Abstract
MicroRNAs (miRNAs) bind to complementary sequences within the 3′ untranslated region (UTR) of mRNAs from hundreds of target genes, leading either to mRNA degradation or suppression of translation. We found that a mutation in the seed region of miR-184 (MIR184) is responsible for familial severe keratoconus combined with early-onset anterior polar cataract by deep sequencing of a linkage region known to contain the mutation. The mutant form fails to compete with miR-205 (MIR205) for overlapping target sites on the 3′ UTRs of INPPL1 and ITGB4. Although these target genes and miR-205 are expressed widely, the phenotype is restricted to the cornea and lens because of the very high expression of miR-184 in these tissues. Our finding highlights the tissue specificity of a gene network regulated by a miRNA. Awareness of the important function of miRNAs could aid identification of susceptibility genes and new therapeutic targets for treatment of both rare and common diseases.
Main Text
Keratoconus (MIM 148300) is a noninflammatory thinning of the central cornea, which results in cone-shaped protrusion of the cornea, alteration in refractive power, reduced visual acuity, and image distortion. It is the most common corneal dystrophy and has an incidence of 1 in 20001 and is a major reason for corneal transplantation.2 Keratoconus is usually inherited as an autosomal-dominant trait with variable expression. Mutations in the visual system homeobox 1 gene (VSX1 [MIM 605020]) have been identified in patients with keratoconus; however, their role in causing disease is controversial.3, 4 The pathological mechanism behind keratoconus is poorly understood, but evidence points toward dysregulation of apoptosis.5, 6
Small (19–25 nucleotide) regulatory strands of RNA, named microRNAs (miRNAs), bind to complementary sequences, usually in the 3′ untranslated region (UTR) of mRNA of target genes, leading to degradation of the mRNA or suppression of translation.7 As part of a RNA-induced silencing complex, each miRNA might target mRNA from hundreds of genes.8 Tissue-specific expression of miRNAs is likely to affect the abundance of proteins in different organs and their component parts.
Within the eye, miR-184 (encoded by MIR184 [MIM 613146]) is expressed in the central corneal epithelial basal and suprabasal cells and in the lens epithelium.9, 10 In both tissues it is the most abundant miRNA. MiR-205 is a widely-expressed epithelial miRNA encoded by MIR205 (MIM 613147). One observed action of miR-184 is the competitive inhibition of the binding of miR-205 to mRNA of the inositol polyphosphate phosphatase-like 1 gene (INPPL1, also known as SHIP2 [MIM 600829]).11 Through this mechanism, miR-184 prevents knock-down by miR-205 and rescues INPPL-1 production. Ultimately, this sustains levels of phosphorylated AKT and phosphorylated BCL-2-associated death promoter (BAD), which regulate apoptosis.11
We investigated the cause of disease in a Northern Irish family in which 18 of 38 individuals from three generations were affected by an autosomal-dominant form of severe anterior keratoconus and early-onset anterior polar cataract. The study was approved by the Northern Ireland Office for Research Ethics Committees and conformed to the tenets of the Declaration of Helsinki. We previously mapped the disease locus in this family to a 5.5 Mb region of chromosome 15q22-q25 and excluded many positional candidate genes by conventional sequencing.12, 13
Recently, we enriched for all genes within the region by sequence capture. A custom sequence capture array (Roche NimbleGen, Madison, Wisconsin, USA) was designed to capture 5 Mb of the 5.5 Mb linkage region at 15q22-q25 (a repetitive gene-devoid 0.5 Mb region was excluded). The array was designed with the Sequence Search and Alignment by Hashing Algorithm and comprised 385,000 unique probes.14 Three study DNA samples were captured: one affected family member and two pooled DNA samples from seven affected and six unaffected family members, respectively. DNA samples were enriched for the targeted sequences with the manufacturer's protocols. Briefly, 21 μg aliquots were fragmented and hybridized to the array followed by ligation-mediated PCR amplification of the enriched, fragmented DNA pool. Five micrograms of amplified enriched DNA underwent massively parallel sequencing on a Genome Analyzer II (Illumina, San Diego, CA, USA) with a single sample per flow cell to generate single-end reads of 40 bp (GATC Biotech, Konstanz, Germany).
