Abstract
There is increasing evidence that epigenetic marks such as DNA methylation contribute to phenotypic variation by regulating gene transcription, developmental plasticity, and interactions with the environment. However, relatively little is known about the relationship between the stability and distribution of DNA methylation within chromosomes and the ability to detect trait loci. Plant genomes have a distinct range of target sites and more extensive DNA methylation than animals. We analyzed the stability and distribution of epialleles within the complex genome of the oilseed crop plant Brassica napus. For methylation sensitive AFLP (MSAP) and retrotransposon (RT) epimarkers, we found a high degree of stability, with 90% of mapped markers retaining their allelic pattern in contrasting environments and developmental stages. Moreover, for two distinct parental lines 97% of epialleles were transmitted through five meioses and segregated in a mapping population. For the first time we have established the genetic position for 17 of the 19 centromeres within this amphidiploid species. Epiloci and genetic loci were distributed within distinct clusters, indicating differential detection of recombination events. This enabled us to identify additional significant QTL associated with seven important agronomic traits in the centromeric regions of five linkage groups.
DNA methylation is a modification of DNA sequence that serves to repress the influence of transposable elements (TEs) and other repetitive sequences within the eukaryote genomes of fungi, plants, and animals (Martienssen and Colot 2001). Cytosine methylation (5mC), catalyzed by DNA methyltransferases, may also lead to heritable and/or reversible changes in chromatin structure and gene expression that do not entail changes in nucleotide sequence (Finnegan et al. 2000; Martienssen and Colot 2001).
DNA methylation in plant nuclear genomes was initially thought to occur predominantly in the cytosines of symmetrical sequences such as CG and CHG, with up to 20–40% of all cytosine residues being methylated (Gruenbaum et al. 1981; Messeguer et al. 1991). However, the level of 5mC varies considerably across plant taxa, with early estimates indicating 6% of cytosines methylated in Arabidopsis (Kakutani et al. 1999), 24% of potential methylation sites in B. oleracea (Kovarik et al. 1997), and up to 25% in maize (Papa et al. 2001). More recently, whole genome bisulfite sequencing at base-pair resolution in Arabidopsis (Lister et al. 2008) has shown that 5.26% of all genomic cytosines are methylated and unevenly distributed in the genome, with the majority in the CG context (55%), followed by CHG (23%), and CHH (22%). Heavy methylation is observed in the centromere and pericentromeric region (Zilberman et al. 2007; Lister et al. 2008; Yan et al. 2010). Transposable elements and sequences flanking insertion sites are typically methylated, suppressing transposon activity, especially in centromeres and pericentromeric regions where abundant TEs insert (Slotkin and Martienssen 2007; Zilberman et al. 2007; Han et al. 2010).
Across most eukaryote taxa, DNA methylation plays an important role in controlling gene expression (Suzuki and Bird 2008) and phenotype, including human disease (Robertson 2005; Watanabe and Maekawa 2010) and plant development (Kalisz and Purugganan 2004; King et al. 2010). In higher plants, genome-wide decreased DNA methylation can alter flowering time (Horváth et al. 2002; Kondo et al. 2006) and specific epialleles may result in phenotype variation in different crops. For example, in tomato, a hypermethylated allele of an SBP-box gene LeSPL reduces its expression in epimutant fruits, leading to inhibition of ripening (Manning et al. 2006). During Arabidopsis seed development >50 genes are imprinted in the female tissue, with corresponding hypomethylation in the endosperm in addition to the well-described imprinted genes FWA, MEA, FIE, and FIS2 (Gehring et al. 2009). Also in B. rapa, the epiallele of SP11 is involved in the dominance interaction that determines the self-incompatibility phenotype (Shiba et al. 2006).
Although there is increasing evidence that phenotype is influenced by epigenetic marks playing a key role in regulating gene transcription, developmental plasticity, and interactions with the environment, relatively little is known about the relationship between the distribution of DNA methylation within chromosomes and the ability to detect trait loci. A first step is to determine the chromosomal distribution of 5mC and how this may affect the detection of recombination events in segregating populations.
The B. rapa genome is estimated to be 480–520 Mb and to contain 40–50% repeat sequences, including TEs in the euchromatin region (14%) and repeat sequences in the large centromeric region (30%) (Johnston et al. 2005; Lim et al. 2005; Hong et al. 2006). A recent cytological study has suggested a distinct distribution of DNA methylation, primarily located in the heterochromatin, when compared with B. oleracea (Braszewska-Zalewska et al. 2010), which diverged from B. rapa ∼3.7 MYA (Inaba and Nishio 2002) and shares a common complex pattern of triplicated segments arising from a series of ancestral duplication events (Yang et al. 2006). A high rate of DNA methylation (52–60%) has been found in different accessions of B. oleracea, detected by the methylation-sensitive amplification polymorphism (MSAP) method, with most of the MSAP fragments stably inherited through meiosis (Salmon et al. 2008).
B. napus is a stable amphidiploid oil crop of major economic significance (canola, oilseed rape), derived from a recent hybridization between B. rapa and B. oleracea (Allender and King 2010). We have previously mapped hundreds of QTL controlling dozens of important agronomic traits on a B. napus reference genetic map (BnTNDH) (Qiu et al. 2006; Long et al. 2007; Shi et al. 2009). Here we report the detection of DNA methylation loci that enabled construction of an epigenetic map, based on MSAP, retrotransposon-specific, and centromeric-specific markers. This enabled the resolution of epi-QTL that were distinct from genetic QTL within the context of an integrated map.
Materials and Methods
Phenotyping and epigenotyping
The reference BnTNDH mapping population has been described previously (Qiu et al. 2006). Phenotypic data for seven traits associated with yield, seed quality, and plant development were collected from between 2 to 10 environments in the field during 2002–2007 (Long et al. 2007; Shi et al. 2009) and used for QTL identification (Table S1). Fully expanded leaves 5–6 from seedlings, and young flower buds at budding stage were sampled to extract genomic DNA.
