibility and results in fully restored fertility or viability, e.g., Hmr (Barbash et al. 2003) and Lhr (Brideau et al. 2006) in D. melanogaster–D. simulans hybrids and Ovd in Bogota–USA hybrids (Phadnis and Orr 2009). Also, Chang and Noor (2010) suggest that the dominance of loci that cause F1 hybrid sterility may be modified by epistatic interaction with other Dobzhansky–Muller partners. If so, this phenomenon would also complicate the mapping of dominant partners involved in postzygotic isolation. This hypothesis would predict, for example, that homozygous USA material at 3001 in an otherwise Bogota background would be sufficient to cause hybrid male sterility even in the absence of the USA material at 2_390, and vice versa. Constructing introgression lines that move the autosomal regions at 2_390 and 3001 from USA into Bogota will allow a test of the hypothesis in this hybridization.
The locus near 2_390 on the second USA chromosome that acts dominantly to cause hybrid sterility in Bogota–USA hybrids is in the same region as a locus that causes hybrid sterility between Drosophila persimilis and D. pseudoobscura Bogota (Chang and Noor 2007). D. persimilis and D. pseudoobscura USA are sympatric species with incomplete reproductive isolation and are known to exchange genes, albeit rarely, in the collinear regions of the genomes (Wang et al. 1997; Noor et al. 2001). Because this locus near marker 2_390 has little or no effect on the sterility of hybrids between D. persimilis and D. pseudoobscura USA, genes in this region may be exchanged between these species with little opposition from natural selection (Noor et al. 2001). These observations raise the possibility that gene flow between D. persimilis and D. pseudoobscura USA may have led them to share alleles at the same gene(s) at 2_390 that cause hybrid sterility when crossed to Bogota (McDermott and Noor 2011). Higher-resolution mapping of this large-effect locus may determine whether the same gene(s) cause hybrid sterility in D. pseudoobscura Bogota–USA hybrids and in D. persimilis–D. pseudoobscura Bogota hybrids.
The study of individual components of hybrid incompatibilities underlying hybrid sterility and inviability has led to significant insights into the likely biological underpinnings of hybrid defects. Yet, studies of individual genes in hybrid incompatibilities have left unanswered important questions about the nature of the Dobzhansky–Muller hybrid incompatibility itself: How many genes are involved and what are their effect sizes? Indeed, given that hybrid incompatibilities accumulate with a snowball effect (Orr 1995; Matute et al. 2010; Moyle and Nakazato 2010), such a comprehensive view may be out of reach for all but the youngest diverged species pairs. My results lay the foundation for fine-mapping experiments to identify all the genes that interact with Ovd to cause male sterility and segregation distortion in hybrid F1 males between the Bogota and USA subspecies of D. pseudoobscura. Since postzygotic isolation between Bogota and USA involves a single incompatibility with a modest number of interacting loci, identification and characterization of these genes will provide important insight into the molecular nature of postzygotic isolation between species.
Acknowledgments
I am grateful to Harmit S. Malik and H. Allen Orr for their generous support for this study. I thank Anna Greenwood and Andrea Sweigart for advice on genotyping and CIM analysis and Dave Begun, Mia Levine, Harmit S. Malik, H. Allen Orr, Katie Peichel, and three anonymous reviewers for providing comments on the manuscript. I also thank Steve Schaeffer for generously providing the D. pseudoobscura Treeline strain. This work was supported by National Institutes of Health grants GM51932 (H. A. Orr) and GM74108 (HSM) and a grant from the Mathers foundation (H. S. Malik). N.P. is a Howard Hughes Medical Institute fellow of the Life Sciences Research Foundation.
