Abstract
The recent progress in sequencing technologies makes possible large-scale medical sequencing efforts to assess the importance of rare variants in complex diseases. The results of such efforts depend heavily on the use of efficient study designs and analytical methods. We introduce here a unified framework for association testing of rare variants in family-based designs or designs based on unselected affected individuals. This framework allows us to quantify the enrichment in rare disease variants in families containing multiple affected individuals and to investigate the optimal design of studies aiming to identify rare disease variants in complex traits. We show that for many complex diseases with small values for the overall sibling recurrence risk ratio, such as Alzheimer’s disease and most cancers, sequencing affected individuals with a positive family history of the disease can be extremely advantageous for identifying rare disease variants. In contrast, for complex diseases with large values of the sibling recurrence risk ratio, sequencing unselected affected individuals may be preferable.
COMMON diseases such as diabetes, cancer, and autism are likely caused by a complex interaction among many genes and environmental factors. Both common and rare genetic variants are expected to play a role. Thus far the available technology has allowed for the identification of common disease susceptibility variants, mostly via genome-wide association studies. However, the common variants detected so far have small effect sizes and overall explain only a small fraction of the estimated trait heritability (Maher 2008; Manolio et al. 2009). The recent advances in next-generation sequencing technologies (Metzker et al. 2010; Tucker et al. 2009) allow for the first time an objective assessment of the importance of rare variants in complex diseases. An increasing number of recent studies on hypertension, schizophrenia, epilepsy, type-1 diabetes, autism, etc. (Ji et al. 2008; Stefansson et al. 2008; Helbig et al. 2009; Nejentsev et al. 2009; Pinto et al. 2010) implicate rare variants in these disorders.
Ongoing sequencing studies are already generating unprecedented amounts of genetic data. The large number of genetic variants in these data sets, most of them with low frequencies (<1%), creates great challenges for statistical analysis. Traditional association testing strategies that have worked well for common variants will have low power to identify rare disease susceptibility variants (Morris and Zeggini 2009; Bansal et al. 2010). To extract the rich information provided by large sequencing data sets, several novel statistical approaches have been proposed, especially designed to identify rare variants that influence disease risk (Li and Leal 2008; Madsen and Browning 2009; Bhatia et al. 2010; Han and Pan 2010; King et al. 2010; Liu and Leal 2010; Price et al. 2010; Ionita-Laza et al. 2011; Neale et al. 2011). The common idea underlying all these methods is to group variants in a region of interest, e.g., a gene, and perform a gene-based test rather than individual tests for each of the variants in a gene.
An important question that has not yet been addressed is the relative power of designs based on affected relatives vs. designs using unselected affected individuals to identify rare disease variants. Since rare disease variants tend to be enriched in families containing multiple affected individuals, family-based designs can play an important role in the identification of rare causal variants. The purpose of this article is to quantify this enrichment and to study its implications for the optimal design of studies that search for rare disease variants in complex traits.
Methods
Effective number of variants in related individuals
By analogy to the concepts of “effective population size” in population genetics (Wright 1931, 1938) and “effective number of markers” in a linkage disequilibrium block (Nyholt 2004), we introduce here a new concept for analysis of rare variants in related individuals: the effective number of variants at a position, i.e., the number of “independent” variants at a specific position in a sample of related individuals. More precisely, the effective number of variants is the number of observed variants corrected for the known familial correlation among the individuals included in a sample. This concept is important as it allows a uniform comparison of designs based on various types of relatives, regardless of the relationship type.
For simplicity of presentation we define the effective number of variants at a position for a pair of individuals. If the individuals are unrelated, then the effective number of variants is equal to the observed number of variants since the two individuals are independent (hence no correction is necessary). However, the effective number of variants in related individuals is less than the total number of observed variants if some of the variants are shared among family members. For example, for a pair of siblings each of whom carries a rare variant in heterozygous state at a position, the effective number of variants will be less than the observed number of variants, i.e., two, due to the high probability that these two variants are shared identical-by-descent (IBD) and hence are not independent. Similarly, for a pair of second cousins that each carry a rare variant in heterozygous state the effective number of variants is less than two, although as shown in the examples below, it is higher than for a pair of siblings due to the lower likelihood that a variant is shared IBD for second cousins compared with siblings. Below we define mathematically the concept of effective number of variants.