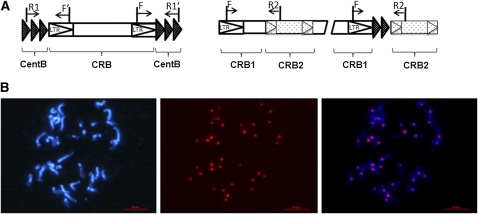
Two sets of epigenetic markers (epimarkers) were used for epigenotyping the DNA methylation status of the mapping population and parents. (1) MSAP markers, which have been used to detect CG and CHG site methylation variation in a range of genomes (Reyna-Lopez et al. 1997; Vuylsteke et al. 1999). For CG site methylation detection, restriction enzyme combinations EcoRI/HpaII and EcoRI/MspI were used, and PstI/MseI for CHG. (2) Two subclasses of retrotransposon-specific markers: (a) genome-wide retrotransposons (RTg) that detect transposons such as TRIM (Kwon et al. 2007) and SINE (Prieto et al. 2005) were detected by a sequence-specific amplification polymorphism (SSAP) approach (Waugh et al. 1997; Ellis et al. 1998) and (b) centromere-specific retrotransposons (RTc) that detect transposons clustered within cetromeric regions. Primers were designed on the basis of centromere-specific sequences within B. rapa (Lim et al. 2007), with insertion–deletion detected by PCR (Figure 1A; Table S2).
Figure 1 .
Centromeric retrotransposons (RTc) amplification and confirmation. (A, left) Single insertion of centromeric retrotransposon (CRB) is amplified from within an array of centromere satellite repeats (CentB). (Right) Nested insertion or doubled insertion of retrotransposons is amplified from within an interretrotransposon region, where the PCR products would be expected either to contain CentB in a doubled insertion interretrotransposon or not contain CentB in nested insertion. (B) BAC–FISH analysis using a Tapidor BAC clone (JBnB029F04) containing the RTc sequence used to design primers for the RTc markers. From left to right are chromosomes stained by 4′,6-diamidino-2-phenylindole in blue, FISH signals derived from BAC clone (red), and merged image. A total of 36 of 38 chromosomes with centromeres are visible in the cell. Major signals were localized in the centromere, indicating that the RTc markers were centromere specific.
Linkage map construction and QTL resolution
Three linkage maps were constructed and designated as epigenetic map (E-map), genetic map (G-map), and integrated map (I-map). Epimarkers were used to construct the E-map, which was superimposed on the framework of the existing well-defined G-map (Shi et al. 2009). This enabled epimarkers to be anchored within the context of stable genetic markers that define the established 19 linkage groups and chromosomes of B. napus. A revised conventional G-map was developed on the basis of genetic markers (genmarkers) that included RFLP, SSR, and STS mostly used in previous studies (Long et al. 2007; Shi et al. 2009). Maps were constructed using JoinMap 4.0 (Van Ooijen 2006) with default parameters and linkage groups distinguished at LOD values between 8 and 19. On the basis of the markers common to the E- and G-maps, BioMercator 2.0 software (http://www.generationcp.org/) was used to generate the I-map.
Phenotypic data from each environment (Long et al. 2007; Shi et al. 2009) were used to identify and map QTL within the three linkage maps (i.e., E-map, G-map, and I-map). Composite interval mapping (CIM) within WinQTLCart 2.5 (Wang et al. 2006) was used with a significance threshold of P = 0.05 using the permutation test method, based on 1000 runs of randomly shuffling the trait values (Churchill and Doerge 1994). QTL identified in different experiments for each trait (primary QTL) were integrated into consensus QTL by the meta-analysis method (Shi et al. 2009) (BioMercator 2.0, http://www.generationcp.org/).
Chromosome fluorescent in situ hybridization analysis
A total of 110 B. napus BAC clones were selected using a B. rapa centromere BAC (AC166739) as the query sequence in BLASTN alignments with TapidorDH BAC-end sequences within GenBank (http://www.ncbi.nlm.nih.gov/sites/entrez?db=nucleotide). Following PCR with RTc marker primers, clone JBnB029F04 was found to contain an identical band as the RTc marker located on linkage group A6. This BAC was used as a probe in BAC–fluorescent in situ hybridization (FISH), as described previously (Lim et al. 2005).
Analysis of transposon insertion divergence using Hydra
Loci containing structural variants (SVs) between the genomes of TapidorDH and Ningyou7DH were detected using Hydra (Quinlan et al. 2010). Solexa GAII paired-end reads (20×) generated from 1-2 kb and 2-3 kb insertion fragments of genomic DNA of Ningyou7DH were mapped to reference sequences of 11 BAC clones of TapidorDH (Cheung et al. 2009) available in GenBank. The software Maq (Abbosh et al. 2008) was used in the mapping step in place of Burrows-Wheeler Alignment tool (Chinnusamy and Zhu 2009). To identify whether the SV was caused by transposon insertion, SV-containing loci were analyzed using repeatMaster (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) with the Brassica retrotransposon database (ftp://ftp.tigr.org/pub/data/TransposableElements/transposon_db.pep) and further validated by sequencing the products using the retrotransposon-based insertion polymorphism (RBIP) method (Flavell et al. 1998). The SV loci associated with transposon insertion were then mapped to linkage map(s) with the PCR markers.
Results
Epimarkers are stable and informative in B. napus
It was important to establish the stability of specific epimarkers, as by their very nature, modifications such as DNA methylation may give rise to constitutive or dynamic behavior in the context of cell lineages through mitosis, ontogeny, response to environment, or transgenerationally through meiosis. We therefore tested the stability of MSAP markers in different tissues, stages of development, and different environments.
