Abstract
We present a systematic review of pleiotropy among SNPs and genes reported to show genome-wide association with common complex diseases and traits. We find abundant evidence of pleiotropy; 233 (16.9%) genes and 77 (4.6%) SNPs show pleiotropic effects. SNP pleiotropic status was associated with gene location (p = 0.024; pleiotropic SNPs more often exonic [14.5% versus 4.9% for nonpleiotropic, trait-associated SNPs] and less often intergenic [15.8% versus 23.6%]), “predicted transcript consequence” (p = 0.001; pleiotropic SNPs more often predicted to be structurally deleterious [5% versus 0.4%] but not more often in regulatory sequences), and certain disease classes. We develop a method to calculate the likelihood that pleiotropic links between traits occurred more often than expected and demonstrate that this approach can identify etiological links that are already known (such as between fetal hemoglobin and malaria risk) and those that are not yet established (e.g., between plasma campesterol levels and gallstones risk; and between immunoglobulin A and juvenile idiopathic arthritis). Examples of pleiotropy will accumulate over time, but it is already clear that pleiotropy is a common property of genes and SNPs associated with disease traits, and this will have implications for identification of molecular targets for drug development, future genetic risk-profiling, and classification of diseases.
Introduction
Pleiotropy occurs when one gene has an effect on multiple phenotypes. The molecular mechanisms of pleiotropy can be dichotomized into multiple molecular functions of a single gene product and multiple consequences of a single molecular function.1, 2, 3 Although pleiotropy in the genetic architecture of complex disease has been proposed,4 to date, evidence for its presence has not been systematically evaluated despite suggestions that this could be useful.5, 6 Previous assessments of a shared genetic basis between multiple phenotypes have been confined to restricted analyses in (1) specific traits, including immune-mediated diseases;7 seven diseases studied by the Wellcome Trust Case Control Consortium genome-wide association study (GWAS)8 including Crohn disease (IBD1 [MIM 266600]);9 blood pressure and selected hematological traits studied by the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium;10 and pancreatic cancer [MIM 260350];11 (2) single-trait GWASs;12, 13 and (3) a recent phenome-wide scan to discover gene-disease associations.14
The National Human Genome Research Institute's (NHGRI) Catalog of Published GWAS15 is a comprehensive resource listing statistically significant SNP-trait associations significant at p < 1 × 10−5 from all GWAS publications that attempt to assay at least 100,000 SNPs. If a study did not report a combined p value, the p value from the largest sample size was recorded in the NHGRI catalog if both discovery and replication samples show an association at p < 1 × 10−5. If a study did not include a replication stage, significant SNPs from the initial stage were recorded in the catalog. We considered that the stringent criteria employed for accepting associations in GWASs, including quality control standards, strict p value thresholds, and a requirement for replication, yielded robust evidence of association and so gave a secure basis from which to study evidence for pleiotropy among genes and common genetic variants.
In this review, by using the open-access NHGRI catalog, we aim to gain insight into the extent and pattern of pleiotropy in the genetics of common, complex disease to characterize pleiotropic genes and SNPs and to describe clusters of diseases and disease traits.
Our prior expectations were that pleiotropic genes would be more common in certain functional groups and pleiotropic SNPs more often located in an upstream or regulatory region than nonpleiotropic genes and SNPs. Furthermore, we anticipated that the network of phenotypes sharing association to common SNPs or genes might give clues to underlying common mechanisms, some of which might be unexpected and suggest hypotheses about shared molecular pathways.
Material and Methods
Identification of Common Variants Showing Association with Complex Phenotypes
Common genetic variants reported to be associated with complex non-Mendelian phenotypes were defined as those contained in the NHGRI catalog (last accessed February 4, 2011). Only SNP-trait associations reporting genome-wide significance (p < 5 × 10−8)16 were considered, and the total number that was included in the analysis was 1687 SNPs. In addition, we considered linkage disequilibrium (LD) blocks-trait association by calculating the LD between all the GWAS hits used in this analysis and collapsing those SNPs that were in high LD into single loci (threshold for high LD r2 > 0.80). Gene nomenclature was standardized by use of the Human Genome Organisation (HUGO) Gene Nomenclature Committee (HGNC) and Ensembl gene Identification numbers (IDs).17, 18, 19 For each SNP-trait association we accepted the NHGRI catalog record of which gene(s) and SNPs were considered by the study authors to mediate the association. This allowed an investigation of pleiotropy at both gene and variant level. Three additional comprehensive and systematic approaches (annotation based on LD, annotation based on a mapped gene feature of the NHGRI catalog and annotation based on taking all genes in the LD block) were adopted to evaluate to what extent the gene author-annotation approach might be subject to bias. In the first approach (denoted here as LD method) a gene was assigned to a SNP if the gene overlapped with that SNP. Otherwise the nearest gene in the same LD block was taken. The second approach (denoted here as NHGRI mapped gene method) employed this feature of the NHGRI catalog in which genes were mapped to the SNP (NCBI) showing the most significant association. If the SNP was located within a gene, that gene was listed. If the SNP was intergenic, both the upstream and downstream genes were listed. When using this method, we (1) restricted the analysis only to those SNPs that were located in a gene and (2) conducted a locus analysis for those SNPs that were intergenic, described the extent of pleiotropy for these loci and added the loci to the genes. The third approach (denoted here as taking all genes in the LD block annotation method) was based on the proposition that it is possible for any locus or gene within an LD block to be associated with the reported SNP and consequently to the phenotype.
Definition of Pleiotropy
We defined pleiotropy as a single gene or variant being associated with more than one distinct phenotypes (disease endpoints or quantitative traits) then systematically applied the following exclusion criteria in order to obtain a conservative estimate of pleiotropy that was not inflated by highly correlated trait and/or disease outcomes: (1) the phenotypes were (patho)-physiological counterparts (e.g., serum uric-acid level and gout), (2) one phenotype was a subset of the other (e.g., Crohn disease [MIM 266600] and inflammatory bowel disease [MIM 266600]), (3) one phenotype is used to calculate the other (e.g., low-density lipoprotein [LDL] cholesterol level and triglyceride levels20), (4) the phenotypes are similar or strongly correlated such that they might be measures of the same genetic effect (e.g., bone mineral density in the spine and hip, serum calcium and phosphorus, serum prostate specific antigen and prostate cancer [MIM 176807], pigmentation traits), (5) one phenotype is a known causal factor for the other (e.g., LDL cholesterol level and myocardial infarction [MIM 608446]).
We categorized genes or variants in the NHGRI catalog as pleiotropic or nonpleiotropic after application of the above definition and criteria. The status in terms of criteria (1–3) was typically self-evident but in terms of criteria (4) and (5) was occasionally uncertain. In these circumstances relationships between the phenotypes associated with the gene or SNP were explored in the current literature. Categorization then took place by consensus after discussion between authors (S.S., H.C., J.F.W.). We estimated the frequency of pleiotropy among the common complex phenotypes studied by GWASs by identifying the number of pleiotropic genes and SNPs and presenting this as a percentage of the overall number of genes and SNPs reported in the NHGRI catalog.
Pleiotropic genes were characterized by comparing the length of pleiotropic genes (in kilobases) to the length of nonpleiotropic genes with Ensembl. We conducted an analysis of biological processes that the genes were associated with (by using Gene Ontology [GO] terms21 via GOrilla22, 23) to investigate evidence for enrichment of biological processes among pleiotropic genes.
We characterized pleiotropic SNPs by their location and consequence for the transcript and compared them to nonpleiotropic SNPs by using standard Ensembl annotation and definitions. The categories for SNP location were intergenic, upstream, 5′ UTR, exon (comprising nonsynonymous coding, synonymous coding, frameshift coding, and STOP gained), intron (SNPs labeled as both intronic and NMDtranscript, intronic and splice site, or intronic and regulatory region are included here), 3′ UTR, downstream, and within a noncoding gene (i.e., within a gene that does not code for a protein). We employed the bioinformatic tools SIFT24 and PolyPhen25 to decide whether nonsynonymous (ns)SNPs were deleterious. These tools predict the nsSNPs likely to affect protein function. We categorized nsSNPs as deleterious if both SIFT and PolyPhen predicted them to be damaging or probably damaging.
