Abstract
The bis(benzimidazol-1-yl)methane molecule of the title compound, C15H12N4·2H2O, displays a trans conformation with a twofold axis running through the methylene C atom. Two adjacent water molecules are bonded to this molecule through O—H⋯N hydrogen bonds, forming a trimer. Adjacent trimers are connected together via C—H⋯O interactions, forming a chain running along the b-axis direction. Two such chains are joined together via π–π interactions [centroid–centroid distance = 3.556 (2) Å], forming double chains, which are connected via the water molecules through C—H⋯O associations, forming a sheet structure. The sheets are stacked on top of each other along the a-axis direction and connected through O—H⋯O and C—H⋯O interactions, forming a three-dimensional ABAB layer network structure.
Related literature
For the use of bridged imidazole derivatives as multidentate N-donor ligands in the construction of functional coordination polymers, see: Chang et al. (2005 ▶); Wen et al. (2006 ▶); Fan et al. (2004 ▶); Abrahams et al. (2002 ▶); Jin et al. (2007 ▶); Ma et al. (2003 ▶). For the synthesis, see: Lavandera et al. (1988 ▶).
Experimental
Crystal data
C15H12N4·2H2O
M r = 284.32
Triclinic,

a = 8.3752 (9) Å
b = 9.2079 (8) Å
c = 10.7199 (10) Å
α = 100.288 (1)°
β = 101.495 (1)°
γ = 116.108 (2)°
V = 693.35 (12) Å3
Z = 2
Mo Kα radiation
μ = 0.09 mm−1
T = 298 K
0.44 × 0.40 × 0.18 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2002 ▶) T min = 0.959, T max = 0.983
3617 measured reflections
2411 independent reflections
1327 reflections with I > 2σ(I)
R int = 0.021
Refinement
R[F 2 > 2σ(F 2)] = 0.050
wR(F 2) = 0.148
S = 1.01
2411 reflections
191 parameters
H-atom parameters constrained
Δρmax = 0.18 e Å−3
Δρmin = −0.18 e Å−3
Data collection: SMART (Bruker, 2002 ▶); cell refinement: SAINT (Bruker, 2002 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811027966/vm2104sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811027966/vm2104Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811027966/vm2104Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1E⋯N2i | 0.85 | 1.99 | 2.841 (3) | 178 |
| O1—H1F⋯O1ii | 0.85 | 2.41 | 2.911 (5) | 118 |
| O2—H2C⋯N4iii | 0.85 | 2.23 | 3.083 (3) | 176 |
| O2—H2D⋯N4iv | 0.85 | 2.12 | 2.968 (3) | 177 |
| C2—H2⋯O1ii | 0.93 | 2.39 | 3.299 (4) | 167 |
| C9—H9⋯O2i | 0.93 | 2.56 | 3.363 (4) | 145 |
| C12—H12⋯O1ii | 0.93 | 2.57 | 3.495 (4) | 173 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Acknowledgments
We gratefully acknowledge financial support by the Education Office Foundation of Zhejiang Province (project No. Y201017321) and the innovation project of Zhejiang A & F University.
supplementary crystallographic information
Comment
Bridged imidazole derivatives can be used as multidentate N-donor ligands in constructing functioned coordination polymers, such as nonlinear optical materials (Chang et al., 2005), novel hybrid inorganic organic photoactive materials (Wen et al., 2006) and novel metal-organic frameworks (Fan et al., 2004; Abrahams et al., 2002). The ligands bearing alkyl spacers are a good choice of a N-donor ligand, and the flexible nature of the spacers allows the ligands to bend and rotate when coordinating to metal centers so as to conform to the coordination geometries of the metal ions. Significant progress has been achieved by us (Jin et al., 2007) and others (Ma et al., 2003) in this area.
However, the archived data on bridged benzimidazole derivatives bearing the methylene spacer have been rare. As an extension of our study in bridged imidazole derivatives, here in this paper, we report the structure of bis(benzimidazol-1-yl)methane dihydrate.
