Abstract
In the title compound, C24H21ClN2O6, the two fused six-membered pyran rings adopt half-chair conformations. The dihedral angle between the pyrimidine ring and the chlorophenyl ring is 51.55 (3)°. In the crystal, molecules are linked by pairs of weak intermolecular C—H⋯O hydrogen bonds, forming inversion dimers. A C—H⋯π interaction is also observed.
Related literature
For biological activity of pyrimidine derivatives, see: Alam et al. (2005 ▶); Kappe (2000 ▶); Condon et al. (1993 ▶); Rovnyak et al. (1995 ▶); Leite et al. (2006 ▶); Sriram et al. (2006 ▶). For related structures, see: Booysen et al. (2011 ▶); Noroozi Pesyan et al. (2009 ▶).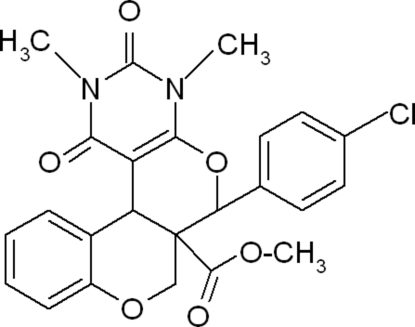
Experimental
Crystal data
C24H21ClN2O6
M r = 468.88
Monoclinic,

a = 10.6177 (5) Å
b = 11.9973 (5) Å
c = 17.5532 (8) Å
β = 99.751 (2)°
V = 2203.69 (17) Å3
Z = 4
Mo Kα radiation
μ = 0.22 mm−1
T = 295 K
0.30 × 0.25 × 0.20 mm
Data collection
Bruker Kappa APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.924, T max = 0.951
29343 measured reflections
7232 independent reflections
4518 reflections with I > 2σ(I)
R int = 0.029
Refinement
R[F 2 > 2σ(F 2)] = 0.052
wR(F 2) = 0.158
S = 1.03
7232 reflections
301 parameters
H-atom parameters constrained
Δρmax = 0.42 e Å−3
Δρmin = −0.60 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681102678X/is2742sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681102678X/is2742Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681102678X/is2742Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg4 is the centroid of the C1–C5/C9 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C16—H16⋯O3i | 0.93 | 2.44 | 3.349 (2) | 166 |
| C19—H19⋯Cg4ii | 0.93 | 2.84 | 3.720 (3) | 158 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
supplementary crystallographic information
Comment
Pyrimidine derivatives are used in the areas of pesticide and pharmaceutical agents (Condon et al., 1993). In addition, pyrimidine-2(1H)-ones/thiones are calcium channel blocker compounds (Rovnyak et al., 1995). They also have other biological activities such as antibacterial, antifungal and antiviral (Kappe, 2000; Alam et al., 2005; Sriram et al., 2006; Leite et al., 2006).
The geometric parameters of the title molecule (Fig. 1) agree well with reported similar structure (Booysen et al., 2011; Noroozi Pesyan et al., 2009). The sum of bond angles around N1 and N2 [359.93 (15) and 358.92°, respectively] indicates the sp2 hybridization state of atoms N1 and N2 in the molecule. The crystal packing is controlled by weak intermolecular C—H···O and C—H···π interactions (Table 1).
Experimental
A mixture of (E)-methyl 2-((2-formylphenoxy)methyl)-3-(4-chlorophenyl)acrylate (0.330 g, 1 mmol) and N,N-dimethylbarbutric acid (0.156 g, 1 mmol) was placed in a round bottom flask and melted at 180 °C for 1 h. After completion of the reaction as indicated by TLC, the crude product was washed with 5 ml of ethylacetate and hexane mixture (1:49 ratio) which successfully provided the pure product methyl-6-(4-chlorophenyl)-2,4-dimethyl-1,3-dioxo- 1,2,3,4,6,6a,7,12octahydrochromeno[4',3',4,5]pyrano[2,3-d]pyrimidine- 6a-carboxylate, as colorless solid in 96% yield.