We dichotomized the parameter consequence for the transcript into two broad categories: very likely to be structurally functional, which consisted of a STOP codon lost or gained, frameshift coding, deleterious nonsynonymous coding, or essential splice site variation and possibly regulatory, which consisted of SNPs located upstream in a 5′ or 3′ UTR, or in an intron or region annotated as regulatory by Ensembl.
In this report we will use the term nonpleiotropic genes or SNPs to denote the group of genes or SNPs in the NHGRI catalog that we have not defined as pleiotropic. In our descriptive analysis of pleiotropic genes and SNPs, we compared pleiotropic genes and SNPs in the NHGRI catalog to this group rather than to all genes and SNPs in the genome because we considered that differences with the latter might be confounded by selection of SNPs for the genotyping arrays or other genic characteristics that are related to detection of an association.
We considered, a priori, that certain disease classes would have higher levels of pleiotropy than others. Figure 1 suggests causes and implications of such overrepresentation. For example, a higher level of pleiotropy might be found in certain disease classes and in these circumstances all characteristics of the genes harboring variants associated with these diseases might appear to be associated with pleiotropy but this might be because of confounding. In this case a stratified analysis by disease category might help to interpret the observed associations with pleiotropy. We expected that variants and genes harboring variants associated with the cluster of immune-mediated phenotypes might form a sizeable proportion of all pleiotropic SNP and genes. We therefore decided to present overall findings and then findings after all immune-mediated phenotypes were coded as one single disease. We thus describe results for immune-mediated pleiotropic (IMP) genes/SNPs and nonimmune-mediated pleiotropic (NIMP) genes/SNPs. Phenotypes coded as immune mediated are presented in Table 1. Additionally, we conducted an analysis to compare the frequency of pleiotropy in three well-defined disease classes—immune-mediated disease, the metabolic syndrome (phenotypes included are presented in Table 1), and cancer.
Figure 1.
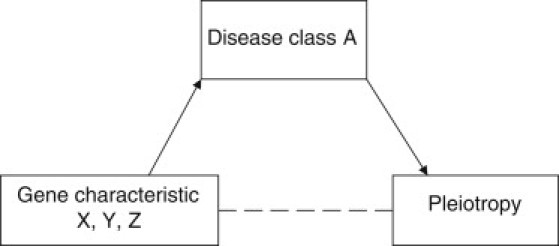
Interpretation of High Levels of Pleiotropy in Particular Disease Classes
If there was a high level of pleiotropy in disease class A, all characteristics of genes associated with disease class A would be associated with pleiotropy (including, but not specifically, the characteristic leading to overrepresentation of disease class A in the pleiotropy category). Therefore, some gene characteristics could be associated with pleiotropy as a result of confounding.
Table 1.
List of Immune-Mediated Diseases and Traits Phenotypes Included in the Metabolic Syndrome Disease Classa
| Immune-Mediated Diseases and Traits | Metabolic Syndrome |
|---|---|
| alopecia areata | adiponectin |
| ankylosing spondylitis | beta-cell function |
| asthma | blood pressure (systolic) |
| atopic dermatitis | blood pressure (diastolic) |
| Behcet's disease | body mass index |
| celiac disease | cholesterol, HDL |
| Crohn disease | cholesterol, LDL |
| eosinophilic esophagitis | cholesterol, total |
| inflammatory bowel disease | c-reactive protein |
| JIA | fasting insulin |
| multiple sclerosis | fasting plasma glucose |
| neonatal lupus | fibrinogen |
| primary biliary cirrhosis | glycated hemoglobin |
| primary sclerosing cholangitis | hypertension |
| psoriasis | hypertension (early onset) |
| psoriatic arthritis | hypertriglyceridaemia |
| rheumatoid arthritis | insulin resistance |
| systemic lupus erythematosus | interleukin 18 |
| systemic sclerosis | lipoprotein a |
| type 1 diabetes | obesity |
| ulcerative colitis | soluble leptin receptor |
| vitiligo | TNFa |
| cd4:cd8 ratio | triglycerides |
| cd8 lymphocyte | two-hour glucose challenge |
| eosinophil count | type 2 diabetes |
| immunoglobulin A | waist circumference |
| immunoglobulin E | waist-hip ratio |
| neutrophil count | weight |
| serum soluble e-selectin | |
| serum soluble p-selectin | |
| soluble ICAM-1 | |
| soluble il-6 receptor | |
| white blood cell count |
Based on International Diabetes Federation.75
Statistical Analysis
The Mann-Whitney U test was used to test for difference in the size (in kb) of pleiotropic and nonpleiotropic genes. To test the difference between pleiotropic and nonpleiotropic genes in terms of enrichment of biological processes using GOrilla, we calculated exact p values by using a hypergeometric model. The threshold α of significance was set to 9.7 × 10−6 after application of the Bonferroni correction for testing 5160 terms. To determine whether there was a statistically significant association between pleiotropic status and the location of the SNP or the consequence for the transcript, we calculated exact p values by using Fisher's exact test.
Probability of Gene Overlaps
We estimated whether the number of observed genetic overlaps between apparently nonrelated diseases or traits is statistically significantly different from that expected by chance. We have used two different methods.
The Independent Model. For each pair of diseases and/or traits A and B, we estimated the probability of having the observed number of overlaps. The chance that exactly i genes harboring variants associated with both traits is based on the probability of drawing i (shared) genes for trait A and the probability of drawing i (shared) genes for trait B. These probabilities are computed as the number of genes harboring variants associated with each trait (na and nb) divided by the total number n of the considered genes. n was equal to either (1) the number of genes in the NHGRI catalog (n = 1380) or (2) the overall number of genes (estimated as n = 20,000). See Supplemental Methods, available online, for an example illustration.
This approach is shown to illustrate the potential for mistakenly identifying correlations as statistically significant (because some publications have reported such correlations in specific disease areas). It employs the simplistic assumptions that (1) the genes act independently, (2) each gene has an equal chance of being associated with a trait, and (3) traits in the pair are unrelated. The approach does not compare the considered phenotype pair with other pairs. More importantly, it focuses only on the overlapping genes and ignores genetic mismatches (i.e., genes harboring variants are associated with only one trait from the pair). When applied to identifying clusters of similar diseases or disease traits, the method favors clusters of more polygenic diseases, which are characterized by greater numbers of the observed overlaps (but also by greater numbers of the observed mismatches).
Degree of Surprise. Our next approach is inspired by outlier detection methods proposed in astronomy in the middle of 19th century26, 27 and recently extended in machine learning.28, 29, 30, 31, 32, 33, 34 When applied to uncovering genetically similar diseases, such approaches aim to determine whether the number of matching genetic causes observed for a considered pair of traits is significantly larger than the number of genetic overlaps between any random pair. We have extended standard methods by assuming that the presence of mismatching genetic causes of two diseases is an indicator of their possible dissimilarity.
To illustrate this idea, consider five uniquely associated genes harboring variants associated with trait A and 10 uniquely associated genes harboring variants associated with trait B. Assume that the traits share two genes in common. In this example, there are two matching and 13 mismatching genes harboring variants associated with the pair {A,B}. In another example, a pair of traits {C,D} shares two genes, and there are no other genes harboring variants associated with either of these traits. The presence of the matching genes in pair {A,B} indicates their possible similarity; however, the pair is less similar than {C,D}, which has no genetic mismatches.