For a pair of relatives we define the effective number of variants, keff, as follows:
By definition, keff|1 = 1 and keff|0 = 0. For keff|2, we show in supporting information, File S1 that
| (1) |
where f is the frequency of the variant, and φ is the kinship coefficient; δ = 0 if the two relatives can share a maximum of one allele IBD (e.g., first cousins) and 1 if they can share two alleles IBD (e.g., siblings). The approximation is based only on the assumption that the variant is rare (i.e., f ≤ 0.01) and is very accurate under this assumption.
When the two individuals are unrelated and each carries a rare variant, φ = 0 and we obtain the expected result that keff|2 = 2. For identical twins φ = 0.5, δ = 1, and keff|2 = 1. For relatives in between, the effective number of variants is between 1 and 2. For two sibs, , δ = 1, and hence for f = 0.01 we obtain keff|2 = 1.17. Similarly for two second cousins, , δ = 0, and hence keff|2 = 1.76. These and other examples are summarized in Table 1. Note that the effective number of variants depends on the frequency f. Hence, as expected, the lower the frequency is, the lower the effective number of variants, reflecting the low probability that these shared variants are independent (and the greater chance they are identical-by-descent).
Table 1. The effective number of variants, keff|2, at a rare variant position in two related individuals that each carry a variant, as defined in Equation 1.
| keff|2 | |||
|---|---|---|---|
| Relationship | φ | f = 0.001 | f = 0.01 |
| Identical twins | 1.00 | 1.00 | |
| Parent–child | 1.11 | 1.17 | |
| Sibs | 1.11 | 1.17 | |
| Half-sibs | 1.22 | 1.34 | |
| Uncle–nephew | 1.22 | 1.34 | |
| First cousins | 1.33 | 1.50 | |
| First cousins-1 (once removed) | 1.44 | 1.64 | |
| Second cousins | 1.55 | 1.76 | |
| Unrelateds | 0 | 2.00 | 2.00 |
φ is the kinship coefficient. Results for two values of the frequency parameter f, 0.001 and 0.01, are shown.
To summarize, for a pair of relatives keff is calculated as follows:
Under the assumption that the variant is not associated with disease, we calculate the expected value of keff for a pair of relatives in File S1 (Equation S7) as
| (2) |
At a rare variant position, for a data set of relative pairs of the same type in N different families, we define , where is the effective number of variants in family i. Then
| (3) |
If the variant is not associated with disease, then the distribution of (as also shown empirically in File S1, Figure S1)
Effective number of variants at a disease locus
At a disease locus, the effective number of variants in two affected relatives is expected to be increased compared to a nondisease locus. We consider here a two-locus genetic heterogeneity model (Risch 1990a), where each locus is an independent cause for disease. We use the two-locus model for mathematical convenience; without loss of generality the second locus can be considered to encompass all the other disease loci that act additively to influence disease risk, in addition to the primary locus under investigation. The effective number of variants we expect to observe at the first disease locus in a pair of affected relatives is
| (4) |
where βR, αR ≥ 1 are derived in File S1 and depend on the type of relationship R (e.g., sibs, first cousins, etc.), the genotype relative risk (GRR) for the first locus, and the overall recurrence risk in siblings as measured by λS (Risch 1990a). The expression in Equation 4 is similar to the expression in Equation 2 for the effective number of variants expected at a nondisease locus. We performed simulations according to this model and show that the empirical estimates for agree very well with the analytical results in (4) (see File S1).