Refinement
H atoms were positioned geometrically and refined using riding model with C—H = 0.93 Å and Uiso(H) = 1.2Ueq(C) for aromatic CH, C—H = 0.98 Å and Uiso(H) = 1.2Ueq(C) for methine CH, C—H = 0.97 Å and Uiso(H) = 1.2Ueq(C) for CH2, C—H = 0.96 Å and Uiso(H) = 1.5Ueq(C) for CH3.
Figures
Fig. 1.
The molecular structure of the title compound, with atom labels and 30% probability displacement ellipsoids for non-H atoms.
Fig. 2.
A packing diagram of the title compound, viewed down the b axis. Hydrogen bonds are shown as dashed lines.
Crystal data
| C24H21ClN2O6 | F(000) = 976 |
| Mr = 468.88 | Dx = 1.413 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 7610 reflections |
| a = 10.6177 (5) Å | θ = 2.2–30.2° |
| b = 11.9973 (5) Å | µ = 0.22 mm−1 |
| c = 17.5532 (8) Å | T = 295 K |
| β = 99.751 (2)° | Block, colourless |
| V = 2203.69 (17) Å3 | 0.30 × 0.25 × 0.20 mm |
| Z = 4 |
Data collection
| Bruker Kappa APEXII CCD diffractometer | 7232 independent reflections |
| Radiation source: fine-focus sealed tube | 4518 reflections with I > 2σ(I) |
| graphite | Rint = 0.029 |
| Detector resolution: 0 pixels mm-1 | θmax = 31.7°, θmin = 2.1° |
| ω and φ scans | h = −15→15 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | k = −15→17 |
| Tmin = 0.924, Tmax = 0.951 | l = −25→25 |
| 29343 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.052 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.158 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0706P)2 + 0.6194P] where P = (Fo2 + 2Fc2)/3 |
| 7232 reflections | (Δ/σ)max = 0.005 |
| 301 parameters | Δρmax = 0.42 e Å−3 |
| 0 restraints | Δρmin = −0.60 e Å−3 |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.76555 (18) | 0.06202 (16) | 0.37637 (10) | 0.0480 (4) | |
| H1 | 0.7599 | 0.1389 | 0.3819 | 0.058* | |
| C2 | 0.8645 (2) | 0.00508 (18) | 0.42011 (11) | 0.0568 (5) | |
| H2 | 0.9268 | 0.0436 | 0.4536 | 0.068* | |
| C3 | 0.87131 (19) | −0.10942 (17) | 0.41423 (11) | 0.0541 (5) | |
| H3 | 0.9383 | −0.1480 | 0.4438 | 0.065* | |
| C4 | 0.77967 (17) | −0.16628 (15) | 0.36499 (10) | 0.0459 (4) | |
| H4 | 0.7837 | −0.2435 | 0.3616 | 0.055* | |
| C5 | 0.68080 (14) | −0.10821 (13) | 0.32016 (9) | 0.0361 (3) | |
| C6 | 0.47962 (14) | −0.12010 (12) | 0.23307 (8) | 0.0338 (3) | |
| H6A | 0.4081 | −0.1706 | 0.2327 | 0.041* | |