Sequence data were converted from Solexa to Sanger standard format by use of Maq 0.7.1 and aligned to the National Center for Biotechnology Information v37 reference sequence with the BWA 0.5.9 short alignment algorithm.15 Sorting, indexing, and removal of duplicates were performed with Samtools 0.1.14.16 We used Picard 1.43 to edit read groups information. We used functions of the Genome Analysis Toolkit (GATK)17 to recalibrate base calls; to realign reads at the sites of possible insertions, duplications, and deletions; and to call polymorphisms (Unified Genotyper). Version 130 of dbSNP and the August 4, 2010, release of Dindel data for Europeans from the 1000 Genomes Project were employed for realignment. VCFtools 0.1.5 was used to compare variant calls from the three sample groups. The sequence was visualized with Integrative Genomics Viewer 1.5.18 We used GATK to annotate the polymorphisms with gene information from the RefSeq Genes track (made by the genome sequencing and analysis group at the Broad Institute from the University of California, Santa Cruz (UCSC) RefSeq Genes Track, which itself was derived from NCBI mRNA reference sequences collection; see Web Resources). Polymorphisms outside gene exons were excluded from further analyses. All variant calls from the three samples were entered into a database, which was queried for instances where the affected individual and the affected pool shared any genetic variation that was not present in the unaffected pool. Mean depth of coverage was 23–28 reads in each sample, with 70%–77% having greater than 15× coverage. Twenty-six variants (25 single nucleotide variants and one deletion of two bases) within exons and untranslated regions of coding and noncoding genes were identified that fitted this pattern (Table 1). We searched dbSNP and the 1000 Genomes Project data release of May 2011 for these polymorphisms. All but two SNPs were already known and were identified in European populations at a frequency of greater than 7.5% and were thus excluded from further consideration. A deletion of two bases was not recorded in dbSNP or the 1000 Genomes Project. Three possible causative mutations were therefore identified (Table 2).
Table 1.
Annotation of Massively Parallel Sequence
| Site of Variant |
Coding Gene |
Noncoding Gene |
||
|---|---|---|---|---|
| Total | Known | Total | Known | |
| Coding sequence | 9 | 9 | 5 | 4 |
| 5′ or 3′ UTR | 12 | 10 | – | – |
Genetic variants detected in both affected individual and affected pool, but not in unaffected pool, are categorized by site.
Table 2.
Novel Exonic Variants
We searched public databases for information about the function and expression of MIR184, DNAJA4, and IREB2. MiR-184 is the most abundantly expressed microRNA in the corneal and lens epithelia and is known to be involved in the regulation of protein levels in those tissues.9 The heterozygous C-to-T transition (r.57c>u) within miR-184 (Figure 1) is in the central nucleotide of the functionally essential seven base miRNA seed region. DNAJA4 is expressed in a wide range of tissues. Its product is a chaperone molecule involved in cholesterol biosynthesis.19 The two bases are deleted from the start codon of an alternatively spliced first exon. The deletion is predicted to prevent formation of isoform 3 of DNAJA4 from this allele. Relatively little is known about this gene, but it is not known to have any important role in the eye. IREB2 (MIM 147582) encodes an iron-responsive element-binding protein that regulates iron metabolism.20 The mutation in IREB2 in the 3′ UTR is not predicted by MicroCosm Targets to interact with any miRNA.21, 22
Figure 1.

Massively Parallel Sequencing Reads Showing Point Mutation in MIR184
Multiple alignment of massively parallel sequence reads spanning the seed region of miR-184 from (A) a single individual affected with familial keratoconus and cataract, (B) a pool of seven affected members of the same family and (C) a pool of six unaffected members of the same family. All nucleotides that vary from the reference sequence, shown below, are indicated. Read depth and heterozygosity are designated at the top of each column.