A total of 627 MSAP markers that reported CHG site methylation variation were screened against homozygous parental lines TapidorDH and Ningyou7DH, from leaves 5–6 at seedling stage in autumn (mean 20.7°), and from buds at floral budding stage in the spring following winter (mean 7.5°), from field-grown plants in Wuhan in the year 2006/2007. A total of 94% of the MSAP markers had a consistent DNA methylation status, of which most (86%) were stable in both parents at both stages/environments. A subset (9%) was stable in only one parent (Table 1). A total of 5% of MSAP markers were variable both between parents and between different developmental stages.
Table 1 . Variation in DNA methylation status for MSAP markers in Brassica napus at different developmental stages.
| Tapidor |
Ningyou7 |
Marker number | Subtotal | Methylated alleles (%) | Mapped on LGs | Subtotal | Marker type (%) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker type | Seedling | Budding | Seedling | Budding | ||||||
| Stable | − | − | + | + | 277 | 566 | 86 | 148 | 280 | 90 |
| + | + | − | − | 289 | 132 | |||||
| Semistable | + | + | − | + | 28 | 61 | 9 | 12 | 24 | 8 |
| − | + | + | + | 22 | 8 | |||||
| − | − | + | − | 6 | 1 | |||||
| + | − | − | − | 5 | 3 | |||||
| Unstable | + | − | − | + | 14 | 33 | 5 | 2 | 6 | 2 |
| − | + | + | − | 19 | 4 | |||||
| Total | 660 | 310 | ||||||||
+, methylated alleles; −, unmethylated alleles.
The heritability of DNA methylation patterns was determined in F1 plants (TapidorDH × Ningyou7DH) with 724 out of 925 (78%) at the seedling stage. A total of 543 (75%) of these loci had identical allelic bands as the dominant allele present in the corresponding parent. Among the dominant loci, the methylated alleles were distributed relatively evenly between the two parents (259:282, χ2 test, P > 0.3).
We checked the stability of MSAP markers in leaves 5–6 from the two parents, and in S5 (self-pollinated fifth generation) lines generated by single-seed descent, grown in the same environment. A total of 36 (97%) of the MSAP markers that were stable between the two developmental stages retained their DNA methylation allelic status through five meioses.
RTg and RTc markers were regarded as stable molecular markers since the assays did not involve the use of methylation sensitive enzymes. The centromeres of 17 linkage groups were identified using 39 RTc markers, with the position of eight centromeres confirmed by both single- and double-insertion RTc markers (Figure 1A; Table S3). The centromeric origin of the marker sequences was confirmed by FISH analysis, using as probes BACs from which the RTc primers had been designed (Figure 1B).
Distribution and density of epiloci within the B. napus genome
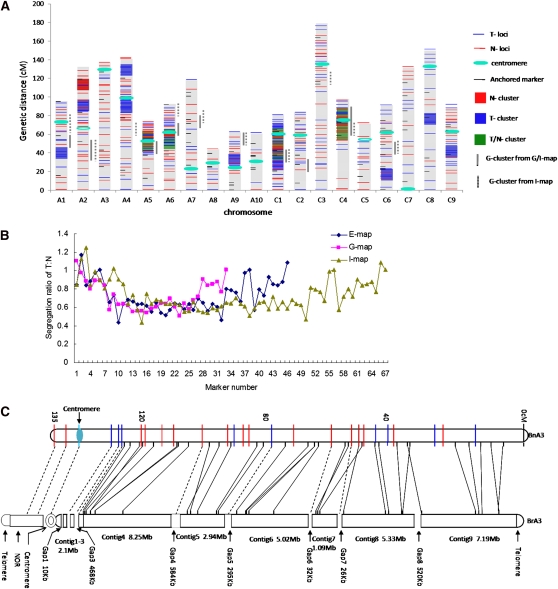
We constructed a new linkage map (E-map) with 484 epimarkers and 84 genetic markers. The latter defined the framework in terms of established linkage groups and provided anchors for subsequent comparison with genomic information (Figure 2A; Table S4). A total of 64% (310) of the 484 MSAP markers placed on the E-map had been confirmed to be consistent either across different developmental stages or environments, and 90% of these (280/310) were stable in both respects (Table 1). Although 36% of mapped MSAP markers were of unknown stability and a few unstable, only those that met the criterion of not influencing the order of non-MSAP genetic markers were retained. This ensured the reliability of the E-map and enabled it to be used for subsequent analysis. Segregation ratios across each chromosome were closely correlated for each type of marker, following similar patterns for the three maps. There appeared to be no bias associated with epimarkers, where 47% had distorted segregation ratios (χ2 test, P < 0.05) compared with 43% for genetic markers, supporting their suitability as reliable genetic markers (Figure 2B). Moreover, there appeared to be no bias introduced into construction of the E-map, with similar numbers of 5mC-derived alleles from each parent line.
Figure 2 .
Epilocus distribution and segregation ratio in the map of Brassica napus. (A) The landscape of the B. napus epigenetic map. Horizontal blue lines represent epialleles from TapidorDH, red lines epialleles from Ningyou7DH, and short black lines represent genmarkers.The red, blue, and green boxes are marker clusters with methylation originating from Ningyou7DH, TapidorDH, or both, respectively. There are 6 N-clusters, 13 T-clusters, and 4 T/N clusters. The vertical gray bars to the right of the LGs indicate genetic-marker clusters, with positions estimated from the common anchored markers in E-map, G-map, and I-map. Centromeres were positioned using RTc markers for 17 of the 19 linkage groups. The locations of the remaining centromeres (in A2 and C5) were estimated by comparison with the corresponding Arabidopsis centromere position (Parkin et al. 2005). (B) Distribution of parental allelic genotype segregation ratio for the three linkage maps in linkage group C1, which contained the highest number of epimarkers. (C) Comparative mapping of linkage group A3 of B. napus (top) and the A3 physical map of B. rapa (below, cited from Mun et al. 2010). The solid lines between the two maps indicate homologous loci detected by comparative mapping using genetic markers derived from the same B. rapa BACs. The dotted lines indicate where epiloci in B. napus are likely to correspond to heterochromatic regions (NOR, centromere and gaps that remained unassembled due to prevalence of repeat sequences) of the B. rapa A3 chromosome.