For a data set of N different families, each consisting of an affected relative pair of the same type, we have
| (5) |
As above, the distribution of (as also shown empirically File S1).
Furthermore, using Equation 4 we can also calculate the effective number of variants we expect at the first disease locus in an affected individual with an affected relative. This is important to evaluate the importance of selecting affected individuals with a positive family history for a disease for inclusion in a sequencing study. We denote by the number of variants at the first locus in an affected individual that is known to have an affected relative. Then using Equation 4,
where βR, αR ≥ 1 are derived in File S1. As above, for a data set of N affected individuals, each with an affected relative. Again it is true that .
Note that the description above pertains to a disease locus with only one disease variant position. However, a disease locus may have multiple disease variant positions (i.e., allelic heterogeneity). The extension to this scenario is straightforward by summing the effective number of variants at each position and using the fact that a sum of independent Poisson random variables is also a Poisson random variable (we assume independence among the different variant positions within a locus, a reasonable approximation given the low frequency of the variants).
Expected P-value at a disease locus
To compare the power of designs on the basis of biologically related cases vs. unrelated cases, we calculate an expected P-value (EPV) (Dempster and Schatzoff 1965; Sackrowitz and Samuel-Cahn 1999). The expected P-value, or expected significance level as originally defined in the pioneering article of Dempster and Schatzoff (1965), has been proposed as a measure of the performance of a test. Unlike the power of a test, the EPV does not depend on any prespecified significance level and is in close connection with the common practice of reporting P-values in applied research. The EPV is a single number that can be used to judge the performance of a test; the smaller the EPV is, the better the test.
For our situation, the effective number of variants under the null hypothesis that neither of the two loci is associated with disease, , follows approximately a Poisson distribution with mean as derived in Equation 3. Similarly, the effective number of variants at the first disease locus in a two-locus disease model, , follows approximately a Poisson distribution with as derived in Equation 5. Then by definition the expected P-value for the first disease locus is
where T1 ∼ Poisson(λ1) and T2 ∼ Poisson(λ2). Since the difference between two independent Poisson random variables follows a Skellam distribution (Skellam 1946), we can calculate the EPV under a disease model only on the basis of the values of λ1 and λ2.
For comparison we also report results based on power, and, as shown below, they are highly correlated with those based on the EPV. Since we assume that the variant frequency in controls is known, the resulting power levels can be considered as achievable as the number of controls grows very large.
Results
Performance of affected relatives vs. unrelated affected individuals
For identifying rare disease variants, it is of great interest to study the circumstances where study designs involving affected individuals from families containing multiple affected individuals are advantageous compared with those involving unselected affected individuals.
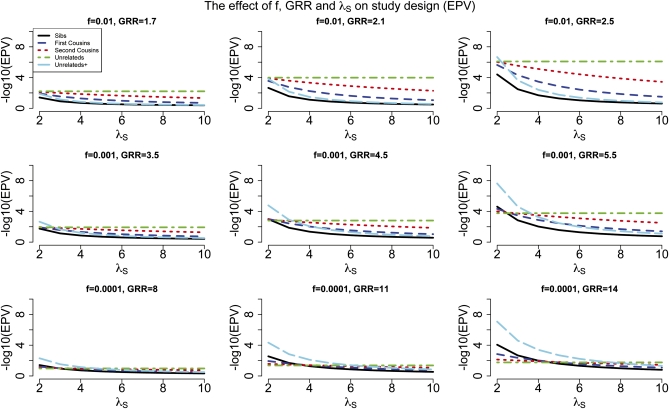
Under the assumption of a complex polygenic model, with many possible disease loci each with a small population attributable risk (Risch and Merikangas 1996), the two-locus disease model can be viewed as consisting of a locus of interest and a second locus accounting for the residual effect. We assume five study designs, each with the same number of affected individuals (i.e., 2000): affected sib-pairs, affected first-cousin pairs, affected second-cousin pairs, unrelated affected individuals, and unrelated affected individuals known to have an affected sibling. We consider a complex trait with prevalence 0.03 and a sibling recurrence risk ratio (λS) between 2 and 10, as observed for many complex traits (Merikangas and Risch 2003). We assume that the variant frequency at the first disease locus is low, between 0.0001 and 0.01, and several possible values for the genotype relative risk with higher GRRs are assumed for lower frequency at the disease locus. In Figure 1 we show expected P-values associated with the first locus as a function of the sibling recurrence risk ratio, for each of the five study designs.