Our degree of surprise (DS) method identifies phenotype pairs with a high number of matching or simultaneously absent genes and a low number of genetic mismatches. The importance weights of the matching and mismatching genes are proportional to the surprise, that is inverse frequencies of their occurrences (see Supplemental Methods Equation 1). In contrast to the independent model, our DS approach does not assume that each gene has an equal chance of being associated with all the phenotypes. Also, the method does not rely on the restrictive assumption that all the traits are unrelated—instead, it makes a much weaker assumption that the genetically related traits are relatively infrequent. DS has a formal probabilistic interpretation and might be viewed as the likelihood ratio of the similarity and dissimilarity models, which allows for an easy accommodation of the noise (false positive and false negative genotype-phenotype associations). The empirical p values of the adjusted DS criterion were found to be useful for identifying both the existing and novel couplings between phenotypes.
In order to identify genetically similar traits we have also considered more conventional approaches (data not shown) based on correlations, cross-entropy, and Jensen's divergence.35 These methods are based on maximizing the degree of genetic overlap or minimizing the degree of genetic mismatch but not both criteria simultaneously. In contrast to the reported degree-of-surprise criterion, these methods were strongly biased to detect either highly polygenic or monogenic traits.
Results
Genes
HGNC and Ensembl IDs were ascertained for 1380 genes out of 1431 distinct genes (96%) contained in the NHGRI catalog. These 1380 genes had been assigned HGNC names and subsequently contributed to the analyses (unless otherwise specified).
Frequency of Pleiotropy
A total of 233 genes were defined as pleiotropic (16.9% of all genes in the catalog). Table 2 shows how the frequency of pleiotropy varies somewhat depending on method of gene assignment in GWASs. All approaches, however, show that pleiotropy is a common property with 13.2%–18.6% of all genes demonstrating pleiotropy as defined in this study. When immune-mediated phenotypes were classified as a single group, 189 genes remained pleiotropic.
Table 2.
Extent of Pleiotropy in Genes of the NHGRI Catalog
| Method | Author Annotation Methoda | LD Methodb | Method Based on NHGRI Mapped Genes(Only Genes)c | Method Based on the NHGRI Mapped Genes (Genes and Loci)d | Method Based on Taking All Genes in the LD Blocke |
|---|---|---|---|---|---|
| Pleiotropic (IMPf and NIMPg) | 233 (16.9%) | 138 (13.2%) | 108 (15.4%) | 166 (14.1%) | 473 (18.6%) |
| Not pleiotropic | 1147 (83.1%) | 909 (86.8%) | 592 (84.6%) | 1008 (85.9%) | 2064 (81.4%) |
| Total | 1380 | 1047 | 700 | 1174 | 2537 |
| Pleiotropic NIMPg | 189 (14.2%) | 101 (10%) | 85 (12.6%) | 127 (11.2%) | 189 (14.2%) |
| Not pleiotropic | 1147 (85.9%) | 909 (90%) | 592 (87.4%) | 1008 (88.8%) | 1147 (85.9%) |
| Total | 1336 | 1010 | 677 | 1135 | 1336 |
Author annotation method is the primary method adopted in this study where a gene was assigned to a SNP based on the decision of the authors of the original study.
For annotation based on LD, a gene was assigned to a SNP if the gene overlapped with that SNP, otherwise the nearest gene in the same LD block (based on HapMap CEU population) was taken.
Annotation based on a mapped gene feature of NHGRI catalog (gene only): genes are mapped to the SNP with the strongest association (NCBI); if the SNP is located within a gene, that gene is listed and if the SNP is intergenic, the upstream and downstream genes are both listed. When using this method we restricted the analysis only to those SNPs that were located in a gene.
Annotation based on a mapped gene feature of NHGRI catalog (gene and locus): we followed the same procedure as above, but for those SNPs that were intergenic we conducted a locus analysis, described the extent of pleiotropy for these loci and added the loci to the genes.
Annotation based ased on taking all genes in the LD block: This approach is based on the proposition that it is possible for any locus or gene within an LD block to be associated with the reported SNP, and consequently to the phenotype.
Genes associated with immune-mediated phenotypes only.
Genes associated with nonimmune-mediated phenotypes.
Characterizing Pleiotropic Genes
Size. The median size of pleiotropic genes was 45.7 kb, compared with a median of 38.7 kb for nonpleiotropic genes (p = 0.072; Mann-Whitney U test). When genes harboring variants associated only with immune-mediated phenotypes were excluded, the median size of pleiotropic genes was 49.2 kb, significantly different than the size of nonpleiotropic genes (p = 0.022). The median size of the pleiotropic genes harboring variants associated only with immune-mediated phenotypes was 32.0 kb. The larger size of pleiotropic compared to nonpleiotropic genes was confirmed when this analysis was repeated employing the three more systematic gene assignment approaches (see Material and Methods and Table S1).
Analysis of GO Terms. GOrilla recognized 1375 of the 1380 genes entered and 1316 were associated with GO terms (see also Figures S1–S4). Of the significantly enriched GO terms among pleiotropic genes (p < 9.7 × 10−6; hypergeometric methods) those most enriched were:
Lipid-Related Processes including macromolecular complex remodeling (out of 15 GWAS genes associated with this term, 11 were pleiotropic), protein-lipid complex remodeling (11 out of 15 were pleiotropic), plasma lipoprotein particle remodeling (11/15), protein-lipid complex subunit organization (11/16), plasma lipoprotein particle organization (11/16), triglyceride metabolic process (11/16), neutral lipid metabolic process (12/18), acylglycerol metabolic process (12/18), and glycerol ether metabolic process (12/19)
Immune system-Related Processes including interferon-gamma-mediated signaling pathway (17/26), cellular response to interferon-gamma (17/27), response to interferon gamma (18/30), regulation of immune effector process (19/39), response to lipopolysaccharide (18/37), and positive regulation of T cell activation (18/37)
We thought that GO terms related to transcription might have been enriched among pleiotropic genes; however, we did not find evidence to support this. The most enriched GO term relating to transcription was regulation of transcription, DNA-dependent, associated with 214 GWAS genes, 54 of which were pleiotropic (p = 7.78 × 10 −4, nonsignificant).
SNPs
Frequency of Pleiotropy
There were 1687 distinct SNPs in the NHGRI catalog, and all were included in the analysis. A total of 77 SNPs were defined as pleiotropic (4.6% of all SNPs in the catalog). Table 3 shows how the frequency of pleiotropy varies somewhat depending on method of SNP assignment in GWASs. Both approaches, however, show that pleiotropy is a relatively common property with 4.6%–7.8% of all SNPs demonstrating pleiotropy as defined in this study. When immune-mediated phenotypes were classified as a single group, 53 SNPs remained pleiotropic.
Table 3.
Extent of Pleiotropy in SNPs of the NHGRI Catalog
| Method | SNP Methoda | LD Block Methodb |
|---|---|---|
| Pleiotropic (IMPc and NIMPd) | 77 (4.6%) | 109 (7.8%) |
| Not pleiotropic | 1610 (95.4%) | 1297 (92.3%) |
| Total | 1687 | 1406 |
| Pleiotropic NIMPd | 53 (3.2%) | 78 (5.7%) |
| Not pleiotropic | 1610 (96.8%) | 1297 (94.3%) |
| Total | 1663 | 1375 |
For author annotation method, we described the extent of pleiotropy in SNPs by using exact matches of SNPs.
For the LD blocks, we calculated the LD between all the GWAS hits used in this analysis and collapsed those SNPs that were in high LD into single loci (threshold for high LD r2 > 0.80).
Genes associated with immune-mediated phenotypes only.
Genes associated with nonimmune-mediated phenotypes.
Characterizing Pleiotropic SNPs
We found a significant association between pleiotropic status and SNP location (p = 0.024; Fisher's exact test). Pleiotropic SNPs were more likely to be exonic than nonpleiotropic SNPs (14% of pleiotropic SNPs were exonic compared to 4.9% of nonpleiotropic SNPs) and were less likely to be intergenic (15.8% versus 23.6%) (Table 4).
Table 4.
