Abstract
Sexual reproduction is a fundamental developmental process that allows for genetic diversity through the control of zygote formation, recombination, and gametogenesis. The correct regulation of these events is paramount. Sexual reproduction in filamentous fungi, including mating strategy (self-fertilization/homothallism or outcrossing/heterothallism), is determined by the expression of mating type genes at mat loci. Aspergillus nidulans matA encodes a critical regulator that is a fungal ortholog of the hSRY/SOX9 HMG box proteins. In contrast to well-studied outcrossing systems, the molecular basis of homothallism and role of mating type genes during a self-fertile sexual cycle remain largely unknown. In this study the genetic model organism, A. nidulans, has been used to investigate the regulation and molecular functions of the matA mating type gene in a homothallic system. Our data demonstrate that complex regulatory mechanisms underlie functional matA expression during self-fertilization and sexual reproduction in A. nidulans. matA expression is suppressed in vegetative hyphae and is progressively derepressed during the sexual cycle. Elevated levels of matA transcript are required for differentiation of fruiting bodies, karyogamy, meiosis, and efficient formation of meiotic progeny. matA expression is driven from both initiator (Inr) and novel promoter elements that are tightly developmentally regulated by position-dependent and position-independent mechanisms. Deletion of an upstream silencing element, matA SE, results in derepressed expression from wild-type (wt) promoter elements and activation of an additional promoter. These studies provide novel insights into the molecular basis of homothallism in fungi and genetic regulation of sexual reproduction in eukaryotes.
SEXUAL reproduction is a central part of the life cycle in most eukaryotic organisms and has a profound impact on the evolution and biology of species. Successful exchange of genetic material requires establishment of sexual identity (male and female function). Sexual identity in animals is governed by genes located on sex chromosomes, whereas in fungi mating type genes residing at mat loci (sexual-identity loci) control cell-type identity and sexual development. The correct regulation and spatiotemporal expression of sex determining genes is crucial during reproduction. Mechanisms underlying gene regulation in eukaryotic sexual development are not well understood (Harley et al. 2003; Fraser and Heitman 2005; Kashimada and Koopman 2010). Mating type genes have been described in numerous heterothallic (cross-fertile) and homothallic (self-fertile) filamentous fungi, where they function as master regulators of sexual reproduction (Kronstad and Staben 1997; Debuchy et al. 2010). The mat loci typically encode transcription factors, either Mat-HMG (high-mobility group box) or Mat-α (alpha box), proteins that coordinate expression of sex-specific genes. Several lines of study indicate that mating type genes in fungi share structural and functional features with the more complex sex chromosomes of algae, plants, and animals (Kronstad and Staben 1997; Marais and Galtier 2003; Fraser and Heitman 2004, 2005; Debuchy et al. 2010). It has also been suggested that Mat-HMG in fungi represents a structural ortholog of the human SRY regulator (sex-determining region Y) (Fraser and Heitman 2004, 2005; Fraser et al. 2005, 2007; Heitman 2006).
The genetic organization of mating type genes at mat loci determines sexual identity and mating strategy in filamentous fungi (Nelson 1996; Coppin et al. 1997; Debuchy et al. 2010). In heterothallic fungi, the mat locus typically encodes one of two nonallelic sequences (idiomorphs) that occupy the same genetic position on homologous chromosomes in two cross-compatible mating strains, termed mat-HMG (+) and mat-α (−). Idiomorphic structure of mat loci governs pheromone signaling and cell-specific gene expression in sexually compatible mating strains. Fertilization occurs exclusively between mat(+) and mat(−) strains and self-fertilization is prohibited (Turgeon 1998). The role of the mating type genes has been well characterized in outcrossing heterothallic systems, where Mat proteins control not only recognition mechanisms leading to cross-fertilization but also later stages of sexual development, including nuclear segregation and coordinated fruiting body differentiation (Metzenberg and Glass 1990; Debuchy and Turgeon 2006). In contrast to heterothallic species, most homothallic fungi have closely linked mat-HMG and mat-α genes present in the haploid genome. This genetic organization confers self-compatible (self-fertile) mating ability (Turgeon 1998; Yun et al. 2000; Debuchy and Turgeon 2006). An exception is self-fertile Neurospora africana, which has only mat A-1 (α box) present in the genome (Glass and Smith 1994). Homothallic species such as Aspergillus nidulans are also cross-fertile. Genetic analysis of outcrosses between self-fertile A. nidulans strains led Pontecorvo et al. (1953) to propose the term “relative heterothallism” to explain the ability of a homothallic species to behave as if it was heterothallic. However, a role for mat loci in discrimination and recognition between mating partners is poorly understood and remains elusive (Pontecorvo et al. 1953; Hoffmann et al. 2001). Several studies suggest that sexual identity in homothallic systems might be regulated by differential expression of mating type genes or the molecular “transformation” of the homothallic mat loci into a heterothallic state by selective epigenetic silencing of one of the two mating type genes (Metzenberg and Glass 1990; Raju and Perkins 2000; Fraser and Heitman 2004; Scazzocchio 2006).
It has been previously demonstrated that the HMG-box mating type transcription factor is necessary for fruiting body development in both heterothallic and homothallic fungi. Mat-HMG is required for correct expression of different sets of target genes directly involved in male and female fertility, fruiting body morphogenesis, fruiting body abundance, and ascospore formation (Coppin et al. 1997; Kronstad and Staben 1997; Debuchy 1999; Debuchy et al. 2010) Surprisingly, very little is known about the mechanisms that govern regulation and transcriptional expression of mating type genes in fungi.
A. nidulans is a homothallic filamentous fungus with unlinked mat loci, matA (HMG box) and matB (α box). In this study we analyzed the regulation and molecular function of the matA mating type gene during the self-fertile sexual cycle. Sexual development in A. nidulans involves coordinated differentiation of three multicellular tissue types: Hülle cells, ascogenous hyphae, and the cleistothecium wall. Hülle cells appear at the onset of the sexual cycle and function as nurse cells to protect unfertilized female organs (protocleistothecia) and provide nutrients for the developing fruiting body (cleistothecium) and ascospores. Sexual conjugation (mating) is believed to occur within clusters of Hülle cells, where cellular equivalents of female and male reproductive structures fuse and combine nuclei within a common cytoplasm during the process of fertilization. After fertilization, parental haploid nuclei divide synchronously in the female organ and are ultimately segregated into dikaryotic cells of proliferating ascogenous hyphae. Dikaryotic cells differentiate by a coordinated series of cell and nuclear divisions to form ascus mother cells, where nuclear fusion (karyogamy) is followed by meiosis. The differentiation of sexual spores (ascospores) occurs within these enclosed structures (asci). Concurrent with development of reproductive hyphae, vegetative aerial hyphae grow in circular fashion surrounding the fertile ascogenous tissue and eventually form the hard cleistothecial wall (Benjamin 1955; Champe et al. 1994; Turgeon 1998; Bruggeman et al. 2003; Pyrzak et al. 2008).
On the basis of gene deletion/complementation studies and the analysis of the transcriptional regulation, we have determined that unique, position-dependent and position-independent mechanisms regulate matA developmental expression and that the level of matA expression is a critical modulator of self-fertility. matA expression is suppressed in vegetative hyphae and is progressively derepressed in the developing sexual reproductive tissue. Vegetative suppression appears to be regulated by an upstream silencer element (SE). Deletion of the silencer, matA SE, resulted in the dramatic derepression/upregulation of the matA transcript in vegetative tissue and approximately threefold depression in developmental tissue. Additionally, we identified a conserved element (29-bp sequence) with striking structural similarity to the silencer of the human SRY gene. However this element did not have an effect on matA regulation. We did not find that vegetative suppression of matA is required for self-fertility. By contrast, an increase in matA transcript during sexual development is critical for efficient fruiting body differentiation and formation of meiotic progeny. Developmental upregulation appears to be driven by novel promoter elements and potential Inr (initiator) sequences located at the sites of transcriptional imitation. These promoter elements do not require other typical upstream promoter elements such as TATA or CAAT boxes, or fungal pyrimidine tracts. Highest matA expression was detected in reproductive tissue and coincided with the stage of rapid differentiation of asci and ascospores. Failure to upregulate matA transcript resulted in aborted formation of ascospores. Therefore, increasing matA transcript abundance is required for progression through specific developmental stages. We found that matA expression is tightly regulated by both position-dependent and position-independent mechanisms. An unusual feature of the position-dependent regulation is that duplication of the 5′ genomic region flanking matA, which includes the silencer element, correlates with position-dependent suppression of matA expression during development and negatively impacts ascospore formation.