| H6B | 0.4906 | −0.1088 | 0.1799 | 0.041* | |
| C7 | 0.44663 (14) | −0.00850 (12) | 0.26651 (8) | 0.0310 (3) | |
| C8 | 0.56484 (14) | 0.06889 (12) | 0.27607 (8) | 0.0328 (3) | |
| H8 | 0.5464 | 0.1355 | 0.3046 | 0.039* | |
| C9 | 0.67411 (15) | 0.00706 (13) | 0.32418 (9) | 0.0351 (3) | |
| C10 | 0.33697 (14) | 0.05058 (13) | 0.21285 (8) | 0.0338 (3) | |
| H10 | 0.3240 | 0.1238 | 0.2350 | 0.041* | |
| C11 | 0.49137 (15) | 0.10189 (13) | 0.13570 (9) | 0.0362 (3) | |
| C12 | 0.58675 (15) | 0.10449 (13) | 0.19648 (9) | 0.0356 (3) | |
| C13 | 0.70722 (17) | 0.15057 (14) | 0.18445 (11) | 0.0438 (4) | |
| C14 | 0.6217 (2) | 0.16628 (16) | 0.04577 (11) | 0.0510 (4) | |
| C15 | 0.21085 (14) | −0.00924 (13) | 0.19859 (9) | 0.0356 (3) | |
| C16 | 0.18358 (16) | −0.09052 (16) | 0.14232 (10) | 0.0464 (4) | |
| H16 | 0.2442 | −0.1090 | 0.1119 | 0.056* | |
| C17 | 0.06716 (18) | −0.14441 (18) | 0.13101 (11) | 0.0529 (5) | |
| H17 | 0.0493 | −0.1992 | 0.0932 | 0.063* | |
| C18 | −0.02226 (16) | −0.11680 (17) | 0.17584 (11) | 0.0482 (4) | |
| C19 | 0.00161 (17) | −0.03659 (19) | 0.23166 (12) | 0.0550 (5) | |
| H19 | −0.0595 | −0.0188 | 0.2619 | 0.066* | |
| C20 | 0.11761 (17) | 0.01751 (16) | 0.24245 (11) | 0.0483 (4) | |
| H20 | 0.1339 | 0.0730 | 0.2798 | 0.058* | |
| C21 | 0.3952 (2) | 0.13297 (19) | −0.00072 (10) | 0.0597 (5) | |
| H21A | 0.3603 | 0.0592 | −0.0081 | 0.090* | |
| H21B | 0.4218 | 0.1577 | −0.0475 | 0.090* | |
| H21C | 0.3312 | 0.1828 | 0.0123 | 0.090* | |
| C22 | 0.8425 (2) | 0.2140 (3) | 0.09356 (16) | 0.0834 (8) | |
| H22A | 0.8659 | 0.1797 | 0.0486 | 0.125* | |
| H22B | 0.9071 | 0.1990 | 0.1376 | 0.125* | |
| H22C | 0.8348 | 0.2931 | 0.0857 | 0.125* | |
| C23 | 0.40621 (15) | −0.02757 (14) | 0.34447 (9) | 0.0364 (3) | |
| C24 | 0.3303 (3) | 0.0581 (2) | 0.44840 (13) | 0.0941 (10) | |
| H24A | 0.3101 | −0.0184 | 0.4569 | 0.141* | |
| H24B | 0.2547 | 0.1028 | 0.4461 | 0.141* | |
| H24C | 0.3943 | 0.0835 | 0.4901 | 0.141* | |
| N1 | 0.50522 (15) | 0.13185 (13) | 0.06206 (8) | 0.0447 (3) | |
| N2 | 0.71979 (16) | 0.16862 (13) | 0.10700 (10) | 0.0523 (4) | |
| O1 | 0.59218 (11) | −0.17168 (9) | 0.27413 (7) | 0.0444 (3) | |
| O2 | 0.37171 (11) | 0.06736 (10) | 0.13744 (6) | 0.0408 (3) | |
| O3 | 0.63502 (17) | 0.19131 (14) | −0.01960 (8) | 0.0738 (5) | |