Figure 1.
The effect of locus frequency, GRR, and overall λS on the relative performance (as measured by the expected P-value) of affected relatives vs. unrelated affected individuals. The three rows correspond to three different frequencies for the disease locus: 0.01, 0.001, and 0.0001. λS is between 2 and 10. The number of affected individuals is 2000: 1000 sib-pairs, 1000 first-cousin pairs, 1000 second-cousin pairs, 2000 unrelateds, and 2000 unrelated individuals known to have an affected sibling (i.e., unrelateds+).
For complex diseases with low values for λS (e.g., 2–4), affected individuals known to have an affected sibling are extremely advantageous to identify rare disease variants of moderate to large effect (e.g., GRR ∼ 5–10, Figure 1). The difference between using affected individuals with an affected sibling vs. unselected affected individuals can be substantial in this case. For example, for a disease with λS = 2, for a locus with frequency 0.001 and a GRR of 5.5 the expected P-value is 10−7.65 for a study with 2000 unrelated affected individuals known to have an affected sibling and only 10−3.77 for a study with 2000 unselected affected individuals. Similarly, for a locus with frequency 0.0001 and a GRR of 14 the corresponding expected P-values are 10−7 and 10−18. For small values of λS and large values of GRR (e.g., λS = 2 and GRR = 14), even a design based on 1000 affected individuals known to have an affected sibling is expected to be more advantageous than a design based on 2000 unselected affected individuals (Figures 1 and 4). Similar results are obtained when the design comparison is based on power rather than expected P-value (Figure S2).
However, for complex diseases with larger values of λS such as autism (λS ≈ 22), the advantage of using family-based designs (either affected relative pairs or affected individuals with a known affected sibling) diminishes, and unselected affected individuals can be more advantageous (Figure 2 and Figure S3). This trend of greater advantage with unselected affected individuals is more pronounced with increasing frequency of disease-causing variants (Figure 1). These results are in concordance with those in Risch (1990b,1992) for linkage analysis, which state that for small values of overall λS and polymorphic information content (PIC) (Botstein et al. 1980) affected sib-pairs are optimal, while for large values of λS and PIC, more distantly related individuals are best.
Figure 2.
Usefulness of sequencing both affected individuals in a pair of affected relatives. The three rows correspond to three different frequencies for the disease locus: 0.01, 0.001, and 0.0001. The number of affected individuals is 2000 for the two affected relatives design, i.e., 1000 sib-pairs and 1000 second-cousin pairs, and only 1000 for the design based on only one affected individual in a pair, i.e., 1000 affected individuals known to have an affected sibling and 1000 affected individuals known to have an affected second cousin.