The distribution of epiloci appeared to be uneven across the E-map (Figure 2A; Table S5). Analysis with the Poisson distribution function demonstrated that 18% (104) were in clusters of more than eight loci within a 10-cM interval (P < 0.001). However, for four epilocus-rich linkage groups (A5, A6, C1, and C4) the clusters were associated with the centromeric region. We identified a larger number of small but distinct clusters that were associated with parent-of-origin DNA methylation, which we termed as ET cluster (methylated alleles derived from the female TapidorDH parent) and EN cluster (from Ningyou7DH). A total of 19 ET/N clusters (182 markers) were identified that contained a combination of ET and EN methylated alleles within the same region. These were distributed in distinct regions across the genome, covering 16% of the linkage map (296 cM) (Table 2; Figure 2A).
Table 2 . Number of epi- and genmarkers and clusters identified within the TNDH I-map.
| Number of markers |
Number of markers |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Epi- |
Gen- |
Total |
Epi- |
Gen- |
Total |
||||
| LG | A genome |
Cluster | LG | C genome |
Cluster | ||||
| A1 | 23 | 44 | 67 | G-cluster (1)a | C1 | 45 | 25 | 70 | E-cluster (3) G-cluster (1) |
| A2 | 35 | 29 | 64 | — | C2 | 20 | 25 | 45 | — |
| A3 | 27 | 36 | 97 | G-cluster (1) | C3 | 36 | 49 | 85 | G-cluster (1) |
| A4 | 37 | 27 | 64 | E-cluster (2) | C4 | 40 | 32 | 72 | E-cluster (2) G-cluster (1) |
| A5 | 29 | 41 | 70 | G-cluster (1) | C5 | 12 | 19 | 31 | — |
| A6 | 40 | 51 | 91 | E-cluster (2) G-cluster (1) | C6 | 22 | 31 | 53 | G-cluster (1) |
| A7 | 16 | 46 | 62 | G-cluster (1) | C7 | 16 | 22 | 38 | — |
| A8 | 9 | 27 | 36 | G-cluster (1) | C8 | 19 | 20 | 39 | — |
| A9 | 18 | 36 | 88 | G-cluster (1) | C9 | 27 | 23 | 50 | — |
| A10 | 4 | 31 | 35 | — | |||||
| Subtotal | 238 | 368 | 674 | 11 | Subtotal | 237 | 246 | 483 | 9 |
| CV | 0.46 | CV | 0.31 | ||||||
CV, coefficient of variation.
Numbers in parentheses indicates the number of clusters detected in each linkage group.
To determine whether this uneven distribution of epiloci was specific to the E-map, the distribution of conventional loci was analyzed within the corresponding BnTNDH G-Map of 648 genetic loci (including the 84 anchoring markers), together with the additional 17 RTc loci (to reveal the position of centromeres) (Table S4). As is common with many genetic maps, there was an uneven distribution of loci in the G-map, with 7 of the 11 clusters identified having a Poisson distribution function located around the centromere.
Since it was difficult to compare the remaining epi- and G-clusters directly, due to lack of common markers within the centromeric regions of the two maps, we generated an integrated map (I-map) using BioMercator 2.0. This consisted of 1157 loci (475 epiloci and 672 genetic loci) covering 2345 cM, with an uneven locus density (Table S4). While there were 1.5 times as many genloci in the A compared with the C genome, there were almost identical numbers of epiloci in each genome. However, the distribution of epiloci on individual linkage groups was extremely uneven, with a notably larger coefficient of variation (0.46) than that of genloci (0.31) (Table 2). A10 had the fewest (4) and lowest density (0.48) of epiloci, while C1 had the most (45) and highest density (7.0), although both linkage groups had a similar number and density of genloci. The I-map retained similar features of cluster distribution for gen- and epiloci as found in the component E- and G-maps (Table S5). Compared with genloci, which were concentrated around the centromeres and formed G-clusters, the epiclusters were distributed throughout the genome. However, there were three overlaps with G-clusters in centromeric regions (Figure 2A). Comparison with the physical map of B. rapa chromosome A3 indicated that ∼40% of epiloci in B. napus linkage group A3 were likely to be located in heterochromatic regions (Figure 2C).
To determine whether the distribution of epiloci was associated with the conserved ancestral blocks arising from sequential ancient duplications within the Brassicaceae, we compared homeologous and paralogous regions within the BnTNDH I-map. The 5mC distribution varied among these homologous blocks. For example, the number of epimarkers in block U (sensu Schranz) varied considerably within the four linkage groups A1 (15), A3 (4), A8 (4), and C7 (7).
DNA sequence properties associated with epiloci
We sequenced MSAP bands associated with 15 epiloci distributed across 10 linkage groups and found that about half of these were from genes coding for various proteins (8 sequences) or putative regulatory sequence (3 sequences) as identified by motif prediction. Most of the remaining sequences (5/15) were retrotransposon related (Figure S1A; Table S6). We confirmed that all 15 MSAP bands contained the methylation sensitive restriction cutting site CTGCAG.
Both RTg markers were sequenced and found to possess the LTR of the retrotransposon that had been used in the primer design, as well as a flanking sequence falling within a genic region, indicating that this type of marker was reliable (Figure S1B). Sequence analysis following cloning of 10 RTc markers confirmed that these were related to known centromere retrotransposons, and all sequences had high identity with the centromere-specific retrotransposon in the original B. rapa BAC (Lim et al. 2007). Three subclasses of centromeric sequences were detected, all possessing centromere retrotransposon LTR sequences, thus confirming that the RTc loci are most likely to be an accurate representation of the centromeric region (Figure S1C). As expected, the polymorphism of RTc markers arose from retrotransposon insertion–deletion events in the centromere region.