| O4 | 0.79552 (13) | 0.17313 (13) | 0.23567 (9) | 0.0621 (4) | |
| O5 | 0.39933 (16) | −0.11577 (11) | 0.37358 (8) | 0.0623 (4) | |
| O6 | 0.37805 (15) | 0.06763 (11) | 0.37639 (7) | 0.0581 (4) | |
| Cl1 | −0.16887 (5) | −0.18537 (6) | 0.16129 (4) | 0.0765 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0536 (10) | 0.0389 (9) | 0.0480 (9) | −0.0044 (8) | −0.0018 (8) | −0.0048 (7) |
| C2 | 0.0545 (11) | 0.0559 (12) | 0.0523 (10) | −0.0036 (9) | −0.0128 (8) | −0.0078 (9) |
| C3 | 0.0480 (10) | 0.0566 (12) | 0.0523 (10) | 0.0083 (9) | −0.0069 (8) | −0.0013 (9) |
| C4 | 0.0447 (9) | 0.0387 (9) | 0.0527 (10) | 0.0080 (7) | 0.0033 (7) | −0.0013 (7) |
| C5 | 0.0350 (7) | 0.0340 (8) | 0.0392 (7) | 0.0002 (6) | 0.0056 (6) | −0.0032 (6) |
| C6 | 0.0363 (7) | 0.0289 (7) | 0.0357 (7) | 0.0003 (6) | 0.0046 (6) | −0.0008 (6) |
| C7 | 0.0342 (7) | 0.0277 (7) | 0.0316 (6) | 0.0001 (5) | 0.0076 (5) | 0.0014 (5) |
| C8 | 0.0373 (7) | 0.0258 (7) | 0.0356 (7) | −0.0010 (6) | 0.0074 (6) | −0.0005 (5) |
| C9 | 0.0371 (8) | 0.0309 (8) | 0.0369 (7) | −0.0015 (6) | 0.0050 (6) | −0.0003 (6) |
| C10 | 0.0382 (7) | 0.0314 (8) | 0.0327 (7) | 0.0036 (6) | 0.0089 (6) | 0.0025 (6) |
| C11 | 0.0462 (8) | 0.0288 (8) | 0.0361 (7) | 0.0018 (6) | 0.0142 (6) | 0.0033 (6) |
| C12 | 0.0397 (8) | 0.0287 (7) | 0.0407 (8) | 0.0012 (6) | 0.0135 (6) | 0.0025 (6) |
| C13 | 0.0435 (9) | 0.0366 (9) | 0.0549 (10) | 0.0011 (7) | 0.0191 (8) | 0.0027 (7) |
| C14 | 0.0686 (12) | 0.0424 (10) | 0.0490 (10) | 0.0040 (8) | 0.0307 (9) | 0.0009 (8) |
| C15 | 0.0343 (7) | 0.0363 (8) | 0.0360 (7) | 0.0054 (6) | 0.0057 (6) | 0.0018 (6) |
| C16 | 0.0396 (8) | 0.0580 (11) | 0.0432 (9) | −0.0004 (8) | 0.0113 (7) | −0.0102 (8) |
| C17 | 0.0461 (10) | 0.0610 (12) | 0.0499 (10) | −0.0055 (8) | 0.0037 (8) | −0.0112 (9) |
| C18 | 0.0334 (8) | 0.0594 (12) | 0.0504 (9) | −0.0006 (7) | 0.0030 (7) | 0.0066 (8) |
| C19 | 0.0386 (9) | 0.0707 (13) | 0.0592 (11) | 0.0044 (9) | 0.0179 (8) | −0.0048 (10) |
| C20 | 0.0430 (9) | 0.0516 (11) | 0.0526 (10) | 0.0039 (8) | 0.0149 (8) | −0.0100 (8) |
| C21 | 0.0799 (14) | 0.0630 (13) | 0.0364 (9) | 0.0073 (11) | 0.0098 (9) | 0.0083 (8) |
| C22 | 0.0676 (15) | 0.099 (2) | 0.0960 (18) | −0.0118 (13) | 0.0491 (14) | 0.0139 (15) |