In light of its role and specific abundance in the cornea and lens, the mutation altering the seed region of miR-184 (r.57c>u) was the compelling candidate of the three previously unknown variants detected within exons. The gene is conserved in all 65 species known to have a copy of miR-184 (Figure 2A). We performed conventional sequencing of MIR184 (Table S1) in 167 unscreened controls and found no mutation within the mature miRNA. The miR-184 (r.57c>u) mutation was not reported in the 1094 individuals in the 1,000 Genomes Project data release of May 2011.23 Predictions of RNA folding with Mfold24 suggest that the mutation does not destabilize the secondary structure of the mir-184 pre-miR (Figure 2B).
Figure 2.
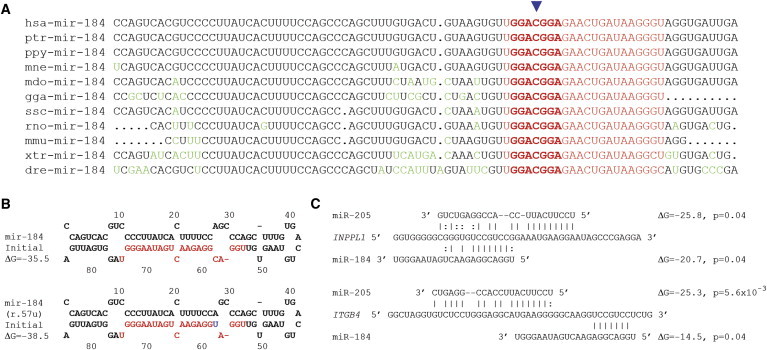
Variants of miR-184 and Conservation across Species, Structure of miR-184, miR-205 and Mutant miR-184, and Interactions of miR-184 and miR-205
(A) Multiple alignment of miR-184 sequences from different vertebrate species. The mutation site (miR-184 r.57c>u, indicated by the blue arrow) is fully conserved across all species. Changes from the human reference sequence are shown in green type; the mature miRNAs are shown in red type, and the seed regions are in bold. The following abbreviations are used: hsa, Homo sapiens; ptr, Pan troglodytes; ppy, Pongo pygmaeus; mne, Macaca nemestrina; mdo, Monodelphis domestica; gga, Gallus gallus; ssc, Sus scrofa; rno, Rattus norvegicus; mmu, Mus musculus; xtr, Xenopus tropicalis; dre, Danio rerio. Fifty-four additional species showed full conservation of the mature miR-184 sequence.
(B) Secondary structures and free energies of wild-type miR-184 and mutant miR-184 (r.57u) predicted by Mfold.
(C) Target sites of miR-184 and miR-205 in the 3′ UTRs of INPPL1 and ITGB4 predicted by MicroCosm with energies shown in kcal/mol.
The known competition between miR-184 and miR-205 for the 3′ UTR of INPPL1 (Figure 2C) facilitated a relatively straightforward functional assessment for miR-184 (r.57c>u). INPPL1 is expressed constitutively by HeLa cells, but neither miR-184 nor miR-205 is expressed.25, 26 We used a computational target prediction (the MicroCosm Targets v.521, 22 implementation of the miRanda algorithm27) to search for other instances where miR-184 and miR-205 had overlapping target sites in a 3′ UTR. This yielded only one other target, integrin beta 4 (ITGB4 [MIM 147557]), which is the main structural protein of hemidesmosomes that connect corneal basal epithelial cells to the basement membrane.28
We adapted Yu et al.'s experiment11 to test the ability of the mutant (r.57u) and wild-type miR-184 to interfere with the miR-205 knockdown of INPPL1 and ITGB4 transcripts in HeLa cells by immunohistochemical staining and by immunoblotting.