We analyzed sequence structural variation between the two parents using Hydra for a 1.8-Mb BAC sequence of TapidorDH (covering 0.7% of the genome) and 1.25 million Solexa reads of Ningyou7DH. A total of 18 structural variants were identified, of which half represented insertions and half represented deletions in Ningyou7DH. Of the nine deletion loci in Ningyou7DH, seven were possibly related to transposon insertion events in TapidorDH. Three of the events were validated by PCR amplification (Figure S1D), and found to be caused by transposon insertion into the TapidorDH genome, mapping to linkage groups A1, A3, and C9.
Detection and features of QTLepi
QTL controlling seven different traits, evaluated in 2–10 different environments, were identified and assigned as QTLepi or QTLdna, dependent upon their detection in either the E-map or G-map, respectively. Within the context of the I-map, where the confidence interval was solely associated with one class of locus, the QTL were further assigned as QTLepi-c (confident for QTLepi) and QTLdna-c (confident for QTLdna), respectively. Conversely, where the confidence interval was associated with a combination of epi- and genloci, the QTL was assigned as QTLepi-l (QTLepi-like) or QTLdna-l (QTLdna-like) according to whether the QTL peak position was closer to the epilocus or genlocus, respectively. For some traits, the same QTL positions could be identified from multiple environments, and these were assigned as “recurring” QTL and integrated into consensus QTL by meta-analysis to avoid overestimating the number. A total of 125 consensus QTLepi were detected from the E-map for the seven traits, compared with 148 (92%) QTLdna detected from the G-map (Table S7). A total of 26% of the QTLepi were distributed in epicluster regions, twice the average (1 QTLepi/17 cM) (χ2 test, P < 0.01). None of the six unstable markers were located within the confidence intervals of QTLepi in either the E-map or I-map.
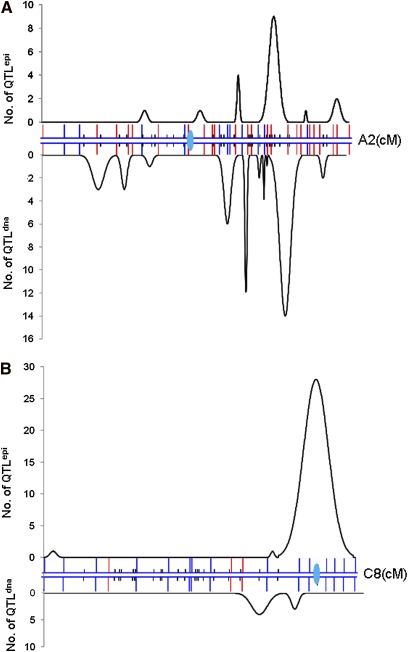
Although fewer QTLepi were detected in the I-map compared with the E-map, these still accounted for 28% of the total detected QTL, with 72% of QTLepi distributed on just nine linkage groups. Three linkage groups (A4, A6, and C8) contained more QTLepi than QTLdna (Table S8; Figure 3A). Conversely, only a few QTLepi were detected on linkage groups A10 and C7, with a very low QTL density in the latter. Linkage group C4 had both the lowest number and density of QTL at both epigenetic and genetic levels.
Figure 3 .
Distribution of consensus QTLepi on the I-map linkage groups. (A) Distribution of QTLepi and QTLdna on linkage group A2. (B) Distribution of QTLepi and QTLdna on linkage group C8. The curves above and below the LG represent QTLepi and QTLdna, regardless of LOD value, and take into account solely the number of QTL. The height of the curve represents the frequency of QTL detected, and the width of the baseline of the curve represents the extent of confidence intervals for the QTL in that region.
We tested the hypothesis of an interaction between QTLepi and environmental specificity. As expected, the number of QTL detected increased with the number of environments, with some QTL being specific to particular environments. Three parameters were calculated for each trait: the average number of QTL identified from one environment; the total integrated consensus QTL detected from all environments; and the index of environmental specificity, which we define as the ratio of consensus QTL to primary QTL for each trait (Table S9). In general, the average number of primary QTLepi detected from one environment was half that of QTLdna (18.3 vs. 39.5). Thus it appears there are no substantial differences between QTLepi and QTLdna in terms of the index of environmental specificity when the combined sets of four types of traits are taken into consideration.
Discussion
We have characterized the inheritance and distribution of two classes of epimarker within the complex B. napus genome. We found 90% of mapped epimarkers retained the same pattern in two contrasting developmental stages and environments. For two distinct parental lines, 97% of alleles were transmitted through five meioses. Epimarkers were distributed nonrandomly in the genome and appeared to affect the ability to detect recombination. A total of 125 QTL were associated with seven agronomic traits in regions having a greater density of DNA methylation markers. Our results will not only facilitate further characterization of epiloci, but also assist assembly of the complex B. napus polyploid genome, currently being sequenced using high throughput sequencing technology.
The centromere is a heterochromatic region where transposons accumulate, with consequent high levels of DNA methylation (Zilberman et al. 2007). Accurate mapping of centromeres is an important step for genomic and epigenomic studies. Although Pouilly et al. (2008) mapped centromere-related transposons on B. napus linkage maps, the definitive centromere position in each linkage group has been uncertain. Our accurate mapping of 17 of 19 centromeres with centromere-specific markers was confirmed by BAC–FISH and a published physical sequence map (A3).
Functional genes have recently been identified within centromeric regions of plants such as rice and wheat (Nagaki et al. 2004; Mutti et al. 2010). Interestingly, we found that the increased map resolution afforded by centromeric and epimarkers enabled us to detect a significant number of QTLepi (14% of total) associated with seven traits in the centromeric regions of five linkage groups. Recombination has been shown in genetic and cytological studies to be severely suppressed in plant centromeric regions (Ma et al. 2007), with reduced recombination frequency detected by conventional markers. Using chromosome orientation FISH (CO-FISH), Jaco et al. (2008) found high mitotic recombination frequency at mouse centromeres and established that DNA methylation was a negative regulator of centromere recombination. Our results suggest epimarkers may provide complementary information to reassess recombination frequency in centromeric regions.