Location of Pleiotropic and Nonpleiotropic SNPs
| Locationa |
Number of SNPs |
|||||
|---|---|---|---|---|---|---|
|
Pleiotropic |
Nonpleiotropic |
|||||
| IMP | NIMP | Overall | Overall Percentage | Number | Percentage | |
| Intergenicb | 3 | 9 | 12 | 15.8 | 380 | 23.6 |
| Upstreamc | 3 | 3 | 6 | 7.9 | 159 | 9.9 |
| 5′ UTRd | 0 | 0 | 0 | 0 | 3 | 0.2 |
| Exon | 3 | 8 | 11 | 14.5 | 79 | 4.9 |
| Intron | 10 | 17 | 27 | 35.5 | 612 | 38.0 |
| 3′ UTRe | 0 | 0 | 0 | 0 | 25 | 1.6 |
| Downstreamf | 5 | 7 | 12 | 15.8 | 151 | 9.4 |
| Within noncoding geneg | 0 | 8 | 8 | 10.5 | 200 | 12.4 |
| Total | 24 | 52 | 76 | 100 | 1609 | 100 |
Annotation provided by Ensembl.
More than 5 kb upstream or downstream of a transcript.
Within 5 kb upstream of the 5′ end of a transcript.
5′ untranslated region.
3′ untranslated region.
Within 5 kb downstream of the 3′ end of a transcript.
Within a gene that does not code for a protein.
We also found a significant association between pleiotropic status and SNP consequence (p = 0.001; Fisher's exact test) (Table 5). Pleiotropic SNPs were more likely to be structurally functional than nonpleiotropic SNPs (5% of pleiotropic SNPs were structurally functional compared to 0.4% of nonpleiotropic SNPs) and pleiotropic SNPs were less likely to be regulatory (42.5% versus 49%).
Table 5.
Consequence to the Transcript Affected by Pleiotropic and Nonpleiotropic SNPs
| Consequence for Transcripta |
Number of SNPs |
|||
|---|---|---|---|---|
| IMP | NIMP | Pleiotropic | Nonpleiotropic | |
| Structurally Functional | ||||
| Deleterious nonsynonymous codingb | 1 | 2 | 3 | 4 |
| Frameshift coding | – | – | – | 1 |
| Stop gained | – | 1 | 1 | 1 |
| Subtotal (%) | 1 | 3 | 4 (5.1%) | 6 (0.4%) |
| Potentially regulatory | ||||
| Upstream | 3 | 3 | 6 | 159 |
| 5′ UTR | – | – | – | 3 |
| Intronic | 10 | 17 | 27 | 612 |
| 3′ UTR | – | – | – | 25 |
| Annotated regulatory | – | 1 | 1 | 1 |
| Subtotal (%) | 13 | 21 | 34 (43.0%) | 800 (49.1%) |
| Other | ||||
| Intergenic, nonessential splice sitec, tolerated nonsynonymous coding, synonymous coding, downstream, within noncoding gene, NMD transcript | 10 | 31 | 41 (51.9%) | 824 (50.6%) |
| Totald | 24 | 55 | 79 | 1630 |
Annotation by Ensembl.
Nonsynonymous Coding SNPs classified as deleterious if predicted to be (probably) damaging by both SIFT and PolyPhen.
Essential splice site: In the first 2 or last 2 base pairs of an intron.
Nonessential splice site: 1–3 bps into an exon or 3–8 bps into an intron.
Twenty SNPs are present in more than one category.
Because the possibly regulatory category was dominated by intronic SNPs, which might not all be regulatory, we repeated the analysis, restricting the regulatory category to only those annotated as regulatory or located upstream or in the 5′ UTR. The association with pleiotropic status remained significant (p = 0.001; Fisher's exact test), with a higher proportion of nonpleiotropic than pleiotropic SNPs being regulatory.
When these analyses were repeated without HLA genes, which are among the most polymorphic in the genome, and also particularly enriched for coding variation, the patterns were similar.
Association of Disease Class with the Extent of Pleiotropy
We found that the extent of pleiotropy is associated with certain disease classes (Table 6). The proportion of pleiotropic genes was significantly increased in all three disease classes studied for both genes and SNPs (aside from cancer SNPs) (Fisher's exact test); 37.7% of genes that harbored variants associated with immune-mediated phenotypes, 34.8% of genes that harbored variants associated with cancer, and 28.5% of genes that harbored variants associated with the metabolic syndrome were pleiotropic, compared to 16.9% of all GWAS genes. In addition, 8.3% of SNPs associated with immune-mediated phenotypes, 4.8% of SNPs associated with cancer, and 8.4% of SNPs associated with the metabolic syndrome were pleiotropic, compared to 4.6% of all GWAS SNPs.
Table 6.
Extent of Pleiotropy in Different Disease Classes
| Disease Class |
Genes |
SNPs |
||||
|---|---|---|---|---|---|---|
| Pleiotropic (%) | Nonpleiotropic (%) | p Valuea | Pleiotropic (%) | Nonpleiotropic (%) | p Valuea | |
| All (comparison group) | 233 (16.9) | 1147 (83.1) | – | 77 (4.6) | 1610 (95.4) | – |
| Immune-mediated phenotypes | 106 (37.7) | 175 (62.3) | <0.0001 | 31 (8.3) | 343 (91.7) | 0.0066 |
| Cancer | 49 (34.8) | 92 (65.2) | <0.0001 | 8 (4.8) | 158 (95.2) | 0.8456 |
| Metabolic syndrome | 79 (28.5) | 198 (71.5) | <0.0001 | 30 (8.4) | 327 (91.6) | 0.0056 |
Fisher's exact test p value.
Clusters of Phenotypes Linked by Pleiotropic Genes
We present several figures illustrating constellations of diseases and disease traits linked by pleiotropic connections (Figures S5–S14). An example is given in Figure 2 showing abundant pleiotropic links for Crohn disease; some of these links, as with leprosy (LPRS1 [MIM 609888]), are unexplained and might be worth further consideration. For a sample of these links we have assessed the likelihood that the patterns seen are due to chance. Results of three different approaches are presented in Table 7, Table 8. In Table 7 we present several examples selected because the links between certain correlations looked interesting based on an independent model, such as between schizophrenia (SCZD [MIM 181500]) and iron status or plasma urate, PSA and several cancers, SLE and blood cell parameters, pigmentation traits and nonmelanoma cancers, and immune-mediated disease and infections or cancer, but in which further analysis by the DS approach showed that these were likely to be represent chance findings.
Figure 2.
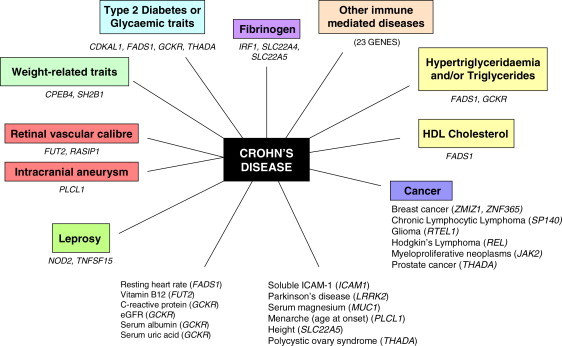
Genes Harboring Variants Associated with Crohn Disease and Other Phenotypes
Genes harboring variants associated with other immune-mediated diseases: variants in BACH2, type 1 diabetes and celiac disease; variants in C11orf30 – atopic dermatitis; variants in CCR6 – RA and vitiligo; variants in DENND1B, asthma; variants in ICOSLG, celiac disease; variants in IL10, type 1 diabetes, ulcerative colitis, and Behcet disease; variants in IL12B, psoriasis and psoriatic arthritis; variants in IL18R1, asthma and celiac disease; variants in IL18RAP, celiac disease; variants in IL1RL1, eosinophil count and celiac disease; variants in IL23R, inflammatory bowel disease, psoriasis, ulcerative colitis, ankylosing spondylitis, and Behcet disease; variants in IL27, early-onset inflammatory bowel disease and type 1 diabetes; variants in IL2RA, multiple sclerosis, rheumatoid arthritis, type 1 diabetes, vitiligo, and alopecia areata; variants in MST1, ulcerative colitis and primary sclerosing cholangitis; variants in ORMDL3, asthma, primary biliary cirrhosis, type 1 diabetes, ulcerative colitis, and white blood cell count; variants in PTPN2, type 1 diabetes and celiac disease; variants in PTPN22, rheumatoid arthritis and type 1 diabetes; variants in REL, Hodgkin lymphoma, celiac disease, psoriasis, rheumatoid arthritis, and ulcerative colitis; variants in SMAD3, asthma; variants in STAT3, multiple sclerosis; variants in TAGAP, celiac disease; variants in TYK2, psoriasis and type 1 diabetes; variants in ZMIZ1, breast cancer, celiac disease, and inflammatory bowel disease.