Our data demonstrate that self-compatible A. nidulans has complex regulatory mechanisms controlling functional expression of the HMG-box mating type gene. Further investigation of mating type gene regulation in A. nidulans may reveal mechanisms controlling recognition between sexually compatible cells and nuclei in the homothallic mating system. Understanding fungal mating type gene regulation may also provide novel insights into those pathways conserved throughout eukaryotic sexual reproduction as well as mechanisms that generate diversity in reproductive strategies.
Materials and Methods
Strains, growth conditions, and genetic manipulations of A. nidulans
A. nidulans strains used in this study are listed in Table 1. Supplemented minimal and complete media were prepared as previously described (Pontecorvo et al. 1953; Kafer 1977; Vallim et al. 2000). Standard genetic techniques were used as described previously (Pontecorvo et al. 1953). Nucleic acid DNA and RNA isolation, Southern blot, Northern blot analysis, and standard molecular techniques were performed as previously described (Miller et al. 1985, 1987; Wu and Miller 1997). DNA-mediated transformation of A. nidulans was performed according to previous protocols (Yelton et al. 1983; Miller et al. 1985). Induction of sexual development and fruiting body analysis in A. nidulans were performed as described by Pyrzak et al. (2008) and Vallim et al. (2000).
Table 1. A. nidulans strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| FGSC A26 | biA1 | Fungal Genetics Stock Center |
| GR5 | pyrG89; wA3; pyroA4 | May, G. S. (M. D. Anderson) |
| UI427 | alcA(p):medA, biA1, pyrG89:matA:pyrG, pabaA1; wA1; argB2; matAΔ::argB, | This study |
| UI461 | yA2, pabaA1, pyroA4; argB2; matA(0)::AfargB; nkuAΔku70::bar | This study |
| UI462 | yA2, pyrG89, pabaA1; nkuAΔku70::bar; argB2; matA(0)::AfargB | This study |
| UI465 | yA2, pyrG89, pabaA1; argB2; matA(0)::AfargB | This study |
| UI465 3.2 | yA2, pyrG89:pWP3−898;pyrG, pabaA1; argB2; matA(0)::AfargB | This study |
| UI465 3.5 | yA2, pyrG89:pWP3−248:pyrG, pabaA1; argB2; matA(0)::AfargB | This study |
| UI465 3.6 | yA2, pyrG89:pWP3−512:pyrG, pabaA1; argB2; matA(0)::AfargB | This study |
| UI467 | yA2, pyrG89:pWP3−170:pyrG, pabaA1; argB2; matA(0)::AfargB, | This study |
| UI468 | yA2, pyrG89:pWP3∆170:pyrG, pabaA1; argB2; matA(0)::AfargB | This study |
| UI472 | yA2, pyG89:pWP3−1001:pyrG, pabaA1; argB2; matA(0)::AfargB | This study |
| FGSC A1153 | yA2, pabaA1, pyroA4; argB2; nkuAΔku70::bar | Hynes, M, Fungal Genetics Stock Center |
| FGSC A1155 | pyrG89, pyroA4; nkuAΔku70::bar | Hynes, M. Fungal Genetics Stock Center |
Light microscopy
Photomicrographs were taken with a Nikon Cool Pix 5400 camera and Zeiss SV8 stereomicroscope. Fruiting bodies were cleaned, suspended in sterile water on glass slides, and crushed under a coverslip. Fruiting body tissue was examined by differential interference contrast (DIC) optics using a Zeiss Axioplan microscope. Photographs were taken with either a Nikon Cool Pix 5400 or a Photometrics CoolSnap ES camera with Metavue software (Universal Imaging).
Comparative RT–qPCR transcript analysis
The relative RT–qPCR method was used to assess the developmental expression of matA transcript. Total RNA was extracted from undifferentiated hyphae and reproductive tissue at different time points. Undifferentiated hyphae tissue was obtained from a submerged culture of A. nidulans grown for 16 hr at 37°. Reproductive tissues were grown on a solid medium as confluent cultures and collected 2, 4, and 6 days postinduction of sexual development. Total RNA was extracted as previously described (Vallim et al. 2000) and treated with DNase-I to remove traces of genomic DNA contamination. To quantify expression, mRNA was reverse transcribed with oligo dT primers using the SuperScript First-Strand Synthesis system for RT–PCR (Invitrogen). Transcript levels were quantitated using the threshold cycle (∆∆CT) method and SYBR green sequence detection was performed using a StepOne real-time PCR system (Applied Biosystems). Relative quantitation, by the (∆∆CT) method, is expressed as a difference in target gene expression with respect to an internal control (actin) in different samples. The actin primers used do not span introns. Nonreverse transcribed controls for every RNA preparation were included to confirm that contaminating genomic DNA did not bias cDNA quantification. Mean CT values and standard deviations were used in ∆∆CT calculations. Primers used in RT–qPCR are listed in Table 2. Each cDNA sample was assayed in triplicate and RNAs were obtained from three separate biological samples. We performed control experiments comparing argB, benA, and actA transcripts using RNAs extending over the developmental time course. These have all been previously used as internal controls for Northern blot analysis in A. nidulans. Our results found that argB and actA appeared to be most constant during development. Actin has been consistently used as internal control in other organisms for RT–qPCR experiments including the filamentous fungi A. fumigatus and Magnaporthe oryzae. General methods have been described previously (Pyrzak et al. 2008).
Table 2. Primers used in this study.
| Primer | Sequence |
|---|---|
| An/AfactinF | GTCAAGATCATTGCTCCTCCTAGA |
| An/AfactinR | TACTCCTGCTTGGAGATCCACAT |
| AnMatAF11 | TGGGAGTGTATCAGCTTCATG |
| AnmatAR11 | TGCCGTATGCTACCTGAG |
| AnmatAF33 | CCGCACGCATCACCGAGCTCC |
| AnmatAR29 | GGTGTGCGCAGAACACGCAGA |
| AnmatAF31 | CATCAGCGAGCGTCCATGTCGGAGGCTTTA |
| AnmatAF32∆matA | CATTGACGACCTAGTTTGGTG |
| AnmatAR35AfargBext | GTATAATTGGGCTGTTGGCGGTGCTACGTATCTAGGACGAACGAGTCAAGT |
| AnmatAF36AfargBext | ACTAAGAGCTCGTTAAGTTGTCCCCTTCCAGGGACGACTGTATGCACTCTC |
| AnmatAF40 | AGTCTTGAGCCATGAGTTGGTAGC |
| AnmatAR5 | CATCATTCGTAGACCCCAGATGACGCAAAG |
| AnmatAR6 | TATCCACGACATTTGTTGGCAAAAGGACTT |
| GeneRacer 5′ primer | CGACTGGAGCACGAGGACACTGA |
| GeneRacer 5′ nested primer | GGACACTGACATGGACTGAAGGAGTA |
| AfargBF1 | TACGTAGCACCGCCAACAGCC |
| AfargBR1 | TGGAAGGGGACAACTTAACGA |
| AnmatAF38 | CACAGTCTTCTGAGACACCATCTC |
| AnmatAR38 | GCTTATTGATTCCGAAAGTGGAAG |
| pWp3matAF1 | CTGTATCGATTGCTATGGAAATCACCAACAC |
| pWp3matAR1 | CAGCCATTTTGGCACTTC |
| AnmatAF30 | GCACCTCGTCTCACTCCAGATTACT |
| AnmatAR27 | CAGAAGGCTTCCGAGGAGTGTAC |
| AnmatSE2F | GTATCCGTCGAGCCTACTCCT |
Mapping of matA mRNA cap sites
RNA start sites were mapped using the Invitrogen Gene Racer kit, L150202. The capped transcripts were identified according to the manufacturer’s instructions. Nested pcr fragments were produced by first using the Gene Racer 5′ primer and AnmatAR6 and then reamplifying the product with the Gene Racer 5′ nested primer and AnmatAR5. The pcr products were separated on a gel and isolated bands were excised. Gel fragments were purified and cloned into a Zero Blunt topo vector. The resulting construct was cloned into TOP10 chemically competent cells and plated on L-ampicillin plates. DNA from individual colonies was sequenced using an Applied Biosystems 3730 sequencer.