| C23 | 0.0387 (8) | 0.0362 (8) | 0.0351 (7) | 0.0014 (6) | 0.0086 (6) | 0.0029 (6) |
| C24 | 0.159 (3) | 0.0829 (18) | 0.0556 (13) | 0.0290 (18) | 0.0609 (16) | 0.0027 (12) |
| N1 | 0.0588 (9) | 0.0422 (8) | 0.0361 (7) | 0.0027 (7) | 0.0168 (6) | 0.0048 (6) |
| N2 | 0.0549 (9) | 0.0483 (9) | 0.0621 (10) | −0.0012 (7) | 0.0342 (8) | 0.0029 (7) |
| O1 | 0.0370 (6) | 0.0292 (6) | 0.0629 (7) | 0.0029 (4) | −0.0030 (5) | −0.0083 (5) |
| O2 | 0.0430 (6) | 0.0459 (7) | 0.0333 (5) | −0.0026 (5) | 0.0057 (4) | 0.0076 (5) |
| O3 | 0.1000 (12) | 0.0798 (11) | 0.0528 (8) | 0.0023 (9) | 0.0455 (8) | 0.0096 (7) |
| O4 | 0.0462 (7) | 0.0716 (10) | 0.0697 (9) | −0.0159 (7) | 0.0130 (7) | 0.0056 (7) |
| O5 | 0.0981 (11) | 0.0404 (8) | 0.0562 (8) | 0.0018 (7) | 0.0359 (8) | 0.0126 (6) |
| O6 | 0.0926 (11) | 0.0442 (7) | 0.0451 (7) | 0.0092 (7) | 0.0330 (7) | −0.0009 (5) |
| Cl1 | 0.0427 (3) | 0.1053 (5) | 0.0794 (4) | −0.0208 (3) | 0.0046 (2) | 0.0005 (3) |
Geometric parameters (Å, °)
| C1—C2 | 1.374 (3) | C13—N2 | 1.405 (2) |
| C1—C9 | 1.384 (2) | C14—O3 | 1.217 (2) |
| C1—H1 | 0.9300 | C14—N2 | 1.364 (3) |
| C2—C3 | 1.380 (3) | C14—N1 | 1.379 (2) |
| C2—H2 | 0.9300 | C15—C16 | 1.383 (2) |
| C3—C4 | 1.369 (3) | C15—C20 | 1.391 (2) |
| C3—H3 | 0.9300 | C16—C17 | 1.379 (3) |
| C4—C5 | 1.388 (2) | C16—H16 | 0.9300 |
| C4—H4 | 0.9300 | C17—C18 | 1.372 (3) |
| C5—O1 | 1.3639 (18) | C17—H17 | 0.9300 |
| C5—C9 | 1.387 (2) | C18—C19 | 1.366 (3) |
| C6—O1 | 1.4280 (18) | C18—Cl1 | 1.7408 (18) |
| C6—C7 | 1.526 (2) | C19—C20 | 1.377 (3) |
| C6—H6A | 0.9700 | C19—H19 | 0.9300 |
| C6—H6B | 0.9700 | C20—H20 | 0.9300 |
| C7—C23 | 1.519 (2) | C21—N1 | 1.464 (2) |
| C7—C10 | 1.541 (2) | C21—H21A | 0.9600 |
| C7—C8 | 1.547 (2) | C21—H21B | 0.9600 |
| C8—C9 | 1.509 (2) | C21—H21C | 0.9600 |
| C8—C12 | 1.516 (2) | C22—N2 | 1.468 (2) |
| C8—H8 | 0.9800 | C22—H22A | 0.9600 |
| C10—O2 | 1.4471 (17) | C22—H22B | 0.9600 |
| C10—C15 | 1.502 (2) | C22—H22C | 0.9600 |
| C10—H10 | 0.9800 | C23—O5 | 1.183 (2) |
| C11—C12 | 1.341 (2) | C23—O6 | 1.328 (2) |
| C11—O2 | 1.3416 (19) | C24—O6 | 1.444 (2) |
| C11—N1 | 1.3734 (19) | C24—H24A | 0.9600 |
| C12—C13 | 1.442 (2) | C24—H24B | 0.9600 |
| C13—O4 | 1.214 (2) | C24—H24C | 0.9600 |
| C2—C1—C9 | 121.17 (17) | O3—C14—N2 | 122.88 (19) |
| C2—C1—H1 | 119.4 | O3—C14—N1 | 121.4 (2) |