Table 7.
Likelihood of Observed Gene Sharing between Selected Pairs of Different Traits and a Comparison of Three Modeling Approaches for Selected Trait Pairs
| Trait | Number of Genes Associated to Trait | Other Trait | Number of Genes Associated to Other Trait | Number of Genes Shared between Traits |
p Values |
||
|---|---|---|---|---|---|---|---|
|
Independent Model |
Degree of Surprise, Accounting for Dissimilaritiesa | ||||||
| Estimated Total Number of Genes, n = 20,000 | Genes Present in the NHGRI Catalog, n = 1380 | ||||||
| Schizophrenia | 16 | iron markers | 7 | 2 | 1.60 × 10−5 | 0.003 | 0.76 |
| Schizophrenia | 16 | uric acid and gout | 16 | 2 | 8.10 × 10−5 | 0.0143 | 0.85 |
| Cancer-related traits | 154 | PSA | 6 | 4 | 1.80 × 10−7 | 0.0043 | 0.98 |
| Cancer-related traits | 154 | pigmentation traits | 19 | 5 | 4.90 × 10−7 | 0.0428 | 0.98 |
| Cancer-related traits | 154 | immune traits | 265 | 19 | 1.00 × 10−12 | 0.0101 | 1 |
| Infections | 29 | immune traits | 265 | 5 | 5.10 × 10−5 | 0.1706 | 0.99 |
| SLE | 26 | corpuscular volume | 29 | 3 | 8.70 × 10−6 | 0.0157 | 0.94 |
Criterion is corrected for the noise in the data, namely the possibility of false positives and false negatives (see Supplemental Data for details). To ensure a fair comparison between the DS approach and the independent model, we assumed that the probabilities of genetic matches were identical for all genes in the GWAS corpus; we also replaced the random sampling of pairs of diseases and/or traits by an exhaustive averaging over all combinations.
Table 8.
Likelihood of Observed Gene Sharing between Pairs of Disease Traits: Ranked by p Value from Degree of Surprise, Accounting for Dissimilarity Approach
| Trait | Number of Genes Associated to Trait | Other Trait(s) | Number of Genes Associated to Other Trait | Genes Shared between Traits |
p Values |
||
|---|---|---|---|---|---|---|---|
|
Independent Model |
Degree of Surprise, Accounting for Dissimilaritiesa | ||||||
| Estimated Total Number of Genes, n = 20,000 | Genes Present in the NHGRI Catalog, n = 1380 | ||||||
| Fetal hemoglobin | 2 | malaria | 1 | HBB | 0.0001 | 0.0014 | 0.0009 |
| Gallstones | 1 | serum campesterol | 2 | ABCG8 | 0.0001 | 0.0014 | 0.0008 |
| Immunoglobin A | 2 | JIA | 1 | HLA-DRB1 | 0.0001 | 0.0014 | 0.0008 |
| Knee osteoarthritis | 3 | narcolepsy | 1 | HLA-DQA2 | 0.0001 | 0.0022 | 0.0009 |
| Venous thromboembolism | 1 | angiotensin-converting enzyme activity | 2 | ABO | 0.0001 | 0.0014 | 0.0011 |
| serum campesterol | 2 | ABO | 0.0001 | 0.0014 | 0.0008 | ||
| serum soluble e-selectin | 1 | ABO | 5.0 × 10−5 | 0.0007 | 0.0004 | ||
| serum soluble P-selectin | 2 | ABO | 0.0001 | 0.0014 | 0.001 | ||
| ICAM-1 | 2 | ABO | 0.0001 | 0.0014 | 0.0011 | ||
| TNFa | 1 | ABO | 5.0 × 10−5 | 0.0007 | 0.0004 | ||
Criterion is corrected for the noise in the data, namely the possibility of false positives and false negatives (see Supplemental Data for details). To ensure a fair comparison between the DS approach and the independent model, we assumed that the probabilities of genetic matches were identical for all genes in the GWAS corpus; we also replaced the random sampling of pairs of diseases and/or traits by an exhaustive averaging over all combinations.
DS measures for selected traits that share genes are shown in Table 8. In the first example, one (HBB [MIM 141900]) out of two genes harboring variants associated with fetal hemoglobin and F cell distribution is also associated with malaria (MIM 611162) (DS value = 0.001) (rs4910742 is associated with levels of fetal hemoglobin and rs11036238 is associated with increased risk of malaria) thus confirming a known biological relationship (as discussed below). In the second example, one (ABCG8 [MIM 605460]) out of two genes harboring variants associated with serum campesterol is also associated with gallstones (GBD1 [MIM 600803]) (DS value = 0.001) (rs41360247 and rs4245791 are associated with levels of campesterol and rs11887534 is associated with increased risk of gallstones). In the third example, one (HLA-DRB1) out of two genes harboring variants associated with immunoglobin A is also associated with juvenile idiopathic arthritis (JIA) (DS value = 0.001) (rs2395148 is associated with increased risk of idiopathic JIA [MIM 604302] and rs2187668 and rs9271366 are associated with levels of immunoglobulin A). These latter two examples generate hypotheses regarding disease etiology. The biological plausibility of the findings is discussed in the following section.
Discussion
Pleiotropy in the Genetics of Complex Disease
With the recent discovery of many common genetic variants influencing disease and disease traits, there is an increasing interest in the detection of pleiotropy.7, 8, 9, 10, 11, 12, 13 We have performed a systematic evaluation of all genetic variants from all GWAS studies published up to January 4, 2011, and associated with more than one different phenotype. Pierce and Ahsan11 describe a pleiotropy scan and illustrate this in the detection of SNPs associated with pancreatic cancer, and other groups have proposed a phenome scan36, 37 that aims to measure all phenotypes—the phenome—related to specified genetic variants (or environmental exposures).
Frequency of Pleiotropy
There is no consensus on the extent of pleiotropy in the human genome.38 It has been proposed that pleiotropy is universal,39 modular,40 or infrequent.41, 42, 43 In this review, we show that pleiotropy is found to be a property of ∼17% of genes and ∼5% of SNPs known to be associated with diseases or disease traits. Analyses adopting three comprehensive systematic approaches confirmed that pleiotropy is common and associated with 13.2%–18.6% of genes and 4.6%–7.8% SNPs and support the conclusion that this substantial level of pleiotropy is not falsely inflated because of an author-annotation bias.
These all are likely to be minimum estimates given the conservative definitions used in this report (in an attempt to avoid inflation of estimates because of highly correlated traits and/or diseases) and because of the rapid rise in reported associations and the large number of common and rare variant associations yet to be found.44
Pleiotropic Genes
We show that pleiotropic genes are longer than nonpleiotropic genes (this is consistent across all analytic methods employed—see Table S1), an effect that might be mediated by two interacting effects. First, longer genes might encode an increased number of protein structural domains, which might in turn give rise to multiple functions; second, longer genes tend to contain more variants with a concomitant rise in the opportunity for some to be involved in different functions. The GO terms significantly enriched among pleiotropic genes reflect the fact that immune-mediated diseases and metabolic syndrome-related traits are overrepresented among pleiotropic gene associations. The GO terms highlighted are either the biological processes most amenable to pleiotropic effects or are due to confounding, and another property of genes in these disease classes causes their pleiotropy (Figure 1).