We have demonstrated that most MSAP markers are transgenerationally stable through five successive meioses for the parent lines, and that polymorphic MSAP alleles still segregate in a Mendelian fashion in the BnTNDH population after three meioses following original fixation by microspore culture. DNA methylation was relatively stable at polymorphic loci. Only 5% of 627 MSAP markers were variable at different developmental stages following growth in contrasting environments. Only 6% of mapped MSAP markers were unstable. In plants, patterns of DNA methylation can be faithfully maintained across generations and transmitted through mitosis and/or meiosis (Martienssen and Colot 2001; Saze 2008). Hypomethylated Arabidopsis epi-recombinant inbred lines have been established varying with high heritability for flowering time and plant height, and stable inheritance of multiple parental DNA methylation variants over at least eight generations (Johannes et al. 2008).
Overall, there was a similar number of epialleles in each of the two parental genomes. However, more were detected in the A genome for Ningyou7DH (127) than TapidorDH (100), with identical numbers (114) in the C genome. We found 1.5 times more informative genmarkers within the A genome compared with the C. This may reflect the breeding pedigree of the Ningyou7DH parents, where B. rapa (AA) contributed as a recent ancestor, and de novo DNA methylation may have arisen during introgression to the A genome of B. napus. Hybrids from interspecific crosses in Arabidopsis (Bushell et al. 2003) and cotton (Keyte et al. 2006) show changes in the DNA methylation pattern compared with parents, and de novo methylation and demethylation occur in interspecific hybrids of beans (Abid et al. 2011).
Epiloci were unevenly distributed between linkage groups, with C1 having the greatest number (45) and density (7.0 per 10 cM), >10 times that within A10. This pattern of differential methylation was also detected between homologous ancestral blocks and may reflect a buffer of gene dosage resulting from formation of the amphidiploid (Paun et al. 2010). In primate genomes, the degree of CpG methylation in homologous genomic regions varies between human, chimpanzee and baboon (Meunier et al. 2005), suggesting the existence of a homology-dependent methylation (HDM) mechanism.
The uneven distribution of epiloci between the A and C genomes, between chromosomes and homologous blocks, may indicate that genome evolution of B. napus has directly or indirectly involved activation or repression of transposons during speciation, cultivar breeding, and possibly arising from cultivation in different environments, which in turn may contribute to the adaptation of different cultivars (Feinberg and Irizarry 2010; Yi et al. 2010)
Conventional QTL analysis relies on detection of recombination using DNA markers where parental alleles cosegregate with heritable phenotypic variation. These have been referred to as QTLdna to distinguish from QTLepi, resolved by detection of recombination using a DNA methylation polymorphism within the corresponding genomic region (Johannes et al. 2008). We used MSAP and retrotransposon-specific markers to reveal QTLepi, where the parental epialleles cosegregate with a trait in the BnTNDH population.
The epimarkers detected additional chiasmata within the population. We took care to ensure that the pattern of segregation and detection of chiasmata was consistent with the conventional genmarkers within each linkage group. The congruent pattern of segregation distortion for epi- and genmarkers along each chromosome also gave us confidence that the epimarkers provide an accurate reflection of recombination within the population. This has consequences for understanding the distribution of recombination frequency in different populations for the ability to detect and resolve QTL and for map-based cloning.
For seven traits in multiple environments, we detected between 2 and 17 QTLepi each, depending on the trait per se and the number of environments in which different traits were scored. The increased ability to resolve previously “cryptic” QTL suggests that studies relying solely on polymorphism of conventional genmarkers may consistently underestimate the quantity and distribution of QTL effects. We found that QTLepi were unevenly distributed among chromosomes, with 8 (12%) on C8 controlling plant development and one seed quality trait, while only 4 (2%) QTLdna were detected on the same chromosome. This suggests that human selection may have a strong impact on specific chromosomes, where the activities of transposons affect agronomic traits, as indicated by Hauben et al. (2009) in B. napus. Within chromosome C4, which contained more epiloci (39) than genloci (32), only 2 QTLdna and no QTLepi were detected. This chromosome may possess more heterochromatin and/or functional genes in regions of reduced polymorphism. Further interpretation of the QTLepi requires establishing a direct causality between the epigenetic status of any functional genes and the trait controlled by the QTLepi.
Retrotransposons have been estimated by FISH and sequence analysis to represent ∼30% of the B. rapa A genome (Lim et al. 2005; Hong et al. 2006). We found a similar proportion in B. napus, on the basis of BAC end sequence analysis (our unpublished data). This major contribution of transposable elements may provide genomic loci for epimarkers that then enable detection of additional QTL. Moreover, these QTLepi had equally as strong environment interaction as those of QTLdna (Table S9). Artificial reduction of DNA methylation has demonstrated effects on a wide range of phenotypic traits (King 1995; Finnegan et al. 1998; Johannes et al. 2008). Our results suggest that naturally occurring variation associated with DNA methylation may also generate epiallelic variation, able to be detected as marker polymorphism in regions where conventional genetic markers are monomorphic. Similar conclusions have been reached by recent selection studies in Brassica by Hauben et al. (2009) and Ni et al. (2009).
Acknowledgments
The authors are grateful to Zaiyun Li of Huazhong Agricultural University for his valuable help with the BAC–FISH technique. Financial support for this work was provided by the National Basic Research and Development Programme (2006CB101600) and the 111 Project (B07041) of China.