| C9—C1—H1 | 119.4 | N2—C14—N1 | 115.75 (15) |
| C1—C2—C3 | 119.86 (17) | C16—C15—C20 | 118.39 (16) |
| C1—C2—H2 | 120.1 | C16—C15—C10 | 121.91 (14) |
| C3—C2—H2 | 120.1 | C20—C15—C10 | 119.70 (15) |
| C4—C3—C2 | 120.16 (17) | C17—C16—C15 | 120.49 (16) |
| C4—C3—H3 | 119.9 | C17—C16—H16 | 119.8 |
| C2—C3—H3 | 119.9 | C15—C16—H16 | 119.8 |
| C3—C4—C5 | 119.75 (17) | C18—C17—C16 | 119.64 (18) |
| C3—C4—H4 | 120.1 | C18—C17—H17 | 120.2 |
| C5—C4—H4 | 120.1 | C16—C17—H17 | 120.2 |
| O1—C5—C9 | 123.41 (14) | C19—C18—C17 | 121.26 (17) |
| O1—C5—C4 | 115.80 (15) | C19—C18—Cl1 | 119.39 (14) |
| C9—C5—C4 | 120.79 (15) | C17—C18—Cl1 | 119.35 (15) |
| O1—C6—C7 | 114.40 (12) | C18—C19—C20 | 118.93 (17) |
| O1—C6—H6A | 108.7 | C18—C19—H19 | 120.5 |
| C7—C6—H6A | 108.7 | C20—C19—H19 | 120.5 |
| O1—C6—H6B | 108.7 | C19—C20—C15 | 121.28 (17) |
| C7—C6—H6B | 108.7 | C19—C20—H20 | 119.4 |
| H6A—C6—H6B | 107.6 | C15—C20—H20 | 119.4 |
| C23—C7—C6 | 109.42 (12) | N1—C21—H21A | 109.5 |
| C23—C7—C10 | 108.69 (12) | N1—C21—H21B | 109.5 |
| C6—C7—C10 | 111.56 (12) | H21A—C21—H21B | 109.5 |
| C23—C7—C8 | 109.90 (12) | N1—C21—H21C | 109.5 |
| C6—C7—C8 | 109.47 (12) | H21A—C21—H21C | 109.5 |
| C10—C7—C8 | 107.78 (11) | H21B—C21—H21C | 109.5 |
| C9—C8—C12 | 115.51 (12) | N2—C22—H22A | 109.5 |
| C9—C8—C7 | 107.36 (12) | N2—C22—H22B | 109.5 |
| C12—C8—C7 | 108.51 (12) | H22A—C22—H22B | 109.5 |
| C9—C8—H8 | 108.4 | N2—C22—H22C | 109.5 |
| C12—C8—H8 | 108.4 | H22A—C22—H22C | 109.5 |
| C7—C8—H8 | 108.4 | H22B—C22—H22C | 109.5 |
| C1—C9—C5 | 118.19 (15) | O5—C23—O6 | 123.51 (15) |
| C1—C9—C8 | 121.53 (14) | O5—C23—C7 | 124.85 (15) |
| C5—C9—C8 | 120.18 (13) | O6—C23—C7 | 111.63 (13) |
| O2—C10—C15 | 105.79 (12) | O6—C24—H24A | 109.5 |
| O2—C10—C7 | 109.87 (11) | O6—C24—H24B | 109.5 |
| C15—C10—C7 | 116.24 (12) | H24A—C24—H24B | 109.5 |
| O2—C10—H10 | 108.2 | O6—C24—H24C | 109.5 |
| C15—C10—H10 | 108.2 | H24A—C24—H24C | 109.5 |
| C7—C10—H10 | 108.2 | H24B—C24—H24C | 109.5 |
| C12—C11—O2 | 125.41 (13) | C11—N1—C14 | 121.08 (15) |
| C12—C11—N1 | 123.77 (15) | C11—N1—C21 | 120.78 (15) |
| O2—C11—N1 | 110.80 (14) | C14—N1—C21 | 118.08 (15) |
| C11—C12—C13 | 117.48 (14) | C14—N2—C13 | 125.05 (15) |
| C11—C12—C8 | 120.89 (13) | C14—N2—C22 | 117.44 (17) |
| C13—C12—C8 | 121.44 (14) | C13—N2—C22 | 116.43 (18) |