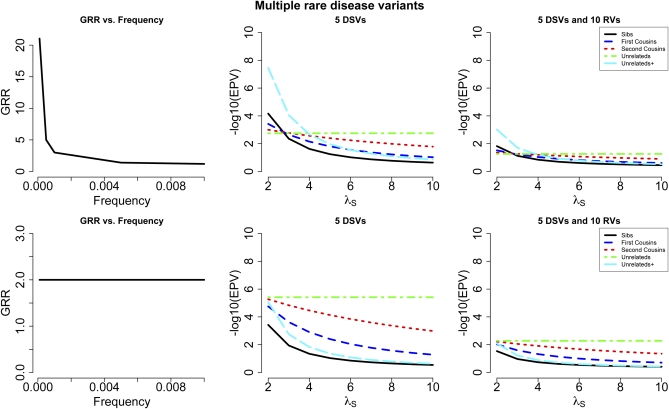
Above we have assumed that the disease locus has only one disease variant position. Although this is clearly a simplified scenario, the results obtained are informative for the more realistic scenario when the locus of interest contains multiple disease variants. In Figure 3 we show the results for the case when 5 different disease variant positions are present at the locus. Two cases are illustrated: (1) the GRRs for the individual variants are a decreasing function of frequency, so that lower-frequency variants have substantially higher GRRs compared to more frequent variants (for example, f = 0.0001 and GRR = 21 vs.f = 0.01and GRR = 1.2), and (2) the GRR is the same for all variants (i.e., 2). As can be seen, for the first case, due to the presence of high-risk low-frequency variants among the 5 disease variants, unrelated affected individuals that are known to have an affected sibling are best for small values of λS (as also shown previously in Figure 1), whereas for the second case where the GRR for low-frequency variants is modest (i.e., 2), distantly related affected individuals, and in particular unselected affected individuals, perform better. Also shown are results when 10 additional random variants are added to the region. In this case, the relative performance of different designs is unchanged, but the performance is reduced for all designs.
Figure 3.
Results are shown for scenarios where multiple (i.e., 5) disease susceptibility variants (DSVs) are present at the disease locus. The relationship between the GRR and disease variant frequency is shown in the first column. In the top panel, the GRR is a decreasing function of variant frequency, while in the bottom panel, the GRR is the same (i.e., 2) regardless of variant frequency. The third column presents results when 10 additional random variants (i.e., not disease variants) are also present.
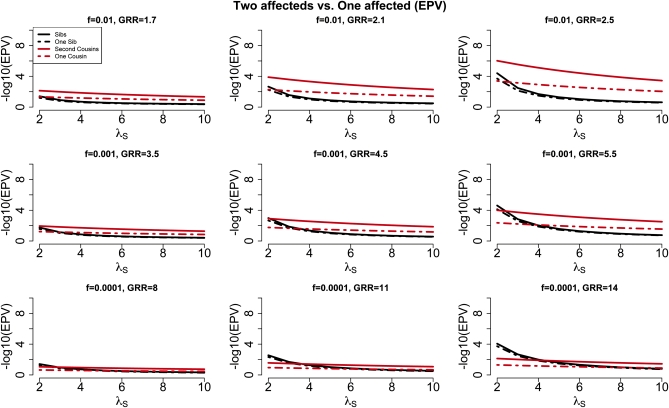
The utility of sequencing both affected individuals in a pair of affected relatives
Sequencing both individuals from a pair of affected relatives doubles the sequencing costs, so it is important to consider the circumstances under which power is increased by this approach. We evaluated the utility of sequencing both affected individuals in an affected relative pair compared to sequencing only one individual from the pair (Figure 4 and Figure S4). We find that for affected sibling pairs, sequencing the second sibling contributes little additional information; hence sequencing only one of two affected siblings is expected to be on average almost as good as sequencing both of them, with the significant advantage of reducing the sequencing cost by half (Figure 4). However, for more distantly related individuals, such as second cousins, sequencing both individuals is expected to be more powerful than sequencing only one of them. The advantage can be significant. For example, when λS = 2, for a locus with frequency 0.001 and a GRR of 5.5 the expected P-value is 10−4 for a study with 1000 affected second-cousin pairs (power at α = 1.6 × 10−6 is 87%) and only 10−2.1 for a study with 1000 affected individuals known to have a second cousin affected (power is 47%). If instead we compare the power of a study with 1000 affected second-cousin pairs with that of a study with 2000 affected individuals known to have a second cousin affected, hence the same sequencing cost for both studies, we find that the power levels are very similar (data not shown).
Figure 4.