Blunt-ended double-stranded miRNA mimics for the predominant isoforms of miR-184 and miR-205 (Figure S1, available online), and for the mutant miR-184 (r.57u) were synthesized by Invitrogen (Table S2). HeLa cells (which express INPPL1 and ITGB4 but not miR-184 or miR-205) were grown to 70% confluence in 24-well plates in DMEM medium without antibiotics and transfected with miRNA mimics (20 nmol/L) with Santa Cruz transfection reagent according to the standard protocol. Cells were cultured for 72 hr before harvesting for immunoblotting and staining with anti-INPPL1 (NEBiolabs; no. 2839), anti-ITGB4 (Abcam; no. 29042) or anti-α-tubulin (Abcam; no. 4074) at a dilution of 1:1000. Donkey anti-rabbit and chicken anti-mouse HRP-linked secondary antibodies were used for immunoblotting and donkey anti-rabbit FITC-linked secondary antibodies (Abcam) with Alexa Fluor-555-labeled phalloidin (Invitrogen) were used for immunofluorescence. Immunoblots were performed for five sets of transfections. Luminescence was detected and quantified with a UVP BioSpectrum AC Imaging System.
Cells from an additional set of transfections were stained with fluorescently labeled antibodies. Immunofluorescence was detected with an Olympus IX51 microscope and Spot Diagnostics V4.1 (Diagnostic Instruments Inc.) software. Semiquantitative analysis was carried out on a minimum of 30 cells for each condition with Adobe Photoshop CS3. Statistical analyses were carried out with PASW statistics v18.0.0 (SPSS Inc., Chicago, IL, USA) and charts plotted with the gplots package within R v2.10.1.29
The levels of INPPL-1 (Figure 3A) and ITGB4 (Figure 3B) in HeLa cells in response to transfection with combinations of synthetic miRNAs was measured by immunoblotting. Knockdown of INPPL-1 was minimal with the miR-184 mimic (4.4%; p = 0.03) and negligible with the mutant miR-184 mimic (1.8%; p = 0.33). Transfection with the miR-205 mimic reduced the amount of INPPL-1 detected to 47% of that in control cells that underwent the process of transfection without any synthetic miRNA (p = 3.5 × 10−5). Transfection with the miR-205 mimic in combination with the wild-type miR-184 mimic resulted in rescue of levels of INPPL-1 to 86% of controls (p = 2.5 × 10−3). However, transfection with the miR-205 mimic in combination with the mutant miR-184 mimic resulted in failure to rescue INPPL-1 levels (44% versus 47%; p = 0.57 compared to miR-205 mimic alone).
Figure 3.

Immunoblot of INPPL-1 and ITGB4
Knockdown by miRNAs was performed in HeLa cells. Signal strengths are shown for INPPL-1 and ITGB4 relative to a sham transfected control, all normalized for α-tubulin loading. One representative image is shown from five replicated experiments. (A) There was no rescue of miR-205 knockdown of INPPL-1 by the mutant miR-184 mimic compared to rescue of miR-205 knockdown of INPPL-1 by wild-type miR-184 mimic. (B) There was no rescue of miR-205 knockdown of ITGB4 by the mutant miR-184 mimic compared to rescue of miR-205 knockdown of ITGB4 by wild-type miR-184 mimic. The error bars represent 95% confidence interval for mean.
Knockdown of ITGB4 was moderate with miR-184 mimic (13%; p = 3.4 × 10−4) and negligible with mutant miR-184 mimic (3%; p = 0.24). Transfection with miR-205 mimic reduced the amount of ITGB4 detected to 47% of that in control cells that underwent the process of transfection without any synthetic miRNA (p = 2.8 × 10−5). Transfection with miR-205 mimic in combination with wild-type miR-184 mimic resulted in rescue of levels of ITGB4 to 80% of controls (p = 4.0 × 10−3). However, transfection with miR-205 mimic in combination with mutant miR-184 mimic resulted in failure to rescue ITGB4 levels (44% versus 47%; p = 0.50 compared to miR-205 mimic alone).