Gene deletion and complementation analysis
Standard gene deletion and complementation analysis were used to elucidate the molecular function and developmental regulation of the matA gene. An A. nidulans matA(0) deletion strain was created by deletion/replacement of the matA entire transcriptional unit (from +2 to +1851 nt) at the resident matA locus on chromosome (chr) III. A fusion PCR approach was used to create a deletion construct according to the protocol developed by Szewczyk et al. (2006). The A. fumigatus argB selectable marker (AfargB) gene was amplified from genomic DNA of A. fumigatus strain (Af293.139) with primers AfargBF1 and AfargBR1 (Table 2). The upstream 5′ and downstream 3′ genomic fragments flanking the matA transcriptional unit were amplified using primers that introduce 30-nt overlapping extensions with the selectable marker, AfargB. The 5′ flank was amplified with primers AnmatAF11 and AnmatAR35AfargBext (Table 2). The 3′ flank was amplified using primers AnmatAF36AfargBext and AnmatAR11 (Table 2). The PCR fragments were subsequently fused into a final matA deletion fragment using AnmatAF11 and AnmatAR11 primers. The final fusion product was directly transformed into Fungal Genetics Stock Center (FGSC) A1153 (nkuAku70Δ) A. nidulans recipient strain and resulted in high frequency of homologous gene targeting. Transformants were recovered on selective media and the complete deletion of the matA transcription unit was confirmed by Southern blot analysis and RT–PCR in >20 individual isolates (data not shown). Plasmid pWP3 carries the coding region of matA flanked by 1-kb upstream and 1.8-kb downstream genomic sequences plus pyrG as a prototrophy marker. pWP3 was constructed by cloning the AnmatA genomic region (primers AnMatAF11 and AnmatAR11) into the ppyrG plasmid (Pyrzak et al. 2008). Deletions of the 5′-upstream regulatory region were accomplished using primers indicated in Table 2 with a site-directed mutagenesis kit (Invitrogen) and pWP3 plasmid as a substrate. The 5′ deletions matA(−898), matA(−512), matA(−248), and matA(−170) used primer AnMatAR11 plus matASE2F, matAF40, matAF31, or AnmatAF32∆matA, respectively (Table 2). The internal deletion (−170) used primers AnmatAF38 and AnmatAR38. Deletion constructs carrying deletions of the upstream genomic region pWP3 or internal deletions of pWP3 (∆170 bp) were used to complement the UI465 matA(0) strain.
5′-fluoroorotic acid selection
A. nidulans strains with a matA transgene integrated via 3′ single integration at the matA(0) locus were used to select rare mitotic recombination events that resulted in excision of the matA(0) allele as well as the pyrG gene and restoration of the wild-type (wt) matA gene structure. Selection of pyrimidine auxotrophic excisants was accomplished by a modification of the 5′-fluoroorotic acid (5′-FOA) resistance scheme described by Boeke et al. (1984). A total of 1 × 106 conidia per plate were spread onto 5% agar plates containing appropriately supplemented minimal media and 5′-fluoroorotic acid (0.1 mg/ml). Plates were incubated at 37° for 2 days. Usually 30–50 viable colonies were recovered and analyzed for correct excision events by Southern blot analysis.
Analysis of the genomic region of the matA locus
The matA locus (locus ID AN4734.3) encoding the HMG box domain mating type protein and flanking neighbor genes sygI (AN 4733.3) and apc5 (AN 4735.3) in A. nidulans are available at the Broad Institute database: (http://www.broadinstitute.org/annotation/genome/aspergillus_terreus/MultiHome.html). Putative start and end of the matA transcription unit were mapped on the basis of DNA sequencing of cDNAs from a sexual cycle library.
Results
Molecular organization of the matA locus and structure of the transcript
The genetic organization of mat genes in homothallic fungi is unique and different between individual species (Turgeon 1998). The A. nidulans matA locus (AN4734.3) is located on chromosome III within a domain that includes three closely linked transcriptional units, (sygI, matA, and apc5) (Figure 1). sygI (suppressor of yeast gpa1) encodes a homolog of a budding yeast signal transduction modulator involved in mating. apc5 (anaphase promoting complex) encodes a protein involved in anaphase initiation during mitosis and meiosis.
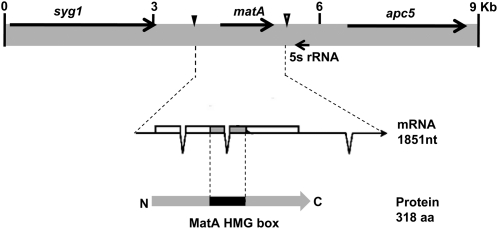
Figure 1.
Genetic context of the matA gene on chromosome III. Black arrows indicate arrangement and orientation of genes (coding regions) flanking the matA locus. The matA transcript and protein structure are shown. Solid triangle indicates the reference transcriptional start site 1 (TSS1), the start of the transcriptional unit. Open triangle indicates the end of the matA transcript.
matA encodes a single transcript (1851 nt) with a short 5′-UTR (134 nt) coding region (958 nt) and a long 3′-UTR (759 nt). The transcript has three introns, two within the coding region and one in the 3′-UTR. Potential transcription start sites and end of the matA transcript have been identified by sequencing full-length cDNA clones. The 5′ end of the longest cDNA was used as the +1 reference for this study and is referred to as transcriptional start site 1 (TSS1) (Figure 1).
Functional complementation of the matA(0) deletion depends upon the chromosomal position of the matA transgene
We created the matA(0) strain UI465, which carries a complete deletion of the matA transcriptional unit. The entire transcription unit, including start of the transcription +1, coding region, and end of the 3′-UTR, was replaced by the A. fumigatus prototrophic marker argB (Figure 2, A and B). Deletion of matA abolishes sexual development without affecting vegetative growth or conidiation (Figure 3, A and B). UI465 is completely sterile and does not differentiate fertile fruiting bodies. Tiny, barren cleistothecia are observed at rare frequency. An additional phenotype of the matA(0) deletion is a massive proliferation of Hülle cells (Figure 3B). In a wild-type A. nidulans strain, Hülle cells appear at the onset of sexual development and form ornamented clusters of tissue surrounding the growing, young fruiting body. Hülle cells become less prevalent when mature fruiting bodies are present. Excessive proliferation of Hülle cells in the UI465 strain might be indicative of unsuccessful fertilization events that prevent further differentiation of fruiting bodies (Figure 3B). Our matA(0) phenotype is similar to, but more severe than the matA deletion previously described by Paoletti et al. (2007).
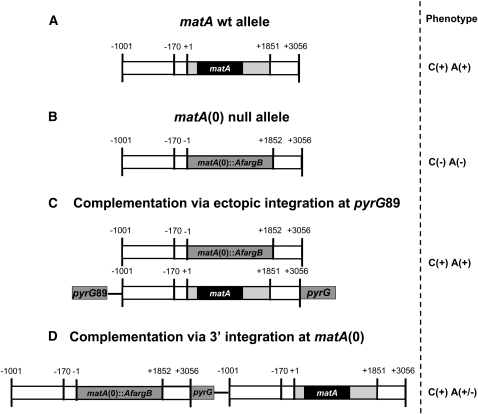
Figure 2.
Schematic representation of the deletion and complementation of matA. (A) Wild type matA allele; solid bar represents coding region (ORF), lightly shaded bars represent matA 5′- and 3′-UTRs, and open bars represent upstream 5′ and downstream 3′ genomic sequences flanking the transcriptional unit. Horizontal solid line represents vector sequence of pWP3. Physical distance is marked (−1001 to +3056 bp). (B) Null matA(0) allele; the matA transcriptional unit was deleted and replaced with the Aspergillus fumigatus argB (AfargB) marker. (C) Complementation via ectopic integration; matA complementing transgene was integrated ectopically at the pyrG89 locus on chromosome I. (D) Complementation via 3′ integration at the matA(0) locus; matA transgene carried on pWP3 was integrated at matA(0) locus via homology between 3′ flanking sequences. The corresponding phenotype is shown: C, cleistothecia; A, ascospores; (+), presence; (−), absence.
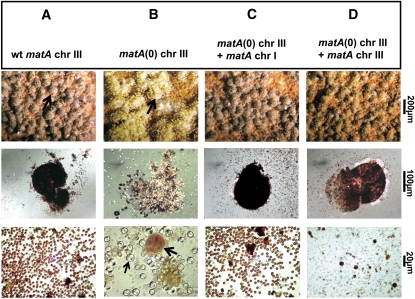
Figure 3.
Sexual development in wild type (wt), matA(0), and complemented A. nidulans strains. Representative strains (A–D) were induced to undergo sexual development as described in Materials and Methods. Abundance of cleistothecia and internal reproductive tissue were examined under specific levels of magnification as indicated. (A, C, and D, top row) Morphology and abundance of mature cleistothecia on rich medium agar plates in wild-type and complemented strains (arrow in A indicates a single cleistothecium). (B, top row) Cluster of hülle cells in the matA(0) strain. (A, C, and D, middle row) Individual mature cleistothecia in wild-type and complemented strains. (B, middle row) Mass of hülle cells in the matA(0) strain. (A, C, and D, bottom row) Efficiency of ascospore production in wild-type and complemented strains, respectively. (B, bottom row) Sexual development in the matA(0) strain is aborted at the stage of unfertilized protocleistothecia, shown as brown spherical structures (thick black arrow) with individual hülle cells (thin black arrow).