Literature Cited
- Barbash D. A., Siino D. F., Tarone A. M., Roote J., 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100: 5302–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W., 1909. Heredity and variation in modern lights, pp. 85–101 Darwin and Modern Science. Cambridge University Press, Cambridge, UK [Google Scholar]
- Benson G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295 [DOI] [PubMed] [Google Scholar]
- Carvalho A. B., Clark A. G., 2005. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 307: 108–110 [DOI] [PubMed] [Google Scholar]
- Chang A. S., Noor M. A., 2007. The genetics of hybrid male sterility between the allopatric species pair Drosophila persimilis and D. pseudoobscura bogotana: dominant sterility alleles in collinear autosomal regions. Genetics 176: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. S., Noor M. A., 2010. Epistasis modifies the dominance of loci causing hybrid male sterility in the Drosophila pseudoobscura species group. Evolution 64: 253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA [Google Scholar]
- Davis A. W., Wu C. I., 1996. The broom of the sorcerer’s apprentice: the fine structure of a chromosomal region causing reproductive isolation between two sibling species of Drosophila. Genetics 143: 1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1937. Genetics and the Origin of Species. Columbia University Press, New York [Google Scholar]
- Dobzhansky T., Hunter A. S., Pavlovsky O., Spassky B., Wallace B., 1963. Genetics of natural populations. XXXI. Genetics of an isolated marginal population of Drosophila pseudoobscura. Genetics 48: 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G. B., Preston C. R., Johnson-Schlitz D. M., Nassif N. A., Phillis R. W., et al. , 1993. Type I repressors of P-element mobility. Genetics 135: 81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1922. Sex-ratio and unisexual sterility in hybrid animals. J. Genet. 12: 8 [Google Scholar]
- Masly J. P., Presgraves D. C., 2007. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5: e243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D. R., Butler I. A., Turissini D. A., Coyne J. A., 2010. A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science 329: 1518–1521 [DOI] [PubMed] [Google Scholar]
- McDermott S. R., Noor M. A., 2011. Genetics of hybrid male sterility among strains and species in the Drosophila pseudoobscura species group. Evolution 65: 1969–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle L. C., Nakazato T., 2010. Hybrid incompatibility “snowballs” between Solanum species. Science 329: 1521–1523 [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution and temperature. Biol. Symp. 6: 71–125 [Google Scholar]
- Naveira H., Fontdevila A., 1986. The evolutionary history of Drosophila buzzatii. Xii. The genetic basis of sterility in hybrids between D. buzzatii and its sibling D. serido from Argentina. Genetics 114: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveira H., Fontdevila A., 1991. The evolutionary history of D. buzzatii. XXII. Chromosomal and genic sterility in male hybrids of Drosophila buzzatii and Drosophila koepferae. Heredity 66(Pt. 2): 233–239 [DOI] [PubMed] [Google Scholar]
- Noor M. A. F., 1995. Incipient sexual isolation in Drosophila pseudoobscura bogotana. Pan-Pac. Entomol. 71: 125–129 [Google Scholar]
- Noor M. A., Grams K. L., Bertucci L. A., Reiland J., 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139: 1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1996. Dobzhansky, Bateson, and the genetics of speciation. Genetics 144: 1331–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Irving S., 2001. Complex epistasis and the genetic basis of hybrid sterility in the Drosophila pseudoobscura Bogota-USA hybridization. Genetics 158: 1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Irving S., 2005. Segregation distortion in hybrids between the Bogota and USA subspecies of Drosophila pseudoobscura. Genetics 169: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barrientos D., Chang A. S., Noor M. A., 2006. A recombinational portrait of the Drosophila pseudoobscura genome. Genet. Res. 87: 23–31 [DOI] [PubMed] [Google Scholar]
- Phadnis N., Orr H. A., 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., 1972. Origin of reproductive isolation in the absence of apparent genic differentiation in a geographic isolate of Drosophila pseudoobscura. Genetics 72: 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180 [DOI] [PubMed] [Google Scholar]
- Richards S., Liu Y., Bettencourt B. R., Hradecky P., Letovsky S., et al. , 2005. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 15: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Schaeffer S. W., Miller E. L., 1991. Nucleotide sequence analysis of Adh genes estimates the time of geographic isolation of the Bogota population of Drosophila pseudoobscura. Proc. Natl. Acad. Sci. USA 88: 6097–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman M., Della Torre A., Powell J. R., 2004. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis. Genetics 167: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Zeng Z. B., Li J., Hartl D. L., Laurie C. C., 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. II. Mapping hybrid male sterility loci on the third chromosome. Genetics 164: 1399–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True J. R., Weir B. S., Laurie C. C., 1996. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142: 819–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. L., Wakeley J., Hey J., 1997. Gene flow and natural selection in the origin of Drosophila pseudoobscura and close relatives. Genetics 147: 1091–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Basten C. J., Zheng Z.-B., 2011. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- Zeng Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]