Cells transfected with miR-205 mimic combined with either mutant or wild-type miR-184 mimic were stained for INPPL-1 and ITGB4. Cells treated with the mutant miR-184 mimic showed a 37% reduction of INPPL-1 compared to cells treated with the wild-type miR-184 mimic (Figures 4A–4C; p = 1.3 × 10−4). Cells treated with the mutant miR-184 mimic showed a 48% reduction of ITGB4 compared to cells treated with wild-type miR-184 mimic (Figure 4D–4F; p = 7.5 × 10−6).
Figure 4.
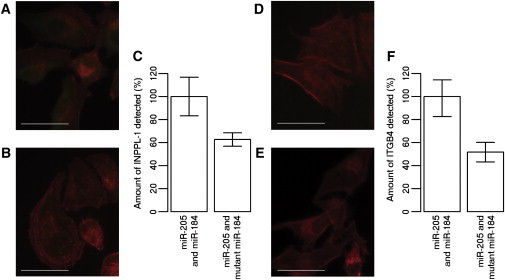
Immunofluorescence Staining of INPPL-1, ITGB4, and Actin
Representative micrographs showing (A and B) INPPL-1 (green) and actin (red) for cells transfected with (A) miR-184 mimic and miR-205 mimic and (B) mutant miR-184 and miR-205. The scale bar represents 50 μm.
(C) Semiquantitative analysis of mutant miR-184 knockdown relative to wild-type miR-184, both in the presence of miR-205. All measurements were standardized for actin. The error bars represent 95% a confidence interval for mean.
Representative micrographs showing (D and E) ITGB4 (green) and actin (red) for cells knocked down with (D) miR-184 mimic and miR-205 mimic and (E) mutant miR-184 and miR-205. The scale bar represents 50 μm.
(F) Semiquantitative analysis of mutant miR-184 knockdown relative to wild-type miR-184, both with miR-205. All measurements were standardized for actin. The error bars represent 95% confidence interval for mean.
The tissue-specific expression of miR-184 is of prime importance in the phenotypic effects that the mutation causes. Within the cornea, miR-184 expression is restricted to central basal and suprabasal epithelial cells,9 under which the stromal thinning occurs in keratoconus. Within the lens, the epithelium lies anteriorly and, paralleling the cornea, has less proliferative capacity centrally.30 This anterior lens epithelium expresses miR-184,9 adjacent to the site of the cataract in the affected family.
In the cornea, ITGB4 forms part of the α6β4 heterodimer, which is the principal component of corneal basal epithelial hemidesmosomes.28 Following corneal injury, hemidesmosomes in the basal epithelial layer are degraded to allow epithelial cell migration and are subsequently rebuilt. Expression of miR-184 is halted at the site of corneal injury and returns after healing.9 Dysregulation of the expression of ITGB4 in the central cornea (the site of miR-184 expression) might therefore also be important. The stromal keratocytes underlying the site of an injury undergo rapid apoptosis as a defense mechanism.6 Therefore, the role of INPPL-1 in regulation of apoptosis might also be vital in the development of keratoconus and cataract.
Expression of as many as 1000 genes might be regulated by miR-184, independently or in competition with other miRNAs, which might lead to complex effects on the levels of a large number of proteins. The harmful effects of mutant miR-184 might be mediated through proteins other than INPPL-1 and ITGB4. Among the predicted targets of miR-184 are a major lens transcription factor (FOXE3 [MIM 601094]) and a major intrinsic protein of eye lens fiber (MIP [MIM 154050]), mutations in both of which cause lens abnormalities in humans.31 The competition between miR-184 and miR-205, identified by Yu et al.11 and confirmed in our study, illustrates the complexity of miRNA action. We do not, at present, know why miR-184 is less effective than miR-205 at knock-down of INPPL-1 and ITGB4. Further studies are required to illuminate the molecular mechanisms involved.