The UI465 matA(0) strain, a pyrG89 auxotroph, was complemented using pWP3, which has the prototrophic marker AnpyrG and a wild-type copy of the matA transcription unit flanked by 1 kb of 5′ and 1.2 kb of 3′ genomic regions. Homologous recombination of the plasmid at matA(0) on chr III resulted in integration of the matA transgene at the resident locus, whereas homologous recombination at the pyrG locus on chr I resulted in the ectopic integration of the matA transgene (Figure 2, C and D). The ectopic matA transgene fully complemented the matA(0) allele on chromosome III (Figures 2C and 3C). The complemented matA(0) strain had a fertile wild-type phenotype, with normal fruiting body abundance, size, and ascospore production. Surprisingly, integration of a single copy of the matA transgene at the resident matA(0) locus only partially restored wild-type phenotype (Figures 2D and 3D). The partially complemented strain differentiated abundant cleistothecia; however, asci and ascospore formation was dramatically reduced by 80% compared to wild-type levels. The complementation phenotypes of the matA(0) strain are consistent with the complementation results for a partial deletion matA∆ strain (K. Y. Miller, W. Czaja, and B. L. Miller, unpublished data). These data suggest that the molecular function of a matA transgene depends on its genetic position and implies complex regulation of the resident matA locus.
Transcriptional expression of the matA transgene at the resident matA(0) locus is suppressed at late stages of sexual development
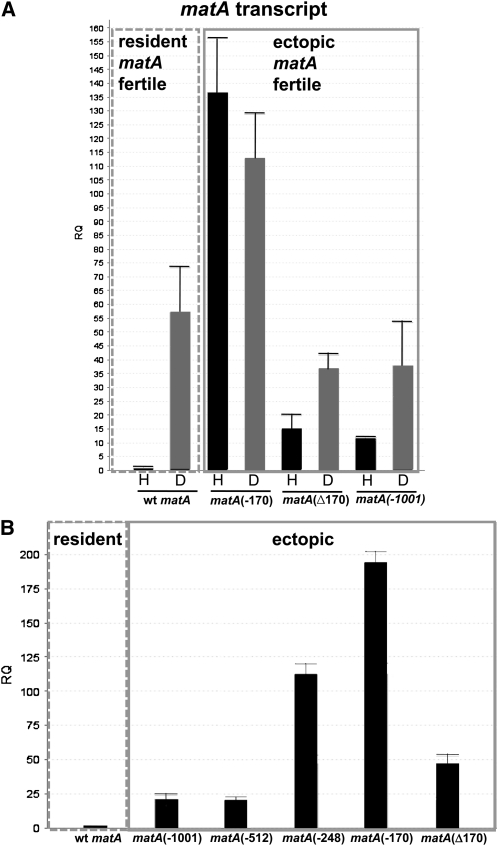
Mating type transcripts are detected on Northern blots at very low abundance. Therefore, we utilized a quantitative RT–qPCR approach to analyze expression of the matA transgene during sexual development. matA expression was analyzed in the growing mycelia (undifferentiated hyphae, 0 days postinduction, dpi) and in the reproductive tissue at 2, 4, and 6 dpi of sexual development. These developmental time points correlate, respectively, with the progressive differentiation of the fruiting body from protocleistothecia with Hülle cells, young pink-walled cleistothecia with ascogenous hyphae and mature dark purple-pigmented cleistothecia with asci and ascospores. We first compared matA expression in undifferentiated hyphae (0 days) and in reproductive tissue (6 days postinduction) in a wild-type strain with a functional matA at the resident locus and the complemented matA(0) strain carrying a functional matA transgene integrated ectopically (Figure 4A). Very low matA transcript abundance transcript was detected in undifferentiated wild-type hyphae, whereas there was a dramatic 62-fold upregulation in reproductive tissue. By contrast, matA expression in the ectopically complemented strain was derepressed 10-fold in undifferentiated hyphae and downregulated by 30–40% in the reproductive tissue relative to wild type (Figure 4A). The difference in expression pattern of the ectopic matA transgene suggests that cis-regulatory elements and trans-acting factors modulate matA expression in both a position-dependent and position-independent manner. However, these differences in developmental matA expression apparently do not affect biological functions of the MatA protein. Elevated expression in hyphae did not promote precocious sexual reproduction in the ectopically complemented strain and reduced levels in developmental tissue were sufficient to drive differentiation of wild-type cleistothecia with mature ascospores (Figure 3C).
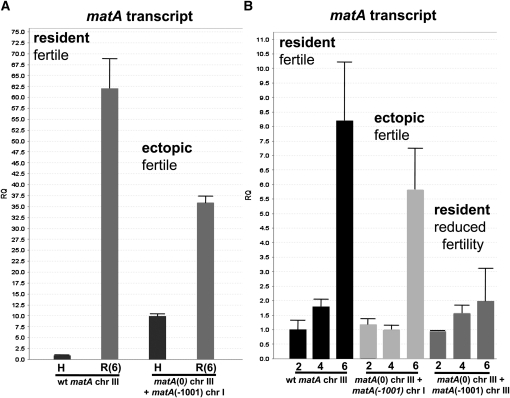
Figure 4.
Comparative transcriptional expression of matA at resident and ectopic loci. Phenotype associated with a specific abundance of matA transcript in the reproductive tissue is indicated. (A) Relative quantitation (RQ) of matA expression from the endogenous matA locus on chromosome (chr) III and from the ectopically integrated matA transgene on chr I. The transgene carries 1000 bp of 5′ flanking upstream sequences. H, undifferentiated hyphae; (R6) reproductive tissue, 6 days postinduction. (B) Relative quantitation of matA expression over a developmental time course at 2, 4, and 6 days postinduction of sexual development. Expression profiles of the endogenous matA, ectopically integrated matA transgene and the matA transgene integrated at resident or ectopic loci are shown.
matA expression was further analyzed in reproductive tissue from different stages of fruiting body development: 2, 4, and 6 dpi of sexual cycle (Figure 4B). In the wild-type strain the highest expression of transcript is detected at 6 days postinduction, which correlates specifically with active meiotic and mitotic divisions and differentiation of mature asci with ascospores. Further progression of sexual development leads to a fully mature cleistothecium and a gradual decrease of matA transcript level (confirmed by Northern blot and RT–qPCR, data not shown).
The ectopically integrated matA transgene had a developmental expression profile (2, 4, and 6 dpi) similar to wild type, with a characteristically significant upregulation of the matA transcript at 6 dpi. Surprisingly, expression of the matA transgene integrated at the resident matA(0) was significantly suppressed (∼25% of wild-type expression) at 6 dpi (Figure 4B). These low levels of matA transcript during the critical later stages of cleistothecium development are apparently insufficient and contribute to failure in ascus and ascospore development.
5′ flanking DNA sequences are dispensable for matA molecular function during sexual reproduction
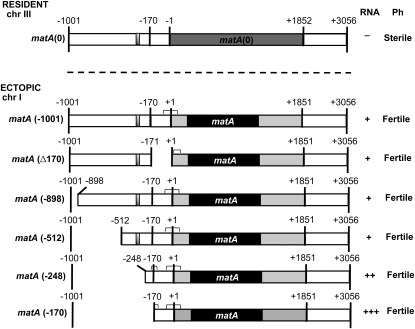
We performed deletion analyses of the 5′ genomic region upstream of TSS1 to identify a putative promoter region and regulatory elements driving functional matA expression. The distal 830 bp of the 5′ flank (from −171 to −1001 bp) upstream of matA was deleted from the pWP3 vector using site-directed mutagenesis. The resulting pWP3−170 deletion construct was transformed into the UI465 matA(0) recipient strain. Transformants carrying single homologous ectopic integrations of pWP3−170 at pyrG89 were analyzed for complementation of the matA(0) phenotype. In all tested transformants, ectopically integrated pWP3−170 fully complemented resident matA(0) deletion and restored wild-type sexual development (Figure 5). Thus, sequence from −1000 to −171 is dispensable for expression levels required for proper matA function during fruiting body development and ascospore differentiation.
Figure 5.