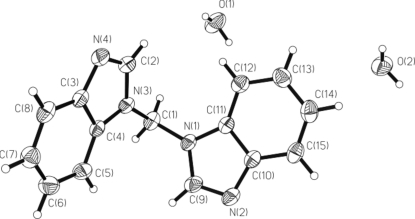
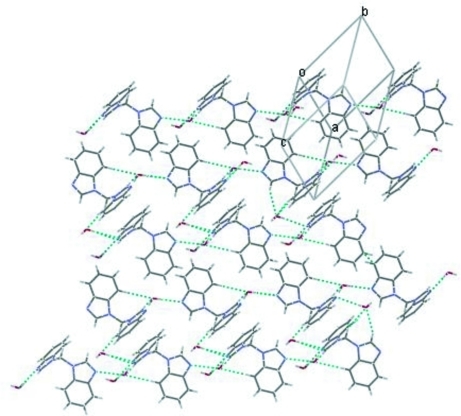
X-ray diffraction analysis indicated that in the title compound there are one bis(benzimidazol-1-yl)methane and two lattice water molecules (Fig. 1). All bond distances and angles are in the normal range. The r.m.s. deviation of the benzimidazole ring bearing the N1 and N2 atoms is 0.0056 Å. The r.m.s. deviation of the benzimidazole ring bearing the N3 and N4 atoms is 0.00123 Å. Both benzimidazole rings make a dihedral angle of 106.9 (3)° with each other. The bis(benzimidazol-1-yl)methane displays trans conformation with a twofold axis running through atom C1. Two water molecules are bonded to the bis(benzimidazol-1-yl)methane molecule through O—H···N hydrogen bonds (Table 1) to form an adduct. These adjacent adducts are connected together via C—H···O interactions to form a one-dimensional chain running along the b axis direction. There are two kinds of C—H···O associations (Table 1), one is arising from the N—CH—N of the benzimidazole moiety, another from the benzene C12—H12. In this chain the bis(benzimidazol-1-yl)methane molecules are parallelly arranged. Two such chains were joined together via the π-π interactions to form a double chain structure (Cg(1)···Cg(2)i distance = 3.556 (2) Å, Cg(1) is the centroid of ring N1,N2, C9-C11, Cg(2) is the centroid of ring C10-C15, symmetry operation: (i) 1 - x, 1 - y, 1 - z). The bis(benzimidazol-1-yl)methane molecules at these two chains are arranged antiparallel. The double chains were connected together via the water molecules through the C—H···O associations to form a two-dimensional sheet extending along the direction forming a dihedral angle of ca 60 ° with the bc plane (Fig. 2). Such sheets were further stacked along the a axis direction via the O—H···O (between two water molecules with O···O separations of 2.911 Å) and C—H···O interactions to form a three-dimensional ABAB layer network structure.
Experimental
The starting material bis(benzimidazol-1-yl)methane was prepared according to the published procedure (Lavandera et al., 1988). A solid of bis(benzimidazol-1-yl)methane (24.8 mg, 0.10 mmol) in 10 ml of 95 percent EtOH was stirred at room temperature to dissolve it, then the solution was filtered into a test tube. The solution was left standing at room temperature for several days, colorless block crystals were isolated after slow evaporation of the solution in air at ambient temperature. The crystals were collected and dried in air to give the title compound.
Refinement
H atoms bonded to the O atoms were located in a difference Fourier map, the O—H distance was kept 0.85 Å and refined isotropically. Other H atoms were positioned geometrically with C—H = 0.93–0.97 Å, and constrained to ride on their parent atoms with Uiso(H) = 1.2 Ueq(C).
Figures
Fig. 1.
The structure of the title compound, showing the atom-numbering scheme. Displacement ellipsoids are drawn at the 30% probability level.
Fig. 2.
Two-dimensional sheet structure formed through hydrogen bonds (blue dashed lines) viewed along the a axis direction.
Crystal data
| C15H12N4·2H2O | V = 693.35 (12) Å3 |
| Mr = 284.32 | Z = 2 |
| Triclinic, P1 | F(000) = 300 |
| Hall symbol: -P 1 | Dx = 1.362 Mg m−3 |
| a = 8.3752 (9) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 9.2079 (8) Å | µ = 0.09 mm−1 |
| c = 10.7199 (10) Å | T = 298 K |
| α = 100.288 (1)° | Block, colorless |
| β = 101.495 (1)° | 0.44 × 0.40 × 0.18 mm |
| γ = 116.108 (2)° |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2411 independent reflections |
| Radiation source: fine-focus sealed tube | 1327 reflections with I > 2σ(I) |
| graphite | Rint = 0.021 |