In some cases, myopia is due to a steeply curving cornea. A genome-wide association study identified a small region encompassing MIR184 as a major myopia locus32 and focused on RASGRF1 (MIM 606600), which is adjacent to MIR184. Our finding that a MIR184 mutation causes keratoconus suggests that this gene warrants further investigation with respect to myopia.
The role of miRNAs in human diseases and possible treatments is a new and expanding field of study. This is the second report of a mutation in a miRNA associated with a human Mendelian disease. The previous report identified two mutations in miR-96 that resulted in hereditary deafness in two Spanish families.33
Our report demonstrates that variation in miRNAs can cause disease that is specific to the tissue in which the miRNA is expressed. This knowledge could open new lines of enquiry for those investigating the causes of diseases and could indeed suggest possible treatments: the therapeutic use of miRNA in eye diseases is a real and noteworthy prospect.
Acknowledgments
This work was supported by Northern Ireland Research and Development Office RRG grant 4.46 and Fight for Sight grant 1787. D.T.B. was funded by the Public Health Agency (Northern Ireland). We are grateful for the longstanding interest of the family who participated in the study and to the clinicians who recruited and cared for the family. Bioinformatics work was conducted on the Queen's University of Belfast High Performance Computing Dell cluster.
Published online: October 13, 2011
Footnotes
Supplemental Data include one figure and two tables and can be found with this article online at http://www.cell.com/AJHG/.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes Project, http://www.1000genomes.org
Burrows-Wheeler Aligner (BWA), http://bio-bwa.sourceforge.net
The Genome Analysis Toolkit (GATK), http://www.broadinstitute.org/gatk
Genomic Annotator Data Tables, http://www.broadinstitute.org/gsa/wiki/index.php/Genomic_Annotator_Data_Tables
Integrative Genomics Viewer (IGV), http://www.broadinstitute.org/software/igv
Mapping and Assembly with Qualities (MAQ), http://maq.sf.net
MicroCosm Targets, http://www.ebi.ac.uk/enright-srv/microcosm
MFold, http://mfold.rna.albany.edu
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
SAMtools, http://samtools.sourceforge.net
VCFtools, http://vcftools.sourceforge.net
Supplemental Data
References
- 1.Kennedy R.H., Bourne W.M., Dyer J.A. A 48-year clinical and epidemiologic study of keratoconus. Am. J. Ophthalmol. 1986;101:267–273. doi: 10.1016/0002-9394(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 2.Ghosheh F.R., Cremona F.A., Rapuano C.J., Cohen E.J., Ayres B.D., Hammersmith K.M., Raber I.M., Laibson P.R. Trends in penetrating keratoplasty in the United States 1980-2005. Int. Ophthalmol. 2008;28:147–153. doi: 10.1007/s10792-007-9177-z. [DOI] [PubMed] [Google Scholar]
- 3.Héon E., Greenberg A., Kopp K.K., Rootman D., Vincent A.L., Billingsley G., Priston M., Dorval K.M., Chow R.L., McInnes R.R., et al. VSX1: A gene for posterior polymorphous dystrophy and keratoconus. Hum. Mol. Genet. 2002;11:1029–1036. doi: 10.1093/hmg/11.9.1029. [DOI] [PubMed] [Google Scholar]
- 4.Dash D.P., George S., O'Prey D., Burns D., Nabili S., Donnelly U., Hughes A.E., Silvestri G., Jackson J., Frazer D., et al. Mutational screening of VSX1 in keratoconus patients from the European population. Eye (Lond.) 2010;24:1085–1092. doi: 10.1038/eye.2009.217. [DOI] [PubMed] [Google Scholar]
- 5.Kim W.J., Rabinowitz Y.S., Meisler D.M., Wilson S.E. Keratocyte apoptosis associated with keratoconus. Exp. Eye Res. 1999;69:475–481. doi: 10.1006/exer.1999.0719. [DOI] [PubMed] [Google Scholar]