Deletion analysis of the 5′ flanking region of matA. The solid bar indicates the matA coding region, lightly shaded bars represent 5′- and 3′-UTRs. The silencer element is indicated as a small shaded box. Chromosomal positions in the genome are indicated (chr III and chr I). Positions of 5′ deletion end points and internal deletions of upstream flanking sequences are indicated by − and Δ, respectively. Transgene deletions were integrated ectopically at the pyrG locus on chr I. Complementation was observed for all transgene constructs. Transcript presence (+) or absence (−) is indicated. Phenotypes associated with each deletion are indicated. Horizontal brackets indicate positions of mRNA cap sites determined for hyphal and developmental RNAs using RACE (refer to Figure 7).
We analyzed the proximal portion of the 5′ flank (−1 to −170 bp) upstream of TSS1 to determine whether these sequences function as a promoter region. A pWP3∆170 deletion construct having this internal 170-bp deletion (see Materials and Methods) was transformed into the UI465 matA(0) recipient strain. A single copy of the pWP3∆170 construct integrated ectopically at pyrG89 fully restored the wild-type sexual phenotype (Figure 5). This observation indicates that the 5′ flanking 170 bp of genomic sequences are not essential to drive functional matA transgene expression. Further, no sequence duplications are detected in the −1 to −170 and −171 to −1001 regions that might suggest the possibility of redundant promoters. Taken together, these data suggest that typical promoter elements do not lie within the upstream region from −1 to −1001 bp relative to matA TSS1. We therefore mapped matA mRNA cap sites in both hyphal and developmental tissues from wild-type and deletion strains to identify potential alternate or novel promoter elements that may lie at, or downstream of TSS1 and are required to drive matA transcription (see below).
Deletion of the 830 bp of 5′ regulatory flank dramatically alters developmentally regulated matA expression without affecting phenotype
To determine whether 5′ flanking sequences had a role in developmentally regulated matA expression, complemented strains with an ectopically integrated matA transgene carrying the −170-bp or internal ∆170-bp deletion were analyzed. matA transcript abundance in the wild-type strain was upregulated ∼55-fold upon transition from undifferentiated hyphae to reproductive tissue (Figure 6A). In the UI465 matA(0) strain there was no detectable level of matA transcript (data not shown). Interestingly, the hyphal level of matA transcript was dramatically derepressed 135-fold in the complemented strain UI467 (−170 bp) compared to wild-type hyphal expression. Expression in reproductive tissue was also derepressed, being 2.5-fold higher relative to wild type and ∼3-fold higher relative to the ectopic control strain (Figure 6A). Therefore, the 830-bp upstream region (from −170 to −1001 bp) has a critical regulatory element/s involved in the developmental repression of matA gene. This dramatic increase of matA transcript in the hyphae and reproductive tissue did not alter sexual development. However, we cannot rule out the possibility that derepressed matA expression results in increased fertilization efficiency or precocious fertilization as these events are not readily detectable in homothallic A. nidulans. By contrast, strain UI468 carrying the 170-bp internal deletion of the proximal portion of 5′ flanking sequences, maintained an expression profile consistent with the ectopic matA transgene having the complete 1 kb of 5′ flanking DNA (Figure 6A). Therefore, the 170 bp proximal to TSS1 does not appear to be necessary for developmentally regulated matA transcription.
Figure 6.
Comparative expression of matA transgene deletions. (A) Developmental expression of matA in transgene strains lacking either 830 bp of upstream sequence or the internal 170 bp proximal to the start of transcription. The level of matA transcript was analyzed in the undifferentiated hyphae, H, and in the reproductive tissue, D, at 4 days postinduction of sexual development. The dashed line box indicates expression of matA at the resident locus in the wild-type strain. The solid line box indicates expression of matA transgenes at the ectopic locus. (B) Expression in additional 5′ deletion strains used to determine position of upstream repressor and promoter elements. Specific 5′ deletion end points or internal deletions are indicated by − or Δ, respectively.
A putative silencer element (matA SE) and novel promoter elements regulate matA expression
Vegetative repression of matA in wild type and derepression in UI467 suggest that a silencer element/s is involved in developmental expression patterns. Analysis of the distal portion of 5′ flank revealed the presence of a short (29 bp) DNA segment localized between −931 and −960 bp that has striking structural similarity to the hSRY silencer (Su and Lau 1993). This 29-bp element and hSRY SE share 83% identity and 93% similarity (data not shown). However, a deletion series between −1001 and −512 indicated that deletion of this sequence does not affect sexual development or ectopic matA expression under our experimental conditions (Figures 5 and 6B). By contrast, we did detect a functional matA SE within sequences between −512 and −170. Compared to the ectopic matA control strain (−1001 bp), matA expression in deletion strains UI465 3.5 (−248 bp) and UI467 (−170 bp) was ∼5-fold and ∼8.5-fold derepressed, respectively (Figure 6B). UI468 (Δ170) also shows a small ∼2-fold derepression in hyphal matA expression (Figure 6B). Located within these sequences are four repeats having the consensus 5′-TRAARSRAARAAYYGRR-3′, which may serve as silencing elements. Two repeats are found between −248 and −512, one between −1 and −109, and a truncated repeat between −170 and −248.
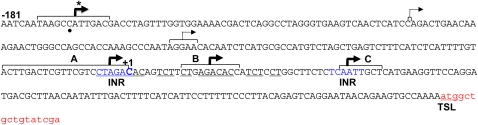
We scanned the 200 bp upstream of TSS1 for canonical TATA box promoter motifs. No A/T rich or other consensus sequences resembling the TATA box were found. Pyrimidine tracts that function in some fungal promoters are also not found upstream of matA TSS1. There are two potential upstream CAAT elements. However, our functional analysis of the proximal 5′ flank (−1 to −170 bp) confirmed that this region does not contain essential promoter elements and was dispensable for developmental regulation of matA. It is possible that transcription is initiated from an initiator element (Inr) located at or near the transcriptional start site. An Inr is functionally similar to a TATA box but can act independently (Smale and Kadonaga 2003). Two putative Inr elements can be identified on the basis of DNA consensus sequences and similarity to the Inr in Drosophila (TCA(+1)G/TTY) and mammals (PyCA(+1)NTYY) (Smale and Kadonaga 2003) (Figure 7). One is located at TSS1 (+1) and the other at +36. matA mRNA cap sites were mapped in hyphal and developmental RNAs to determine functionality of these elements. Cap sites for wild-type and ectopically integrated constructs were also compared to determine whether any deletion resulted in aberrant transcriptional initiation. We found that transcription is initiated approximately equally from three zones in both hyphal and reproductive tissue (Figure 7). Zone A corresponds to the putative Inr at TSS1 while zone C corresponds to the downstream Inr at +36 relative to TSS1. However, a third distinct cluster of transcriptional start sites was centered at +18 relative to TSS1. Sequence at this position does not resemble an Inr or any other recognized promoter element. Closer inspection of sequences around the three clusters of mRNA cap sites revealed that zone A is probably not an Inr. Rather zones A and B share very similar sequences, with most cap sites centered within two overlapping repeats having the consensus 5′ YKAGACACMRTCTYCT 3′. This sequence represents a novel promoter element not previously identified. matA mRNA cap sites for hyphal and developmental RNAs from wild-type, ectopic control, and deletion strains all initiated from these three zones except for UI468 (Δ170), which lacked zone A. Transcription in this strain was initiated only from zones B and C. Transcription in developmental tissue from overexpression strains UI465 3.5 (−248) and UI467 (−170) was initiated from these same three zones, similar to wild type. However, we observed an additional, novel cluster of cap sites in hyphal tissues from these two strains. This cluster was centered at −169 relative to TSS1 (Figure 7). Surprisingly, this indicates that transcription in hyphae of UI467 was initiated at the very end of the deletion and without any matA upstream sequences. There is no sequence resembling an Inr or any other known promoter element located near −170, indicating that this may represent another novel promoter element that recruits the transcriptional apparatus to this site, or that an unknown downstream promoter element directs transcriptional initiation upstream near −170. Regardless, the matA SE identified above normally suppresses expression from this cryptic promoter element in hyphae.
Figure 7.
Identification of both Inr and novel promoter elements that regulate matA transcription. Mapping of mRNA cap sites identified three zones of transcriptional initiation in hyphae and developmental tissue from wild-type and deletion strains (A–C). Each zone is indicated by brackets. An asterisk marks a fourth zone near position −170 observed in hyphal tissue from derepressed strains matA (−248) and matA (−170). The position of the majority of cap sites within a zone is indicated by a thick arrow. Thin arrows mark two minor sites of mRNA start sites. The reference +1 represents the 5′ end of the largest cDNA previously identified from a sexual development library. Potential Inrs are indicated in blue; novel promoter elements are underlined. The translational start site (TSL) and beginning of the ORF are in red text. A black dot below the text marks the −170 position for reference.