| O4—C13—N2 | 119.60 (16) | C5—O1—C6 | 119.44 (12) |
| O4—C13—C12 | 124.74 (16) | C11—O2—C10 | 116.93 (12) |
| N2—C13—C12 | 115.66 (16) | C23—O6—C24 | 116.01 (16) |
| C9—C1—C2—C3 | −2.2 (3) | O2—C10—C15—C20 | −141.61 (15) |
| C1—C2—C3—C4 | 0.0 (3) | C7—C10—C15—C20 | 96.17 (18) |
| C2—C3—C4—C5 | 0.9 (3) | C20—C15—C16—C17 | −0.8 (3) |
| C3—C4—C5—O1 | −178.52 (17) | C10—C15—C16—C17 | 179.57 (16) |
| C3—C4—C5—C9 | 0.5 (3) | C15—C16—C17—C18 | 0.2 (3) |
| O1—C6—C7—C23 | −68.32 (16) | C16—C17—C18—C19 | 0.1 (3) |
| O1—C6—C7—C10 | 171.38 (12) | C16—C17—C18—Cl1 | −179.90 (16) |
| O1—C6—C7—C8 | 52.18 (16) | C17—C18—C19—C20 | 0.3 (3) |
| C23—C7—C8—C9 | 65.52 (15) | Cl1—C18—C19—C20 | −179.69 (16) |
| C6—C7—C8—C9 | −54.69 (15) | C18—C19—C20—C15 | −1.0 (3) |
| C10—C7—C8—C9 | −176.20 (11) | C16—C15—C20—C19 | 1.3 (3) |
| C23—C7—C8—C12 | −169.00 (12) | C10—C15—C20—C19 | −179.14 (17) |
| C6—C7—C8—C12 | 70.80 (14) | C6—C7—C23—O5 | −1.3 (2) |
| C10—C7—C8—C12 | −50.71 (15) | C10—C7—C23—O5 | 120.72 (18) |
| C2—C1—C9—C5 | 3.5 (3) | C8—C7—C23—O5 | −121.56 (18) |
| C2—C1—C9—C8 | 179.82 (17) | C6—C7—C23—O6 | 179.19 (14) |
| O1—C5—C9—C1 | 176.26 (16) | C10—C7—C23—O6 | −58.77 (17) |
| C4—C5—C9—C1 | −2.7 (2) | C8—C7—C23—O6 | 58.96 (17) |
| O1—C5—C9—C8 | −0.1 (2) | C12—C11—N1—C14 | 0.7 (3) |
| C4—C5—C9—C8 | −178.98 (15) | O2—C11—N1—C14 | −177.63 (15) |
| C12—C8—C9—C1 | 93.78 (18) | C12—C11—N1—C21 | −176.51 (17) |
| C7—C8—C9—C1 | −145.05 (15) | O2—C11—N1—C21 | 5.2 (2) |
| C12—C8—C9—C5 | −90.03 (17) | O3—C14—N1—C11 | 178.87 (17) |
| C7—C8—C9—C5 | 31.15 (19) | N2—C14—N1—C11 | −0.2 (2) |
| C23—C7—C10—O2 | −178.60 (12) | O3—C14—N1—C21 | −3.9 (3) |
| C6—C7—C10—O2 | −57.87 (15) | N2—C14—N1—C21 | 177.04 (16) |
| C8—C7—C10—O2 | 62.33 (15) | O3—C14—N2—C13 | 173.81 (18) |
| C23—C7—C10—C15 | −58.56 (16) | N1—C14—N2—C13 | −7.1 (3) |
| C6—C7—C10—C15 | 62.17 (16) | O3—C14—N2—C22 | 6.2 (3) |
| C8—C7—C10—C15 | −177.63 (12) | N1—C14—N2—C22 | −174.76 (19) |
| O2—C11—C12—C13 | −176.39 (15) | O4—C13—N2—C14 | −167.39 (18) |
| N1—C11—C12—C13 | 5.6 (2) | C12—C13—N2—C14 | 13.1 (3) |
| O2—C11—C12—C8 | −1.2 (2) | O4—C13—N2—C22 | 0.4 (3) |
| N1—C11—C12—C8 | −179.28 (14) | C12—C13—N2—C22 | −179.16 (18) |
| C9—C8—C12—C11 | 142.93 (15) | C9—C5—O1—C6 | −6.3 (2) |
| C7—C8—C12—C11 | 22.38 (19) | C4—C5—O1—C6 | 172.70 (14) |