- 6.Wilson S.E., Chaurasia S.S., Medeiros F.W. Apoptosis in the initiation, modulation and termination of the corneal wound healing response. Exp. Eye Res. 2007;85:305–311. doi: 10.1016/j.exer.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 8.Meola N., Gennarino V.A., Banfi S. microRNAs and genetic diseases. Pathogenetics. 2009;2:7. doi: 10.1186/1755-8417-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryan D.G., Oliveira-Fernandes M., Lavker R.M. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol. Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 10.Karali M., Peluso I., Gennarino V.A., Bilio M., Verde R., Lago G., Dollé P., Banfi S. miRNeye: A microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715. doi: 10.1186/1471-2164-11-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J., Ryan D.G., Getsios S., Oliveira-Fernandes M., Fatima A., Lavker R.M. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc. Natl. Acad. Sci. USA. 2008;105:19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes A.E., Dash D.P., Jackson A.J., Frazer D.G., Silvestri G. Familial keratoconus with cataract: Linkage to the long arm of chromosome 15 and exclusion of candidate genes. Invest. Ophthalmol. Vis. Sci. 2003;44:5063–5066. doi: 10.1167/iovs.03-0399. [DOI] [PubMed] [Google Scholar]
- 13.Dash D.P., Silvestri G., Hughes A.E. Fine mapping of the keratoconus with cataract locus on chromosome 15q and candidate gene analysis. Mol. Vis. 2006;12:499–505. [PubMed] [Google Scholar]
- 14.Ning Z., Cox A.J., Mullikin J.C. SSAHA: A fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robichon C., Varret M., Le Liepvre X., Lasnier F., Hajduch E., Ferré P., Dugail I. DnaJA4 is a SREBP-regulated chaperone involved in the cholesterol biosynthesis pathway. Biochim. Biophys. Acta. 2006;1761:1107–1113. doi: 10.1016/j.bbalip.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Meyron-Holtz E.G., Ghosh M.C., Rouault T.A. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science. 2004;306:2087–2090. doi: 10.1126/science.1103786. [DOI] [PubMed] [Google Scholar]
- 21.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright Lab. (2011). MicroCosm Targets Version 5, http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/.
- 23.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barad O., Meiri E., Avniel A., Aharonov R., Barzilai A., Bentwich I., Einav U., Gilad S., Hurban P., Karov Y., et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prasad N.K., Decker S.J. SH2-containing 5′-inositol phosphatase, SHIP2, regulates cytoskeleton organization and ligand-dependent down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 2005;280:13129–13136. doi: 10.1074/jbc.M410289200. [DOI] [PubMed] [Google Scholar]
- 27.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishida T., Saika S. In: Cornea. Krachmer J.H., editor. Mosby/Elsevier; St. Louis, Mo: 2011. Cornea and Sclera: Anatomy and Physiology; p. 17. [Google Scholar]
- 29.R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2009. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 30.Dai E., Boulton M.E. In: Ophthalmology. Yanoff M., Duker J.S., editors. Mosby/Elselvier; Edinburgh: 2009. Basic Science of the lens; p. 381. [Google Scholar]
- 31.Brémond-Gignac D., Bitoun P., Reis L.M., Copin H., Murray J.C., Semina E.V. Identification of dominant FOXE3 and PAX6 mutations in patients with congenital cataract and aniridia. Mol. Vis. 2010;16:1705–1711. [PMC free article] [PubMed] [Google Scholar]
- 32.Hysi P.G., Young T.L., Mackey D.A., Andrew T., Fernández-Medarde A., Solouki A.M., Hewitt A.W., Macgregor S., Vingerling J.R., Li Y.J., et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat. Genet. 2010;42:902–905. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mencía A., Modamio-Høybjør S., Redshaw N., Morín M., Mayo-Merino F., Olavarrieta L., Aguirre L.A., del Castillo I., Steel K.P., Dalmay T., et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