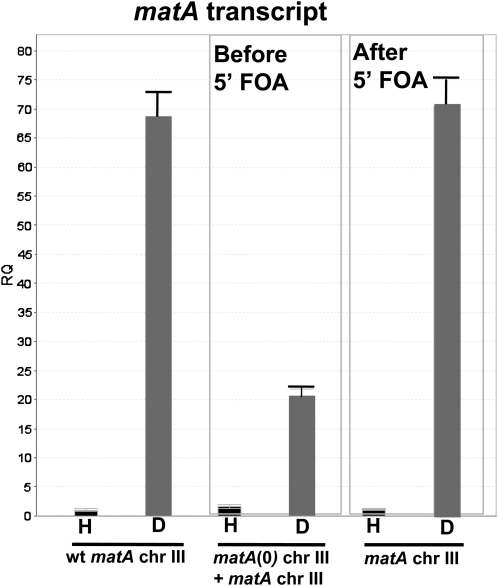
Removal of the matA(0) allele and duplicated flanking sequences from the resident matA locus restores wild-type expression of the matA transgene and wild-type phenotype of the complemented strain
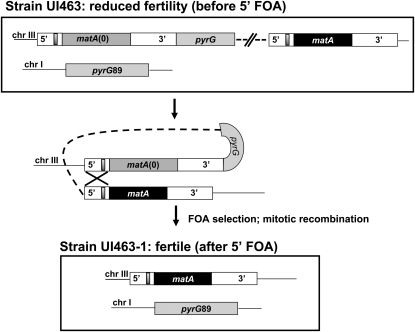
Integration of a functional matA transgene at the matA(0) locus on chromosome III resulted in greatly reduced developmental transcript abundance and reduced fertility. Integration of the pWP3 plasmid with the functional matA transgene at the matA(0) locus duplicates matA 5′ and 3′ flanking sequences, which could potentially alter the distance and/or copy number of important regulatory elements that control functional matA expression. To further understand the basis of altered expression of the matA transgene at matA(0), we deleted the entire matA(0) allele along with its flanking 1-kb upstream and 1.2-kb downstream genomic regions. This leaves a single copy of the intact matA gene at the resident locus on chromosome III. This was accomplished using 5′-FOA selection to isolate mitotic recombinants that had looped out the matA(0) copy including the pyrG gene plus plasmid sequences of pWP3 (Figure 8). All recombinant strains recovered after 5′-FOA treatment had restored wild-type phenotype with abundant cleistothecia and ascospore formation. We confirmed that functional matA expression had also been restored in these strains by analyzing expression in undifferentiated hyphae (0 days) and 6-dpi reproductive tissue in three strains: wild type (matA resident copy), UI463 [matA(0) + matA transgene at resident locus], and UI463-1 (reconstructed matA resident copy). matA expression was restored to wild-type levels upon deletion of the matA(0) allele and its associated flanking 5′ and 3′ sequences from the resident matA locus (Figure 9). Therefore, we excluded the possibility that undetected secondary mutations alter sexual development. Instead, our data suggest that the physical presence of the duplicated flanking DNA sequences at the resident mat locus interferes with the functional expression of the complementing matA gene.
Figure 8.
Schematic depiction of matA flanking sequence duplication and restoration of wild-type matA gene structure at the endogenous locus. Genetic organization of the matA locus after 3′ homologous integration of pWP3 carrying the matA transgene (solid box). 5′ and 3′ flanking regions are shown with the silencer element (shaded box). The DNA fragment containing pyrG and the matA transgene allows alignment of duplicated 5′ flanking regions and rearrangement during mitotic recombination. The matA(0) allele and pWP3 carrying pyrG are excised and wild-type matA allele is restored at chromosome III. Recombinant events containing wild-type matA and the pyrG89 mutation (chr I) were selected after plating on media supplemented with 5′-FOA.
Figure 9.
Developmental expression of matA at the resident locus. Expression of matA was analyzed in undifferentiated hyphae, H, and in reproductive tissue 6 days postinduction of sexual development, D. Expression of the matA transgene at the matA(0) locus is suppressed in the reproductive tissue (before 5′-FOA). Wild-type expression of matA is restored at the resident locus (after 5′-FOA) upon removal of matA(0) and plasmid sequences and the recovery of wild-type genomic structure.
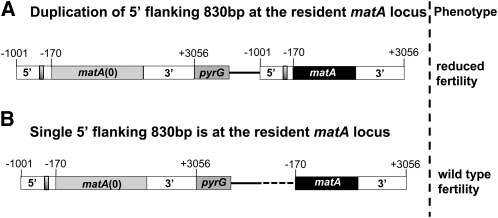
Duplication of 5′ flanking sequences containing the matA SE interferes with the expression of the matA transgene at the resident mat locus
Partial functionality of the matA transgene at the resident matA(0) locus could be the result of duplication of the upstream and/or downstream noncoding regions that flank both matA transgene and matA(0) allele. To test this possibility, we analyzed a strain with a single integrated copy of a matA transgene lacking the distal −830 bp at the resident matA(0). This 5′ truncated transgene fully complemented the deletion and restored full development of wild-type cleistothecia with mature ascospores. Therefore, suppression of the matA transgene at the matA(0) that we previously observed is correlated with duplication of the 830 bp containing the putative matA SE silencer element/s described above. We also analyzed a single integration of the pWP3∆170 construct at the resident matA(0) locus. This integrated transgene retains the matA SE and resulted in reduced fertility and only partial complementation of the matA(0) phenotype. Taken together, these results confirm that duplication of the distal 830 bp of the 5′ flanking region containing the matA SE interferes with the expression and function of matA at the resident locus (Figure 10, A and B).
Figure 10.
Duplication of 830 bp of matA 5′ flanking sequences at the resident matA locus interferes with the expression and molecular function of the matA transgene. Structure of the complemented endogenous matA(0) locus and associated phenotype are shown. Bars represent matA with flanking regions and pyrG/pyrG89 gene. Solid line represents vector sequence of pWP3; dashed line represents deleted 5′ flanking sequences from −1001 to −171. Shaded box represents silencer element. (A) Integration of the pWP3 plasmid via 3′ homology introduces a duplication of 830 bp of 5′ flanking sequences at the resident matA locus and interferes with fertility. (B) Integration of the pWP3 (∆830 bp), via 3′ homology at the resident matA locus results in wild-type fertility.
Discussion
The genetic organization of mating type genes at mat loci determines sexual compatibility and a self-fertile (homothallic) and/or cross-fertile (heterothallic) mating strategy in filamentous fungi. Heterothallic fungi have an idiomorphic organization of mat genes that governs recognition between mating partners and promotes obligate cross-fertility. Conversely, homothallic species usually have both HMG box and α box mat genes present in a haploid genome. Mechanisms of sexual identity and self-/nonself-recognition during mating are not understood in these species (Yun et al. 2000; Rydholm et al. 2007; Debuchy et al. 2010). However, the existence of self-/nonself-recognition mechanisms in homothallic species has been well documented in A. nidulans and has been termed relative heterothallism (Pontecorvo et al. 1953; Hoffmann et al. 2001). Several lines of study suggest that sexual recognition in a homothallic system might be achieved by complex developmental regulation and differential expression of mating type genes. We have analyzed the developmental regulation and molecular function of A. nidulans matA to elucidate the molecular basis of sexual identity in a homothallic mating system.
Position-dependent and independent mechanisms control matA expression and function
A strain with matA deleted from chromosome III, matA(0), was unable to initiate a sexual cycle resulting in the absence of fruiting bodies. This self-sterile phenotype is consistent with previous studies in A. nidulans (Paoletti et al. 2007) and deletion of the mat-HMG gene in other homothallic fungi such as Gymnoderma zea and Sordaria macrospora (Lee et al. 2003; Desjardins et al. 2004; Poggeler et al. 2006).
Complementation studies revealed that matA expression is dependent upon its chromosomal position and suggest that the resident mat locus has complex regulatory mechanisms. The matA(0) deletion is fully complemented by a matA transgene ectopically integrated on chromosome I. Surprisingly, the same matA transgene integrated homologously at the resident matA(0) locus resulted in only a partial complementation phenotype with a dramatic 80% decrease in ascospore numbers relative to wild type.
Transcription of the resident matA gene in the wild-type strain is tightly regulated developmentally, being highly suppressed in undifferentiated hyphae and progressively upregulated in reproductive tissue. This observation indicates that MatA protein is essential throughout sexual development, but is dispensable for vegetative growth and conidiation. This is consistent with the matA(0) phenotype. Developmental expression of A. nidulans matA reported here confirms earlier observations (Paoletti et al. 2007; Pyrzak et al. 2008). Similarly, tight genetic regulation of mating type genes has been proposed for heterothallic fungi (Leubner-Metzger et al. 1997; Coppin and Debuchy 2000).