Pleiotropic SNPs
We show that pleiotropic SNPs are more likely to be exonic and structurally functional than nonpleiotropic SNPs. Our data does not support our a priori hypothesis that pleiotropic SNPs would be more likely to be present in regulatory regions that nonpleiotropic SNPs. We repeated these analyses separately for immune-mediated phenotypes because of concerns about possible confounding (see Figure 1) but the findings were unchanged.
Clusters of Phenotypes Defined by Pleiotropic Links
By systematically evaluating all gene-trait and SNP-trait associations in the NHGRI catalog and observing the various constellations of diseases and traits that arose, it is possible to contribute evidence to the understanding of disease etiology and pathogenesis by (1) suggesting hypotheses and adding evidence to hypotheses for which there is currently a paucity of data and (2) adding evidence to established hypotheses surrounding the etiology of disease. We note that if a phenotype pair is, after further research, shown to have a causal relationship, then the gene associated with both these phenotypes should no longer strictly be considered as pleiotropic, because this would now be an example of variation in one phenotype causing variation in the second phenotype. However, until etiological hypotheses are shown to be correct and a causal relationship accepted, we will describe genes harboring variants associated with two correlated phenotypes as pleiotropic. We present examples of constellations of diseases and traits in Figures S5–S14.
We demonstrate that calculation of simple probabilities (by using the independent model) would wrongly assign many pleiotropic associations as statistically significant. For example, a relationship between schizophrenia and serum iron markers45, 46, 47 and between schizophrenia and serum uric acid levels48, 49 have been suggested. A simple probability calculation (Table 7) suggests that these pleiotropic associations are unlikely to be due to chance and might suggest further research on this topic. However, our more stringent DS criterion, which takes into account (1) only genes harboring variants associated with one or more traits to date (i.e., not all of the more than 20,000 genes), (2) the number of mismatching genes as the marker of dissimilarity between traits (and therefore our criterion does not overestimate similarities between polygenic traits as other common approaches do), (3) false negatives (true gene-trait associations that did not reach a genome-wide significance level), and (4) false positives (although unlikely, given the stringent p values used in GWAS) and (5) accommodates the binary nature of the present/absent genetic overlaps, clearly shows this gene sharing as likely to be due to chance.
We have listed in Table 8 the top ranked correlations between diseases and/or traits with this approach. The top ranked correlation is that between fetal hemoglobin and malaria risk, which confirms a known biological relationship. The protective properties of high concentrations of fetal hemoglobin in erythrocytes during the first few months of life have been known since the 1970s to offer resistance to infection with Plasmodium falciparum,50 and the physiological mechanisms have been described.51 This demonstrates the potential of this method of studying similarity of genetic causes between two traits to correctly identify shared disease mechanisms and motivate functional research.
The second ranked finding was the ABCG8 pleiotropic association between serum campesterol and gallstones risk. Gallstones often have a high content of cholesterol, and cholesterol oversaturation is one of the main risk factors in gallstone etiology.52, 53 Cholesterol absorption and secretion into bile is affected by phytosterols (including campesterol) but also by genetic variants in ABCG8.54, 55 Thus, it is biologically plausible that serum campesterol and gallstones share a common pathway and this merits further study.
Our approach also identified the HLA-DRB1 pleiotropic associations with JIA and immunoglobulin A (IgA) as very unlikely to be due to chance. High IgA levels have been reported in a patient with JIA.56 Gilliam et al.57 have shown that JIA patients with joint erosions and joint-space narrowing had significantly elevated levels of IgA. The etiology of JIA is unclear, but because of similarity in genetic influences on IgA and JIA, it is possible that these traits share a common pathophysiological pathway and our findings support further investigation of the involvement of IgA in JIA etiology.
The biological significance of the potentially significant overlap between the pathophysiological pathways of narcolepsy and osteoarthritis is less clear. Osteoarthritis is a chronic degenerative disorder related to aging, but studies have also shown a strong genetic component. However, pathophysiological mechanisms have not been clarified.58 Narcolepsy is caused by the deficiency of hypocretin.59 An autoimmune etiology has been suggested because of the very strong association with the HLA subtype DQB1∗0602.60, 61 The same rationale might suggest an autoimmune component to osteoarthritis etiology. However, at this point the basis for the shared genetic influences in these conditions is not clear.
We identified a significant genetic sharing (through ABO [MIM 110300]) between angiotensin-converting enzyme activity, venous thromboembolism [MIM 188050] and serum campesterol, and between TNFa and soluble adhesion molecules E-selectin, P-selectin, and ICAM-1. Many of these associations have been previously reported in the literature,62, 63, 64, 65, 66 and our findings suggest that these merit further study.
Implications of Findings
It is likely that there was a degree of misclassification of genes and/or SNPs into pleiotropic and nonpleiotropic categories. We attempted to limit this by adopting several approaches to gene and SNP assignment including systematic approaches that should not be affected by observer bias that could have been present in author-annotated genes (for example, a tendency to assign associations to larger genes or to genes for whom the function is known). Furthermore, we defined and systematically employed a series of exclusion criteria in order to obtain a conservative definition of pleiotropy and to attempt to limit the degree of subjectivity in the definition of pleiotropy (for example to avoid the recognized upward bias in pleiotropy estimates that occurs if trait correlations are not taken into account as in the exclusion criteria in this study). The majority of available data at present relate to Northern American and European white populations, and so it is not possible to assess whether these findings might vary in other ethnic groups.
As Wagner and Zhang67 have noted, we can only assess pleiotropy in the context of which characters have been already been studied. Therefore labeling a gene or SNP as nonpleiotropic is always subject to further research. In this study we have investigated the findings for three illustrative disease groups for which there are sufficient data. In future it might be possible to extend this analysis more generally to other disease groups. We studied and reported these separately because we considered that certain disease classes might exhibit a higher frequency of pleiotropy. It is possible that this finding could simply be due to an artifact because these diseases have been explored by GWASs in greater depth. When all disease classes have been thoroughly examined, apparent differences in the frequency of pleiotropy might not appear so pronounced. We have accounted for a small probability of false positive and false negative GWAS associations in the adjusted degree-of-surprise method. However, mislabeling a gene as pleiotropic could have occurred because of reporting bias. Finally, in many cases only markers in linkage disequilibrium with the true causal variant are recorded in the NHGRI catalog, rather than the true causal variant itself.
Notwithstanding these limitations, our results suggest that pleiotropy is common in the genetics of complex disease, and 16.9% of genes and 4.6% of SNPs recorded in the NHGRI catalog are defined as pleiotropic. Additionally, we have characterized pleiotropic genes as being larger than nonpleiotropic genes and pleiotropic SNPs as more likely to be structurally functional and located exonically and downstream than nonpleiotropic SNPs.
The impact of pleiotropy on genetic testing for common, complex diseases in clinical and research settings has been recognized and described with APOE [MIM 107741] as an exemplar68, 69, 70 but has perhaps not received sufficient emphasis. Companies offering direct-to-consumer testing and consumers taking this up should be aware that any variants about which they gain information could, in the future, be found to be associated with additional (potentially more stigmatizing or untreatable) diseases. An understanding of pleiotropic effects is also of key importance for drug development. For example, although statins inhibit HMG-CoA reductase, they also have multiple other molecular actions with effects beyond cholesterol reduction,71 and this has been proposed as the basis for their efficacy in the reduction of cardiovascular outcomes.72, 73 Similarly, selective serotonin reuptake inhibitors have been found to be effective across different psychiatric disorders because they act on a pathway common to these differing disorders.36 When a gene has been shown to have opposing effects on different common diseases (as has been recently described)74 then this is likely to greatly complicate drug development and marketing, although knowledge of pleiotropic associations could help to predict side effects. These issues are likely to gain in importance as the full extent of pleiotropy in the genome becomes apparent.
More generally, gaining insight into the level of genetic connectivity between different diseases and disease traits gives the opportunity to deduce whether our current classification and categorization of diseases is valid genetically or whether genetic similarities traverse current divisions. This insight might be particularly useful when considering diseases that have a clinically derived diagnosis and lack validated diagnostic tests (such as a number of psychiatric disorders).