Literature Cited
- Abbosh P. H., Wang M., Eble J. N., Lopez-Beltran A., Maclennan G. T., et al. , 2008. Hypermethylation of tumor-suppressor gene CpG islands in small-cell carcinoma of the urinary bladder. Mod. Pathol. 21: 355–362 [DOI] [PubMed] [Google Scholar]
- Abid G., Muhoviski Y., Jacquemin J. M., Mingeot D., Sassi K., et al. , 2011. Changes in DNA-methylation during zygotic embryogenesis in interspecific hybrids of beans (Phaseolus ssp.). Plant Cell Tissue Organ Cult. 105: 383–393 [Google Scholar]
- Allender C. J., King G. J., 2010. Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braszewska-Zalewska A., Bernas T., Maluszynska J., 2010. Epigenetic chromatin modifications in Brassica genomes. Genome 53: 203–210 [DOI] [PubMed] [Google Scholar]
- Bushell C., Spielman M., Scott R. J., 2003. The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15: 1430–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F., Trick M., Drou N., Lim Y. P., Park J. Y., et al. , 2009. Comparative analysis between because genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell 21: 1912–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J. K., 2009. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T. H., Poyser S. J., Knox M. R., Vershinin A. V., Ambrose M. J., 1998. Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol. Gen. Genet. 260: 9–19 [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Irizarry R. A., 2010. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc. Natl. Acad. Sci. USA 107: 1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E. J., Genger R. K., Peacock W. J., Dennis E. S., 1998. DNA methylation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 223–247 [DOI] [PubMed] [Google Scholar]
- Finnegan E. J., Peacock W. J., Dennis E. S., 2000. DNA methylation, a key regulator of plant development and other processes. Curr. Opin. Genet. Dev. 10: 217–223 [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Knox M. R., Pearce S. R., Ellis T. H., 1998. Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J. 16: 643–650 [DOI] [PubMed] [Google Scholar]
- Gehring M., Bubb K. L., Henikoff S., 2009. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Naveh-Many T., Cedar H., Razin A., 1981. Sequence specificity of methylation in higher plant DNA. Nature 292: 860–862 [DOI] [PubMed] [Google Scholar]
- Han Y., Wang G., Liu Z., Liu J., Yue W., et al. , 2010. Divergence in centromere structure distinguishes related genomes in Coix lacryma-jobi and its wild relative. Chromosoma 119: 89–98 [DOI] [PubMed] [Google Scholar]
- Hauben M., Haesendonckx B., Standaert E., Van Der Kelen K., Azmi A., et al. , 2009. Energy use efficiency is characterized by an epigenetic component that can be directed through artificial selection to increase yield. Proc. Natl. Acad. Sci. USA 106: 20109–20114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C. P., Plaha P., Koo D. H., Yang T. J., Choi S. R., et al. , 2006. A survey of the Brassica rapa genome by BAC-end sequence analysis and comparison with Arabidopsis thaliana. Mol. Cells 22: 300–307 [PubMed] [Google Scholar]
- Horváth E., Szalai G., Janda T., Páldi E., Rácz I., et al. , 2002. Effect of vernalisation and azacytidine on the DNA methylation. Acta Biologica Szegediensis 46: 35–36 [Google Scholar]
- Inaba R., Nishio T., 2002. Phylogenetic analysis of Brassiceae based on the nucleotide sequences of the S-locus related gene, SLR1. Theor. Appl. Genet. 105: 1159–1165 [DOI] [PubMed] [Google Scholar]
- Jaco I., Canela A., Vera E., Blasco M. A., 2008. Centromere mitotic recombination in mammalian cells. J. Cell Biol. 181: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F., Colot V., Jansen R. C., 2008. Epigenome dynamics: a quantitative genetics perspective. Nat. Rev. Genet. 9: 883–890 [DOI] [PubMed] [Google Scholar]
- Johnston J. S., Pepper A. E., Hall A. E., Chen Z. J., Hodnett G., et al. , 2005. Evolution of genome size in Brassicaceae. Ann. Bot. (Lond.) 95: 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakutani T., Munakata K., Richards E. J., Hirochika H., 1999. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics 151: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisz S., Purugganan M. D., 2004. Epialleles via DNA methylation: consequences for plant evolution. Trends Ecol. Evol. 19: 309–314 [DOI] [PubMed] [Google Scholar]
- Keyte A. L., Percifield R., Liu B., Wendel J. F., 2006. Infraspecific DNA methylation polymorphism in cotton (Gossypium hirsutum L.). J. Hered. 97: 444–450 [DOI] [PubMed] [Google Scholar]
- King G. J., 1995. Morphological development in brassica-oleracea is modulated by in-vivo treatment with 5-azacytidine. J. Hortic. Sci. 70: 333–342 [Google Scholar]
- King G. J., Amoah S., Kurup S., 2010. Exploring and exploiting epigenetic variation in crops. Genome 53: 856–868 [DOI] [PubMed] [Google Scholar]
- Kondo H., Ozaki H., Itoh K., Kato A., Takeno K., 2006. Flowering induced by 5-azacytidine, a DNA demethylating reagent in a short-day plant, Perilla frutescens var. crispa. Physiol. Plant. 127: 130–137 [Google Scholar]
- Kovarik A., Matyasek R., Leitch A., Gazdova B., Fulnecek J., et al. , 1997. Variability in CpNpG methylation in higher plant genomes. Gene 204: 25–33 [DOI] [PubMed] [Google Scholar]
- Kwon S. J., Kim D. H., Lim M. H., Long Y., Meng J. L., et al. , 2007. Terminal repeat retrotransposon in miniature (TRIM) as DNA markers in Brassica relatives. Mol. Genet. Genomics 278: 361–370 [DOI] [PubMed] [Google Scholar]
- Lim K. B., de Jong H., Yang T. J., Park J. Y., Kwon S. J., et al. , 2005. Characterization of rDNAs and tandem repeats in the heterochromatin of Brassica rapa. Mol. Cells 19: 436–444 [PubMed] [Google Scholar]
- Lim K. B., Yang T. J., Hwang Y. J., Kim J. S., Park J. Y., et al. , 2007. Characterization of the centromere and peri-centromere retrotransposons in Brassica rapa and their distribution in related Brassica species. Plant J. 49: 173–183 [DOI] [PubMed] [Google Scholar]