By contrast, developmental expression of the ectopically integrated matA transgene is less tightly regulated, being derepressed in undifferentiated hyphae and less upregulated in reproductive tissue. Therefore, we propose that regulated matA expression includes regional, position-dependent cis elements, that may influence chromatin organization and that lie outside the matA locus.
Interestingly, the altered expression profile of the ectopic matA transgene, with elevated vegetative mRNA abundance, did not have an observable effect on the sexual development program. However, early fertilization events in A. nidulans are difficult to observe and we cannot rule out an effect upon precocious development or altered mating efficiency. Transcript abundance in the reproductive tissue, though less than at the resident locus, was sufficient to drive sexual development. Apparently, this level of expression meets the critical threshold level that we have previously shown to be required to complete a fertile sexual cycle (Pyrzak et al. 2008).
Remarkably, expression of the same matA transgene integrated at the resident matA(0) locus was significantly reduced during later stages of sexual development. The reduced level of matA transcript was strongly correlated with a dramatic reduction in ascospore formation, but without an impact upon fruiting body differentiation. To our knowledge this is the first example, where the presence of a matA(0) deletion at the resident mat locus “interferes” with the functional expression of an adjacent complementing transgene. This observation suggests mat locus-specific, position-dependent requirements regarding distance and/or dosage of regulatory elements involved in developmental matA expression. It is possible that interference is due to the presence of duplicated copies of the matA SE silencing element when the intact matA transgene is integrated at the resident locus, and the ability of trans factors at this silencer to act at distances of at least 9–10 kb. This is supported by two observations. 5′-FOA eviction and deletion of duplicated sequences having matA SE restored wild-type gene structure and wild-type ascospore numbers. Integration of a matA transgene lacking matA SE at the matA(0) locus also fully complemented the deletion and restored a wild-type phenotype.
Upstream silencing elements and novel promoter elements regulate position-independent matA expression
Neither the proximal region (−1 to −170 bp) nor distal region (−171 to −1001 bp) were required for full complementation by the matA transgene. Sequence analysis of this region did not reveal the presence of potentially redundant promoter structure or typical TATA-like promoter elements. Two CAAT-like elements located within the proximal 170 bp can be deleted without effect. Taken together, these results suggest that matA transcription does not require upstream promoter elements for proper initiation, but is initiated directly from three closely spaced promoter elements located at the major starts of transcription. The most downstream element resembles an Inr, on the basis of core sequence conservation and homology to promoter elements described in Drosophila and mammals. More importantly, the two upstream elements represent novel promoter elements not previously described. These elements may represent specialized promoter sequences that presumably would be bound by TFIID-like complexes containing either a sexual cycle-specific TATA-binding protein-associated factor (TAF) or a sexual cycle-specific TATA-binding protein-related factor (TRF). This would be analogous to cell- and tissue-specific gene expression described in animals (Levine and Tjian 2003). Our results provide a molecular basis for the observation of Wirsel et al. (1998) that truncation of the 5′ noncoding region of Cochliobolus heterostrophus MAT1-2-1 HMG, including the major transcriptional start site, resulted in normal formation of pseudothecia and ascospores. Interestingly, studies of promoter activity for the porcine sex-determining gene, SRY(HMG-box), revealed that sequences located downstream of the transcriptional start site were important for the promoter function. (Pilon et al. 2003). Therefore, it is intriguing that the structure and regulation of the matA gene may be a conserved feature of sex-determining genes.
Although 5′ flanking sequences are not required for matA transcriptional initiation, we observed that deletion of sequences between −512 and −170 resulted in dramatic overexpression of matA in the hyphae and reproductive tissue compared to wild type. Several repeated elements are located in this region that may function as a silencer of matA expression. The matA SE element together with associated trans factor(s) represent a new master regulator of matA expression. A related observation has been made for the heterothallic fungus Podospora anserina. Repression of SMR2(HMG) transcription during the vegetative phase requires an upstream cis-acting element located between 1.4 and 4.7 kb upstream of the SMR2 translational start (Coppin and Debuchy 2000). Analysis of the DNA sequence in the distal region of 5′ flank (from −1001 to −811 bp) revealed a conserved element similar to the mammalian MHC class silencer and with striking structural similarity to the silencer identified upstream of hSRY (Weissman and Singer 1991a; Su and Lau 1993). In line with our studies, intriguing parallels in the negative regulation governing yeast mating type genes and genes of the mammalian major histocompatibility complex have been previously reported (Weissman and Singer 1991a,b). Structural and functional similarity between regulatory elements of fungal and human origin might reflect conservation of regulatory pathways during sexual reproduction.
Elimination of developmental suppression and overexpression of matA did not have an observable impact upon fruiting body morphogenesis or ascospore formation. This result suggests that developmental repression of the matA gene may modulate some aspects of the sexual cycle but is not essential for MatA functions that are directly required for the basic process of sexual differentiation. Coppin and Debuchy (2000) also found that overexpression of SMR2 and its derepression in vegetative hyphae, when driven by the constitutive gpd promoter, did not impact sexual reproduction in P. anserina. However, this does raise questions as to why mat-HMG transcription is so highly regulated and to what extent other posttranscriptional mechanisms might play a role in matA expression. Analyses of mating type genes in C. heterostrophus and S. macrospora indicate that mat genes are both transcriptionally and post-transcriptionally regulated (Leubner-Metzger et al. 1997; Poggeler and Kuck 2000). In general, elevated levels of matA transcript were correlated with higher abundance of cleistothecia and better overall efficiency of sexual reproduction in A. nidulans, whereas low levels during later developmental stages interfered specifically with asci and ascospore development. These data are consistent with our earlier studies (Pyrzak et al. 2008) and suggest that critical spatiotemporal thresholds of matA transcript must be met to drive meiosis and ascospore formation. Similar requirements for mat expression have been proposed for P. anserina (Debuchy 1999; Coppin and Debuchy 2000).
Concluding remarks
Our studies provide novel insights into mechanisms underlying the regulation and molecular functions of the matA gene, an essential modulator of sexual processes in homothallic A. nidulans. Collectively, our data demonstrate that intricate mechanisms govern functional expression of this HMG-box mating type gene during self-fertile reproduction. Analyses of the developmental pattern of matA expression reveals unique and distinctive features, but also shows parallels to mating systems in other fungi and, intriguingly, to certain aspects of the regulation of mammalian sex determination. Manipulation of the matA regulatory region resulted in abundant expression of matA in both vegetative hyphae and sexual tissue, which resembles the constitutive expression pattern of mating type genes in some heterothallic fungi such as N. crassa and P. anserina (Ferreira et al. 1998; Coppin and Debuchy 2000). Therefore, it is tempting to speculate that conversion between homothallic and relative heterothallic reproductive mode in A. nidulans could be accomplished by changes in the expression pattern of mat genes. Molecular dissection of the regulation of mating type genes may also provide important insights into the genetic basis and molecular mechanisms that control sex determination and sex chromosome evolution in eukaryotes. Further studies of the developmental regulation of matA and other fungal mating type genes should provide significant insights into the origin, evolution, and choice of homothallic and heterothallic reproductive lifestyles, particularly in those species capable of both self-fertility and cross-fertility.