For complex traits with a large value for λS (e.g., λS = 22 for autism) we show expected P-values for a disease locus with frequency f = 0.001 as a function of the GRR for five study designs, each with 2000 affected individuals: 1000 sib-pairs, 1000 first-cousin pairs, 1000 second-cousin pairs, 2000 unrelateds, and 2000 unrelated individuals known to have an affected sibling (i.e., unrelateds+).
Discussion
In complex diseases characterized by extensive genetic heterogeneity, each disease locus is likely to be responsible for a very small fraction of affected individuals in a population. The choice of the affected individuals to be included in a study greatly influences the power to identify disease loci. The framework developed here, based on the effective number of variants in a pair of affected relatives, makes it feasible to quantify the enrichment in rare disease variants in family-based vs. population-based samples and to investigate the optimal study design for identifying rare disease variants in complex traits.
We have shown here that for diseases with small values for the sibling recurrence risk ratio, as observed with many complex traits, sequencing affected individuals with an affected close relative, such as a sibling, can be an extremely powerful strategy for identifying rare disease variants with moderate to large GRRs. This finding is in concordance with previous findings for breast cancer and common variants (Antoniou and Easton 2003). However, we find that the advantage of using affected individuals with a positive family history declines with increasing values of λS. For complex diseases with large values of λS, such as autism (λS ≈ 22), unselected affected individuals may be preferable. The main explanation for these results is that for a complex disease involving many contributing disease loci, one single locus is likely to explain less of the overall familial aggregation when λS is large than when λS is small. Hence when λS is large, focusing on affected relatives (or individuals with an affected relative) may enrich the sample in rare disease loci that are less likely to be shared among multiple families, and therefore unselected affected individuals may be preferable.
Designs based on affected relative pairs are commonly employed in linkage analysis (Weeks and Lange 1988). Although here we have not attempted to examine the performance of linkage analysis, we note that the results we obtained agree well with those for linkage analysis (Risch 1990b, 1992), which state that for small values of overall λS and disease locus frequency affected sib-pairs are optimal, while for large values of λS and disease locus frequency, more distantly related individuals are best.
The framework developed here is important for the statistical analysis of rare variant data. We have introduced here the concept of effective number of variants at a position in a set of relatives, which allows for a unified treatment of both biologically related and unrelated affected individuals and makes feasible natural extensions of statistical tests recently developed for population-based designs (such as the weighted-sum statistic in Madsen and Browning 2009) to general designs based on affected relative pairs and/or unrelated affected individuals. One simple statistic for affected relative pairs would be a weighted sum of the effective number of variants at different positions. Such extensions are currently under development.
The main goal of this article is to introduce a framework to assess the enrichment of rare disease mutations in affected relative pairs of different types. Such an enrichment analysis is of relevance for any association testing method, and therefore the results presented are expected to be valid in general for association testing. We have applied this framework to the problem of optimal study design for association studies with rare variants.
Software implementing the described methods is available in File S2, as well as on the first author’s webpage at http://www.columbia.edu/~ii2135/, and should be helpful in the design of association studies with rare variants.
Acknowledgments
We are grateful to Mitchell Gail and Joseph Buxbaum for useful discussions. We also thank the associate editor and two reviewers for their comments that helped improve our article. This work was supported by grants R03HG005908-2 (to I.I.-L.) and RC2 NS070344 (to R.O.) from the National Institutes of Health and DMS-1100279 (to I.I.-L.) from the National Science Foundation.