- Lister R., O'Malley R. C., Tonti-Filippini J., Gregory B. D., Berry C. C., et al. , 2008. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y., Shi J., Qiu D., Li R., Zhang C., et al. , 2007. Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 177: 2433–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Wing R. A., Bennetzen J. L., Jackson S. A., 2007. Plant centromere organization: a dynamic structure with conserved functions. Trends Genet. 23: 134–139 [DOI] [PubMed] [Google Scholar]
- Manning K., Tor M., Poole M., Hong Y., Thompson A. J., et al. , 2006. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Martienssen R. A., Colot V., 2001. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293: 1070–1074 [DOI] [PubMed] [Google Scholar]
- Messeguer R., Ganal M. W., Steffens J. C., Tanksley S. D., 1991. Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol. Biol. 16: 753–770 [DOI] [PubMed] [Google Scholar]
- Meunier J., Khelifi A., Navratil V., Duret L., 2005. Homology-dependent methylation in primate repetitive DNA. Proc. Natl. Acad. Sci. USA 102: 5471–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun J. H., Kwon S. J., Seol Y. J., Kim J. A., Jin M., et al. , 2010. Sequence and structure of Brassica rapa chromosome A3. Genome Biol. 11: R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti J. S., Sandhu D., Sidhu D., Gill K. S., 2010. Dynamic nature of a wheat centromere with a functional gene. Mol. Breed. 26: 177–187 [Google Scholar]
- Nagaki K., Cheng Z., Ouyang S., Talbert P. B., Kim M., et al. , 2004. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36: 138–145 [DOI] [PubMed] [Google Scholar]
- Ni Z., Kim E. D., Ha M., Lackey E., Liu J., et al. , 2009. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa C. M., Springer N. M., Muszynski M. G., Meeley R., Kaeppler S. M., 2001. Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13: 1919–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin I. A., Gulden S. M., Sharpe A. G., Lukens L., Trick M., et al. , 2005. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O., Bateman R. M., Fay M. F., Hedren M., Civeyrel L., et al. , 2010. Stable epigenetic effects impact adaptation in allopolyploid orchids (Dactylorhiza: Orchidaceae). Mol. Biol. Evol. 27: 2465–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouilly N., Delourme R., Alix K., Jenczewski E., 2008. Repetitive sequence-derived markers tag centromeres and telomeres and provide insights into chromosome evolution in Brassica napus. Chrom. Res. 16: 683–700 [DOI] [PubMed] [Google Scholar]
- Prieto J. L., Pouilly N., Jenczewski E., Deragon J. M., Chevre A. M., 2005. Development of crop-specific transposable element (SINE) markers for studying gene flow from oilseed rape to wild radish. Theor. Appl. Genet. 111: 446–455 [DOI] [PubMed] [Google Scholar]
- Qiu D., Morgan C., Shi J., Long Y., Liu J., et al. , 2006. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor. Appl. Genet. 114: 67–80 [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Clark R. A., Sokolova S., Leibowitz M. L., Zhang Y., et al. , 2010. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 20: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna-Lopez G. E., Simpson J., Ruiz-Herrera J., 1997. Differences in DNA methylation patterns are detectable during the dimorphic transition of fungi by amplification of restriction polymorphisms. Mol. Gen. Genet. 253: 703–710 [DOI] [PubMed] [Google Scholar]
- Robertson K. D., 2005. DNA methylation and human disease. Nat. Rev. Genet. 6: 597–610 [DOI] [PubMed] [Google Scholar]
- Salmon A., Clotault J., Jenczewski E., Chable V., Manzanares-Dauleux M. J., 2008. Brassica oleracea displays a high level of DNA methylation polymorphism. Plant Sci. 174: 61–70 [Google Scholar]
- Saze H., 2008. Epigenetic memory transmission through mitosis and meiosis in plants. Semin. Cell Dev. Biol. 19: 527–536 [DOI] [PubMed] [Google Scholar]
- Shi J., Li R., Qiu D., Jiang C., Long Y., et al. , 2009. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba H., Kakizaki T., Iwano M., Tarutani Y., Watanabe M., et al. , 2006. Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat. Genet. 38: 297–299 [DOI] [PubMed] [Google Scholar]
- Slotkin R. K., Martienssen R., 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Suzuki M. M., Bird A., 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9: 465–476 [DOI] [PubMed] [Google Scholar]
- Van Ooijen J. W., 2006. JoinMap4, software for the calculation of genetic linkage maps in experimental populations. Kyazma B. V., Wageningen, The Netherlands [Google Scholar]
- Vuylsteke M., Mank R., Antonise R., Bastiaans E., Senior M. L., et al. , 1999. Two high-density AFLP linkage maps of Zea mays L.: analysis of distribution of AFLP markers. Theor. Appl. Genet. 99: 921–935 [Google Scholar]
- Wang S. C., Bastern J., Zeng Z. B., 2006 Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC: (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). [Google Scholar]
- Watanabe Y., Maekawa M., 2010. Methylation of DNA in cancer. Adv. Clin. Chem. 52: 145–167 [DOI] [PubMed] [Google Scholar]
- Waugh R., McLean K., Flavell A. J., Pearce S. R., Kumar A., et al. , 1997. Genetic distribution of Bare-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol. Gen. Genet. 253: 687–694 [DOI] [PubMed] [Google Scholar]
- Yan H., Kikuchi S., Neumann P., Zhang W., Wu Y., et al. , 2010. Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J. 63: 353–365 [DOI] [PubMed] [Google Scholar]
- Yang T.-J., Kim J. S., Kwon S.-J., Lim K.-B., Choi B.-S., et al. , 2006. Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C. X., Zhang S. L., Liu X. K., Bui H. T. N., Hong Y., 2010. Does epigenetic polymorphism contribute to phenotypic variances in Jatropha curcas L.? BMC Plant Biol. 10: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R. K., Ballinger T., Henikoff S., 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39: 61–69 [DOI] [PubMed] [Google Scholar]