| C9—C8—C12—C13 | −42.1 (2) | C7—C6—O1—C5 | −21.03 (19) |
| C7—C8—C12—C13 | −162.66 (14) | C12—C11—O2—C10 | 11.6 (2) |
| C11—C12—C13—O4 | 168.86 (17) | N1—C11—O2—C10 | −170.11 (13) |
| C8—C12—C13—O4 | −6.3 (3) | C15—C10—O2—C11 | −168.67 (13) |
| C11—C12—C13—N2 | −11.6 (2) | C7—C10—O2—C11 | −42.47 (17) |
| C8—C12—C13—N2 | 173.23 (14) | O5—C23—O6—C24 | −3.5 (3) |
| O2—C10—C15—C16 | 38.0 (2) | C7—C23—O6—C24 | 176.02 (19) |
| C7—C10—C15—C16 | −84.24 (19) |
Hydrogen-bond geometry (Å, °)
| Cg4 is the centroid of the C1–C5/C9 ring. |
| D—H···A | D—H | H···A | D···A | D—H···A |
| C16—H16···O3i | 0.93 | 2.44 | 3.349 (2) | 166 |
| C19—H19···Cg4ii | 0.93 | 2.84 | 3.720 (3) | 158 |
Symmetry codes: (i) −x+1, −y, −z; (ii) x−1, y, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IS2742).
References
- Alam, O., Imran, M. & Khan, S. A. (2005). Indian J. Heterocycl. Chem. 14, 293–296.
- Booysen, I., Muhammed, I., Soares, A., Gerber, T., Hosten, E. & Betz, R. (2011). Acta Cryst. E67, o1592. [DOI] [PMC free article] [PubMed]
- Bruker (2004). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Condon, M. E., Brady, T. E., Feist, D., Malefyt, T., Marc, P., Quakenbush, L. S., Rodaway, S. J., Shaner, D. L. & Tecle, B. (1993). Brighton Crop Protection Conference–Weeds, pp. 41–46.
- Kappe, C. O. (2000). Acc. Chem. Res. 33, 879–888. [DOI] [PubMed]
- Leite, A. C. L., Lima, R. S., Moreira, D. R. M., Cardoso, M. V. O., Brito, A. C. G., Santos, L. M. F., Hernandes, M. Z., Kiperstok, A. C., Lima, R. S. & Soares, M. B. P. (2006). Bioorg. Med. Chem. 14, 3749–3757. [DOI] [PubMed]
- Noroozi Pesyan, N., Rastgar, S. & Hosseini, Y. (2009). Acta Cryst. E65, o1444. [DOI] [PMC free article] [PubMed]
- Rovnyak, G. C., Kimball, S. D., Beyer, B., Cucinotta, G., DiMarco, J. D., Gougoutas, J., Hedberg, A., Malley, M., McCarthy, J. P., Zhang, R. & Moreland, S. (1995). J. Med. Chem. 38, 119–129. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Sriram, D., Yogeeswari, P. & Devakaram, R. V. (2006). Bioorg. Med. Chem. 14, 3113–3118. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681102678X/is2742sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681102678X/is2742Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681102678X/is2742Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