Acknowledgments
We acknowledge contributions from Jose G. Alcocer for his technical assistance and providing experimental input supporting functionality of the silencer element. This work was supported by grant no. 0318801 from the National Science Foundation and grant no. P20 RR015587 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
Literature Cited
- Benjamin C. R., 1955. Ascocarps of Aspergillus and Penicillium, pp. 669–687 in Mycologia. Mycological Society of America, Washington, DC [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R., 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197: 345–346 [DOI] [PubMed] [Google Scholar]
- Bruggeman J., Debets A. J., Wijngaarden P. J., deVisser J. A., Hoekstra R. F., 2003. Sex slows down the accumulation of deleterious mutations in the homothallic fungus Aspergillus nidulans. Genetics 164: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champe S. P., Nagle D. L., Yager L. N., 1994. Sexual sporulation. Prog. Ind. Microbiol. 29: 429–454 [PubMed] [Google Scholar]
- Coppin E., Debuchy R., 2000. Co-expression of the mating-type genes involved in internuclear recognition is lethal in Podospora anserina. Genetics 155: 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppin E., Debuchy R., Arnaise S., Picard M., 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61: 411–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R., 1999. Internuclear recognition: a possible connection between euascomycetes and homobasidiomycetes. Fungal Genet. Biol. 27: 218–223 [DOI] [PubMed] [Google Scholar]
- Debuchy R., Turgeon B. G., 2006. Mating-type structure, evolution, and function in Euascomycetes, pp. 293–323 in The Mycota I. Springer-Verlag, Berlin [Google Scholar]
- Debuchy R., Berteaux-Lecellier V., Silar P., 2010. Mating systems and sexual morphogenesis in Ascomycetes, pp. 501–535 in Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. ASM Press, Washington, DC [Google Scholar]
- Desjardins A. E., Brown D. W., Yun S. H., Proctor R. H., Lee T., et al. , 2004. Deletion and complementation of the mating type (MAT) locus of the wheat head blight pathogen Gibberella zeae. Appl. Environ. Microbiol. 70: 2437–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A. V., An Z., Metzenberg R. L., Glass N. L., 1998. Characterization of mat A-2, mat A-3 and deltamatA mating-type mutants of Neurospora crassa. Genetics 148: 1069–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. A., Heitman J., 2004. Evolution of fungal sex chromosomes. Mol. Microbiol. 51: 299–306 [DOI] [PubMed] [Google Scholar]
- Fraser J. A., Heitman J., 2005. Chromosomal sex-determining regions in animals, plants and fungi. Curr. Opin. Genet. Dev. 15: 645–651 [DOI] [PubMed] [Google Scholar]
- Fraser J. A., Giles S. S., Wenink E. C., Geunes-Boyer S. G., Wright J. R., et al. , 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437: 1360–1364 [DOI] [PubMed] [Google Scholar]
- Fraser J. A., Stajich J. E., Tarcha E. J., Cole G. T., Inglis D. O., et al. , 2007. Evolution of the mating type locus: insights gained from the dimorphic primary fungal pathogens Histoplasma capsulatum, Coccidioides immitis, and Coccidioides posadasii. Eukaryot. Cell 6: 622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N. L., Smith M. L., 1994. Structure and function of a mating-type gene from the homothallic species Neurospora africana. Mol. Gen. Genet. 244: 401–409 [DOI] [PubMed] [Google Scholar]
- Harley V. R., Clarkson M. J., Argentaro A., 2003. The molecular action and regulation of the testis-determining factors, SRY (sex-determining region on the Y chromosome) and SOX9. [SRY-related high-mobility group (HMG) box 9] Endocr. Rev. 24: 466–487 [DOI] [PubMed] [Google Scholar]
- Heitman J., 2006. Sexual reproduction and the evolution of microbial pathogens. Curr. Biol. 16: R711–R725 [DOI] [PubMed] [Google Scholar]
- Hoffmann B., Eckert S. E., Krappmann S., Braus G. H., 2001. Sexual diploids of Aspergillus nidulans do not form by random fusion of nuclei in the heterokaryon. Genetics 157: 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer E., 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19: 33–131 [DOI] [PubMed] [Google Scholar]
- Kashimada K., Koopman P., 2010. Sry: the master switch in mammalian sex determination. Development 137: 3921–3930 [DOI] [PubMed] [Google Scholar]
- Kronstad J. W., Staben C., 1997. Mating type in filamentous fungi. Annu. Rev. Genet. 31: 245–276 [DOI] [PubMed] [Google Scholar]
- Lee J., Lee T., Lee Y. W., Yun S. H., Turgeon B. G., 2003. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol. Microbiol. 50: 145–152 [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G., Horwitz B. A., Yoder O. C., Turgeon B. G., 1997. Transcripts at the mating type locus of Cochliobolus heterostrophus. Mol. Gen. Genet. 256: 661–673 [DOI] [PubMed] [Google Scholar]
- Levine M., Tjian R., 2003. Transcription regulation and animal diversity. Nature 424: 147–151 [DOI] [PubMed] [Google Scholar]
- Marais G., Galtier N., 2003. Sex chromosomes: how X-Y recombination stops. Curr. Biol. 13: R641–R643 [DOI] [PubMed] [Google Scholar]
- Metzenberg R. L., Glass N. L., 1990. Mating type and mating strategies in Neurospora. Bioessays 12: 53–59 [DOI] [PubMed] [Google Scholar]
- Miller B. L., Miller K. Y., Timberlake W. E., 1985. Direct and indirect gene replacements in Aspergillus nidulans. Mol. Cell. Biol. 5: 1714–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B. L., Miller K. Y., Roberti K. A., Timberlake W. E., 1987. Position-dependent and -independent mechanisms regulate cell-specific expression of the SpoC1 gene cluster of Aspergillus nidulans. Mol. Cell. Biol. 7: 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. A., 1996. Mating systems in ascomycetes: a romp in the sac. Trends Genet. 12: 69–74 [DOI] [PubMed] [Google Scholar]
- Paoletti M., Seymour F. A., Alcocer M. J., Kaur N., Calvo A. M., et al. , 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 17: 1384–1389 [DOI] [PubMed] [Google Scholar]
- Pilon N., Daneau I., Paradis V., Hamel F., Lussier J. G., et al. , 2003. Porcine SRY promoter is a target for steroidogenic factor 1. Biol. Reprod. 68: 1098–1106 [DOI] [PubMed] [Google Scholar]
- Poggeler S., Kuck U., 2000. Comparative analysis of the mating-type loci from Neurospora crassa and Sordaria macrospora: identification of novel transcribed ORFs. Mol. Gen. Genet. 263: 292–301 [DOI] [PubMed] [Google Scholar]
- Poggeler S., Nowrousian M., Ringelberg C., Loros J. J., Dunlap J. C., et al. , 2006. Microarray and real-time PCR analyses reveal mating type-dependent gene expression in a homothallic fungus. Mol. Genet. Genomics 275: 492–503 [DOI] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J. A., Hemmons L. M., MacDonald K. D., Bufton A. W., 1953. The Genetics of Aspergillus nidulans. Adv. Genet. 5: 141–238 [DOI] [PubMed] [Google Scholar]
- Pyrzak W., Miller K. Y., Miller B. L., 2008. Mating type protein Mat1–2 from asexual Aspergillus fumigatus drives sexual reproduction in fertile Aspergillus nidulans. Eukaryot. Cell 7: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju N. B., Perkins D. D., 2000. Programmed ascospore death in the homothallic ascomycete Coniochaeta tetraspora. Fungal Genet. Biol. 30: 213–221 [DOI] [PubMed] [Google Scholar]
- Rydholm C., Dyer P. S., Lutzoni F., 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot. Cell 6: 868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C., 2006. Aspergillus genomes: secret sex and the secrets of sex. Trends Genet. 22: 521–525 [DOI] [PubMed] [Google Scholar]
- Smale S. T., Kadonaga J. T., 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72: 449–479 [DOI] [PubMed] [Google Scholar]
- Su H., Lau Y. F., 1993. Identification of the transcriptional unit, structural organization, and promoter sequence of the human sex-determining region Y (SRY) gene, using a reverse genetic approach. Am. J. Hum. Genet. 52: 24–38 [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E., Nayak T., Oakley C. E., Edgerton H., Xiong Y., et al. , 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1: 3111–3120 [DOI] [PubMed] [Google Scholar]
- Turgeon B. G., 1998. Application of mating type gene technology to problems in fungal biology. Annu. Rev. Phytopathol. 36: 115–137 [DOI] [PubMed] [Google Scholar]
- Vallim M. A., Miller K. Y., Miller B. L., 2000. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol. Microbiol. 36: 290–301 [DOI] [PubMed] [Google Scholar]
- Weissman J. D., Singer D. S., 1991a A complex regulatory DNA element associated with a major histocompatibility complex class I gene consists of both a silencer and an enhancer. Mol. Cell. Biol. 11: 4217–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman J. D., Singer D. S., 1991b Striking similarities between the regulatory mechanisms governing yeast mating-type genes and mammalian major histocompatibility complex genes. Mol. Cell. Biol. 11: 4228–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsel S., Horwitz B., Yamaguchi K., Yoder O. C., Turgeon B. G., 1998. Single mating type-specific genes and their 3′ UTRs control mating and fertility in Cochliobolus heterostrophus. Mol. Gen. Genet. 259: 272–281 [DOI] [PubMed] [Google Scholar]
- Wu J., Miller B. L., 1997. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell. Biol. 17: 6191–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelton M. M., Hamer J. E., de Souza E. R., Mullaney E. J., Timberlake W. E., 1983. Developmental regulation of the Aspergillus nidulans trpC gene. Proc. Natl. Acad. Sci. USA 80: 7576–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S. H., Arie T., Kaneko I., Yoder O. C., Turgeon B. G., 2000. Molecular organization of mating type loci in heterothallic, homothallic, and asexual Gibberella/Fusarium species. Fungal Genet. Biol. 31: 7–20 [DOI] [PubMed] [Google Scholar]