Literature Cited
- Antoniou A. C., Easton D. F., 2003. Polygenic inheritance of breast cancer: implications for design of association studies. Genet. Epidemiol. 25: 190–202 [DOI] [PubMed] [Google Scholar]
- Bansal V., Libiger O., Torkamani A., Schork N. J., 2010. Statistical analysis strategies for association studies involving rare variants. Nat. Rev. Genet. 11: 773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia G., Bansal V., Harismendy O., Schork N. J., Topol E. J., et al. , 2010. A covering method for detecting genetic associations between rare variants and common phenotypes. PLoS Comput. Biol. 6: e1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W., 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32: 314–331 [PMC free article] [PubMed] [Google Scholar]
- Dempster A. P., Schatzoff M., 1965. Expected significance level as a sensitivity index for test statistics. J. Am. Stat. Assoc. 60: 420–436 [Google Scholar]
- Han F., Pan W., 2010. A data-adaptive sum test for disease association with multiple common or rare variants. Hum. Hered. 70: 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig I., Mefford H. C., Sharp A. J., Guipponi M., Fichera M., et al. , 2009. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 41: 160–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I., Buxbaum J., Laird N. M., Lange C., 2011. A new testing strategy to identify rare variants with either risk or protective effect on disease. PLoS Genet. 7: e1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Foo J. N., O’Roak B. J., Zhao H., Larson M. G., et al. , 2008. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat. Genet. 40: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C. R., Rathouz P. J., Nicolae D. L., 2010. An evolutionary framework for association testing in resequencing studies. PLoS Genet. 6: e1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Leal S. M., 2008. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. J., Leal S. M., 2010. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 6(10): e1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen B. E., Browning S. R., 2009. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 5: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher B., 2008. Personal genomes: the case of the missing heritability. Nature 456: 18–21 [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas K. R., Risch N., 2003. Genomic priorities and public health. Science 302: 599–601 [DOI] [PubMed] [Google Scholar]
- Metzker M. L., 2010. Sequencing technologies—the next generation. Nat. Rev. Genet. 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Morris A. P., Zeggini E., 2009. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet. Epidemiol. 34: 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale B. M., Rivas M. A., Voight B. F., Altshuler D., Devlin B., et al. , 2011. Testing for an unusual distribution of rare variants. PLoS Genet. 7: e1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejentsev S., Walker N., Riches D., Egholm M., Todd J. A., 2009. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324: 387–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt D. R., 2004. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet. 74: 765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Pagnamenta A. T., Klei L., Anney R., Merico D., et al. , 2010. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Kryukov G. V., de Bakker P. I., Purcell S. M., Staples J., et al. , 2010. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86: 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N., 1990a Linkage strategies for genetically complex traits. I. Multilocus models. Am. J. Hum. Genet. 46: 222–228 [PMC free article] [PubMed] [Google Scholar]
- Risch N., 1990b Linkage strategies for genetically complex traits. III. The effect of marker polymorphism on analysis of affected relative pairs. Am. J. Hum. Genet. 46: 242–253 [PMC free article] [PubMed] [Google Scholar]
- Risch N., 1992. Corrections to “Linkage strategies for genetically complex traits. III. The effect of marker polymorphism on analysis of affected relative pairs”. Am. J. Hum. Genet. 51: 673–675 [PMC free article] [PubMed] [Google Scholar]
- Risch N., Merikangas K., 1996. The future of genetic studies of complex human diseases. Science 273: 1516–1517 [DOI] [PubMed] [Google Scholar]
- Sackrowitz H. B., Samuel-Cahn E., 1999. P-values as random variables: expected P-values. Am. Stat. 53: 326–331 [Google Scholar]
- Skellam J. G.1946. The frequency distribution of the difference between two Poisson variates belonging to different populations. J. R. Stat. Soc. Ser. A, 109: 296. [PubMed] [Google Scholar]
- Stefansson H., Rujescu D, Cichon S, Pietilinen O. P, Ingason A, et al. , 2008. Large recurrent microdeletions associated with schizophrenia. Nature 455: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker T., Marra M., Friedman J. M., 2009. Massively parallel sequencing: the next big thing in genetic medicine. Am. J. Hum. Genet. 85: 142–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks D. E., Lange K., 1988. The affected-pedigree-member method of linkage analysis. Am. J. Hum. Genet. 42: 315–326 [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1938. Size of population and breeding structure in relation to evolution. Science 87: 430–431 [Google Scholar]