Acknowledgments
E.T. is funded by Cancer Research UK Fellowship C31250/A10107.
Published online: November 10, 2011
Footnotes
Supplemental Data include Supplemental Methods, 14 figures, and one tables and can be found with this article online at http://www.cell.com/AJHG/.
Web Resources
The URLs for data presented herein are as follows:
National Human Genome Research Institute, Catalog of Genome-Wide Association Studies, http://www.genome.gov/gwastudies/
Online Mendelian Inheritance in Man, http://www.omim.org
Supplemental Data
References
- 1.Grüneberg H. An analysis of the “pleiotropic” effects of a new lethal mutation in the rat (Mus norvegicus) Proc. R. Soc. Lond. B Biol. Sci. 1938;125:123–144. [Google Scholar]
- 2.Dudley A.M., Janse D.M., Tanay A., Shamir R., Church G.M. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100004. 2005, 0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He X., Zhang J. Toward a molecular understanding of pleiotropy. Genetics. 2006;173:1885–1891. doi: 10.1534/genetics.106.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker K.G. The common variants/multiple disease hypothesis of common complex genetic disorders. Med. Hypotheses. 2004;62:309–317. doi: 10.1016/S0306-9877(03)00332-3. [DOI] [PubMed] [Google Scholar]
- 5.Hudson M., Rojas-Villarraga A., Coral-Alvarado P., López-Guzmán S., Mantilla R.D., Chalem P., Baron M., Anaya J.M., Canadian Scleroderma Research Group. Colombian Scleroderma Research Group Polyautoimmunity and familial autoimmunity in systemic sclerosis. J. Autoimmun. 2008;31:156–159. doi: 10.1016/j.jaut.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Karasik D., Kiel D.P. Evidence for pleiotropic factors in genetics of the musculoskeletal system. Bone. 2010;46:1226–1237. doi: 10.1016/j.bone.2010.01.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhernakova A., van Diemen C.C., Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat. Rev. Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 8.Huang W., Wang P., Liu Z., Zhang L. Identifying disease associations via genome-wide association studies. BMC Bioinformatics. 2009;10(Suppl 1):S68. doi: 10.1186/1471-2105-10-S1-S68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lees C.W., Barrett J.C., Parkes M., Satsangi J. New IBD genetics: Common pathways with other diseases. Gut. 2011 doi: 10.1136/gut.2009.199679. in press. Published online February 7, 2011. [DOI] [PubMed] [Google Scholar]
- 10.Huang J., Johnson A.D., O'Donnell C.J. PRIMe: A method for characterization and evaluation of pleiotropic regions from multiple genome-wide association studies. Bioinformatics. 2011;27:1201–1206. doi: 10.1093/bioinformatics/btr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pierce B.L., Ahsan H. Genome-wide “pleiotropy scan” identifies HNF1A region as a novel pancreatic cancer susceptibility locus. Cancer Res. 2011;71:4352–4358. doi: 10.1158/0008-5472.CAN-11-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada Y., Takahashi A., Ohmiya H., Kumasaka N., Kamatani Y., Hosono N., Tsunoda T., Matsuda K., Tanaka T., Kubo M., et al. Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum. Mol. Genet. 2011;20:1224–1231. doi: 10.1093/hmg/ddq551. [DOI] [PubMed] [Google Scholar]
- 13.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. MAGIC investigators. GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: Demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hindorff, L.A., Junkins, H.A., Hall, P.N., Mehta, J.P., and Manolio, T.A. (2011). A Catalog of Published Genome-Wide Association Studies. www.genome.gov/gwastudies.
- 16.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 17.Seal R.L., Gordon S.M., Lush M.J., Wright M.W., Bruford E.A. genenames.org: The HGNC resources in 2011. Nucleic Acids Res. 2011;39(Database issue):D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flicek P., Amode M.R., Barrell D., Beal K., Brent S., Chen Y., Clapham P., Coates G., Fairley S., Fitzgerald S., et al. Ensembl 2011. Nucleic Acids Res. 2011;39(Database issue):D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.HUGO Gene Nomenclature Committee (2011). HGNC Database. http://www.genenames.org/.
- 20.Friedewald W.T., Levy R.I., Fredrickson D.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 21.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eden E., Lipson D., Yogev S., Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peirce B. Criterion for the Rejection of Doubtful Observations. Astron. J. 1852;II:161–163. [Google Scholar]
- 27.Peirce B. On Peirce's criterion. Proc. Am. Acad. Arts Sci. 1877;13:348–351. [Google Scholar]
- 28.Bishop C.M. Novelty detection and Neural Network validation. Proceedings of the IEE Conference on Vision, Image and Signal Processing. 1994;141:217–222. [Google Scholar]
- 29.Chandola V., Banerjee A., Kumar V. Anomaly detection: A survey. ACM Comput. Surv. 2009;41 doi: 10.1145/1541880.1541882. [DOI] [Google Scholar]
- 30.Gao J., Cheng H., Tan P.-N. Semi-supervised outlier detection. Proceedings of the SAC ‘06 ACM Symposium on Applied Computing. 2006 doi: 10.1145/1141277.1141421. [DOI] [Google Scholar]
- 31.Markou M., Singh S. Novelty detection: A review, part 1: Statistical approaches. Signal Processing. 2003;83:2481–2497. [Google Scholar]
- 32.McEliece R.J. Addison-Wesley; Cambridge: 1977. The Theory of Information and Coding. [Google Scholar]
- 33.Rousseeuw P., Leroy A. Third Edition. John Wiley & Sons; Hoboken, NJ: 1996. Robust Regression and Outlier Detection. [Google Scholar]
- 34.Tarassenko, L. (1995). Novelty detection for the identification of masses in mammograms. Proceedings of the 4th IEE International Conference on Artificial Neural Networks. 4, 442–447.
- 35.Bishop C.M. Springer; New York: 2006. Pattern Recognition and Machine Learning. [Google Scholar]
- 36.Freimer N., Sabatti C. The human phenome project. Nat. Genet. 2003;34:15–21. doi: 10.1038/ng0503-15. [DOI] [PubMed] [Google Scholar]
- 37.Jones R., Pembrey M., Golding J., Herrick D. The search for genenotype/phenotype associations and the phenome scan. Paediatr. Perinat. Epidemiol. 2005;19:264–275. doi: 10.1111/j.1365-3016.2005.00664.x. [DOI] [PubMed] [Google Scholar]
- 38.Stearns F.W. One hundred years of pleiotropy: A retrospective. Genetics. 2010;186:767–773. doi: 10.1534/genetics.110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright S. Volume 1. University of Chicago Press; Chicago: 1968. (Evolution and the genetics of populations). [Google Scholar]
- 40.Welch J.J., Waxman D. Modularity and the cost of complexity. Evolution. 2003;57:1723–1734. doi: 10.1111/j.0014-3820.2003.tb00581.x. [DOI] [PubMed] [Google Scholar]
- 41.Albert A.Y., Sawaya S., Vines T.H., Knecht A.K., Miller C.T., Summers B.R., Balabhadra S., Kingsley D.M., Schluter D. The genetics of adaptive shape shift in stickleback: Pleiotropy and effect size. Evolution. 2008;62:76–85. doi: 10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- 42.Brem R.B., Yvert G., Clinton R., Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 43.Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Insel B.J., Schaefer C.A., McKeague I.W., Susser E.S., Brown A.S. Maternal iron deficiency and the risk of schizophrenia in offspring. Arch. Gen. Psychiatry. 2008;65:1136–1144. doi: 10.1001/archpsyc.65.10.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGrath J., Brown A., St Clair D. Prevention and schizophrenia—the role of dietary factors. Schizophr. Bull. 2011;37:272–283. doi: 10.1093/schbul/sbq121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sørensen H.J., Nielsen P.R., Pedersen C.B., Mortensen P.B. Association between prepartum maternal iron deficiency and offspring risk of schizophrenia: Population-based cohort study with linkage of Danish national registers. Schizophr. Bull. 2011;37:982–987. doi: 10.1093/schbul/sbp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buie L.W., Oertel M.D., Cala S.O. Allopurinol as adjuvant therapy in poorly responsive or treatment refractory schizophrenia. Ann. Pharmacother. 2006;40:2200–2204. doi: 10.1345/aph.1H222. [DOI] [PubMed] [Google Scholar]
- 49.Dickerson F.B., Stallings C.R., Origoni A.E., Sullens A., Khushalani S., Sandson N., Yolken R.H. A double-blind trial of adjunctive allopurinol for schizophrenia. Schizophr. Res. 2009;109:66–69. doi: 10.1016/j.schres.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 50.Pasvol G., Weatherall D.J., Wilson R.J., Smith D.H., Gilles H.M. Fetal haemoglobin and malaria. Lancet. 1976;1:1269–1272. doi: 10.1016/s0140-6736(76)91738-4. [DOI] [PubMed] [Google Scholar]
- 51.Shear H.L., Grinberg L., Gilman J., Fabry M.E., Stamatoyannopoulos G., Goldberg D.E., Nagel R.L. Transgenic mice expressing human fetal globin are protected from malaria by a novel mechanism. Blood. 1998;92:2520–2526. [PubMed] [Google Scholar]
- 52.Koivusalo A.I., Pakarinen M.P., Sittiwet C., Gylling H., Miettinen T.A., Miettinen T.E., Nissinen M.J. Cholesterol, non-cholesterol sterols and bile acids in paediatric gallstones. Dig. Liver Dis. 2010;42:61–66. doi: 10.1016/j.dld.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Marschall H.U., Einarsson C. Gallstone disease. J. Intern. Med. 2007;261:529–542. doi: 10.1111/j.1365-2796.2007.01783.x. [DOI] [PubMed] [Google Scholar]
- 54.Jakulj L., Vissers M.N., Tanck M.W., Hutten B.A., Stellaard F., Kastelein J.J., Dallinga-Thie G.M. ABCG5/G8 polymorphisms and markers of cholesterol metabolism: Systematic review and meta-analysis. J. Lipid Res. 2010;51:3016–3023. doi: 10.1194/jlr.M008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sudhop T., Sahin Y., Lindenthal B., Hahn C., Lüers C., Berthold H.K., von Bergmann K. Comparison of the hepatic clearances of campesterol, sitosterol, and cholesterol in healthy subjects suggests that efflux transporters controlling intestinal sterol absorption also regulate biliary secretion. Gut. 2002;51:860–863. doi: 10.1136/gut.51.6.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yildiz B., Kural N. IgG1 deficiency and high IgA level with juvenile idiopathic arthritis. Eur. J. Pediatr. 2007;166:1179–1180. doi: 10.1007/s00431-006-0360-4. [DOI] [PubMed] [Google Scholar]
- 57.Gilliam B.E., Chauhan A.K., Low J.M., Moore T.L. Measurement of biomarkers in juvenile idiopathic arthritis patients and their significant association with disease severity: A comparative study. Clin. Exp. Rheumatol. 2008;26:492–497. [PubMed] [Google Scholar]
- 58.Valdes A.M., Spector T.D. The contribution of genes to osteoarthritis. Rheum. Dis. Clin. North Am. 2008;34:581–603. doi: 10.1016/j.rdc.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 59.De la Herrán-Arita A.K., Guerra-Crespo M., Drucker-Colín R. Narcolepsy and orexins: An example of progress in sleep research. Front Neurol. 2011;2:26. doi: 10.3389/fneur.2011.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hallmayer J., Faraco J., Lin L., Hesselson S., Winkelmann J., Kawashima M., Mayer G., Plazzi G., Nevsimalova S., Bourgin P., et al. Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat. Genet. 2009;41:708–711. doi: 10.1038/ng.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hor H., Kutalik Z., Dauvilliers Y., Valsesia A., Lammers G.J., Donjacour C.E., Iranzo A., Santamaria J., Peraita Adrados R., Vicario J.L., et al. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nat. Genet. 2010;42:786–789. doi: 10.1038/ng.647. [DOI] [PubMed] [Google Scholar]
- 62.Blann A.D., Nadar S.K., Lip G.Y. The adhesion molecule P-selectin and cardiovascular disease. Eur. Heart J. 2003;24:2166–2179. doi: 10.1016/j.ehj.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 63.Dandona P., Dhindsa S., Ghanim H., Chaudhuri A. Angiotensin II and inflammation: The effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockade. J. Hum. Hypertens. 2007;21:20–27. doi: 10.1038/sj.jhh.1002101. [DOI] [PubMed] [Google Scholar]
- 64.Das U.N. Is angiotensin-II an endogenous pro-inflammatory molecule? Med. Sci. Monit. 2005;11:RA155–RA162. [PubMed] [Google Scholar]
- 65.Leeuwenberg J.F., Smeets E.F., Neefjes J.J., Shaffer M.A., Cinek T., Jeunhomme T.M., Ahern T.J., Buurman W.A. E-selectin and intercellular adhesion molecule-1 are released by activated human endothelial cells in vitro. Immunology. 1992;77:543–549. [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt-Ott K.M., Kagiyama S., Phillips M.I. The multiple actions of angiotensin II in atherosclerosis. Regul. Pept. 2000;93:65–77. doi: 10.1016/s0167-0115(00)00178-6. [DOI] [PubMed] [Google Scholar]
- 67.Wagner G.P., Zhang J. The pleiotropic structure of the genotype-phenotype map: The evolvability of complex organisms. Nat. Rev. Genet. 2011;12:204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- 68.Cooper Z.N., Nelson R.M., Ross L.F. Informed consent for genetic research involving pleiotropic genes: An empirical study of ApoE research. IRB. 2006;28:1–11. [PubMed] [Google Scholar]
- 69.Wachbroit R. The question not asked: The challenge of pleiotropic genetic tests. Kennedy Inst. Ethics J. 1998;8:131–144. doi: 10.1353/ken.1998.0013. [DOI] [PubMed] [Google Scholar]
- 70.Wade C.H., Wilfond B.S. Ethical and clinical practice considerations for genetic counselors related to direct-to-consumer marketing of genetic tests. Am. J. Med. Genet. C. Semin. Med. Genet. 2006;142C:284–292. doi: 10.1002/ajmg.c.30110. discussion 293. [DOI] [PubMed] [Google Scholar]
- 71.Takemoto M., Liao J.K. Pleiotropic effects of 3-hydroxy-3-methylglutaryl coenzyme a reductase inhibitors. Arterioscler. Thromb. Vasc. Biol. 2001;21:1712–1719. doi: 10.1161/hq1101.098486. [DOI] [PubMed] [Google Scholar]
- 72.McFarlane S.I., Muniyappa R., Francisco R., Sowers J.R. Clinical review 145: Pleiotropic effects of statins: Lipid reduction and beyond. J. Clin. Endocrinol. Metab. 2002;87:1451–1458. doi: 10.1210/jcem.87.4.8412. [DOI] [PubMed] [Google Scholar]
- 73.Wolfrum S., Jensen K.S., Liao J.K. Endothelium-dependent effects of statins. Arterioscler. Thromb. Vasc. Biol. 2003;23:729–736. doi: 10.1161/01.ATV.0000063385.12476.A7. [DOI] [PubMed] [Google Scholar]
- 74.Wang K., Baldassano R., Zhang H., Qu H.Q., Imielinski M., Kugathasan S., Annese V., Dubinsky M., Rotter J.I., Russell R.K., et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum. Mol. Genet. 2010;19:2059–2067. doi: 10.1093/hmg/ddq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.The International Diabetes Federation. (2006). The IDF consensus worldwide definition of the metabolic syndrome, http://www.idf.org/webdata/docs/IDF_Meta_def_final.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