| phi and ω scans | θmax = 25.0°, θmin = 2.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2002) | h = −9→9 |
| Tmin = 0.959, Tmax = 0.983 | k = −10→10 |
| 3617 measured reflections | l = −10→12 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.050 | H-atom parameters constrained |
| wR(F2) = 0.148 | w = 1/[σ2(Fo2) + (0.0658P)2 + 0.0465P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max < 0.001 |
| 2411 reflections | Δρmax = 0.18 e Å−3 |
| 191 parameters | Δρmin = −0.18 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.005 (4) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.4209 (3) | 0.3324 (3) | 0.2893 (2) | 0.0459 (6) | |
| N2 | 0.7039 (3) | 0.5622 (3) | 0.3488 (3) | 0.0575 (7) | |
| N3 | 0.2169 (3) | 0.0593 (3) | 0.1258 (2) | 0.0451 (6) | |
| N4 | 0.1831 (3) | −0.2003 (3) | 0.0636 (2) | 0.0573 (7) | |
| O1 | −0.0360 (3) | 0.1203 (3) | 0.5805 (2) | 0.0937 (8) | |
| H1E | 0.0616 | 0.2167 | 0.6024 | 0.112* | |
| H1F | −0.0050 | 0.0476 | 0.5969 | 0.112* | |
| O2 | 0.7756 (3) | 0.5078 (3) | 0.9768 (2) | 0.0843 (8) | |
| H2C | 0.8861 | 0.5915 | 1.0009 | 0.101* | |
| H2D | 0.7822 | 0.4171 | 0.9644 | 0.101* | |
| C1 | 0.2299 (4) | 0.2083 (3) | 0.2103 (3) | 0.0511 (8) | |
| H1A | 0.1780 | 0.2595 | 0.1549 | 0.061* | |
| H1B | 0.1557 | 0.1745 | 0.2694 | 0.061* | |
| C2 | 0.1760 (4) | −0.0886 (4) | 0.1540 (3) | 0.0556 (8) | |
| H2 | 0.1458 | −0.1089 | 0.2303 | 0.067* | |
| C3 | 0.2331 (4) | −0.1197 (3) | −0.0318 (3) | 0.0454 (7) | |
| C4 | 0.2538 (3) | 0.0428 (3) | 0.0055 (3) | 0.0409 (7) | |
| C5 | 0.2971 (4) | 0.1488 (4) | −0.0736 (3) | 0.0531 (8) | |
| H5 | 0.3109 | 0.2568 | −0.0481 | 0.064* | |
| C6 | 0.3189 (4) | 0.0870 (4) | −0.1916 (3) | 0.0630 (9) | |
| H6 | 0.3456 | 0.1536 | −0.2485 | 0.076* | |
| C7 | 0.3019 (4) | −0.0730 (4) | −0.2280 (3) | 0.0610 (9) | |
| H7 | 0.3204 | −0.1096 | −0.3078 | 0.073* | |
| C8 | 0.2590 (4) | −0.1780 (4) | −0.1502 (3) | 0.0547 (8) | |
| H8 | 0.2475 | −0.2850 | −0.1759 | 0.066* | |
| C9 | 0.5360 (4) | 0.4721 (3) | 0.2616 (3) | 0.0553 (8) | |
| H9 | 0.4989 | 0.5015 | 0.1868 | 0.066* | |
| C10 | 0.6988 (4) | 0.4744 (3) | 0.4420 (3) | 0.0457 (7) | |
| C11 | 0.5243 (4) | 0.3304 (3) | 0.4066 (3) | 0.0431 (7) | |
| C12 | 0.4831 (4) | 0.2208 (4) | 0.4834 (3) | 0.0533 (8) | |
| H12 | 0.3658 | 0.1250 | 0.4595 | 0.064* | |
| C13 | 0.6236 (5) | 0.2602 (4) | 0.5962 (3) | 0.0643 (9) | |
| H13 | 0.6016 | 0.1883 | 0.6493 | 0.077* | |
| C14 | 0.7974 (4) | 0.4042 (4) | 0.6334 (3) | 0.0658 (9) | |
| H14 | 0.8887 | 0.4272 | 0.7112 | 0.079* | |
| C15 | 0.8376 (4) | 0.5132 (4) | 0.5586 (3) | 0.0571 (8) | |
| H15 | 0.9542 | 0.6104 | 0.5848 | 0.069* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0420 (13) | 0.0347 (13) | 0.0527 (15) | 0.0176 (11) | 0.0083 (12) | 0.0057 (11) |
| N2 | 0.0515 (16) | 0.0387 (14) | 0.0686 (17) | 0.0166 (12) | 0.0115 (14) | 0.0092 (13) |
| N3 | 0.0420 (13) | 0.0337 (13) | 0.0507 (14) | 0.0156 (10) | 0.0099 (11) | 0.0066 (11) |
| N4 | 0.0621 (16) | 0.0388 (14) | 0.0582 (16) | 0.0194 (12) | 0.0095 (13) | 0.0112 (13) |
| O1 | 0.0680 (15) | 0.0786 (17) | 0.1061 (19) | 0.0124 (12) | 0.0326 (14) | 0.0224 (15) |
| O2 | 0.0832 (17) | 0.0511 (14) | 0.112 (2) | 0.0364 (12) | 0.0122 (15) | 0.0216 (13) |
| C1 | 0.0415 (16) | 0.0475 (17) | 0.0584 (19) | 0.0239 (14) | 0.0094 (14) | 0.0044 (15) |
| C2 | 0.0492 (18) | 0.0466 (18) | 0.0557 (19) | 0.0133 (14) | 0.0107 (15) | 0.0159 (16) |
| C3 | 0.0386 (15) | 0.0358 (15) | 0.0510 (18) | 0.0164 (12) | 0.0035 (13) | 0.0060 (14) |
| C4 | 0.0328 (14) | 0.0348 (15) | 0.0471 (17) | 0.0155 (12) | 0.0047 (13) | 0.0068 (13) |
| C5 | 0.0533 (18) | 0.0411 (17) | 0.066 (2) | 0.0240 (14) | 0.0182 (16) | 0.0187 (16) |
| C6 | 0.064 (2) | 0.061 (2) | 0.065 (2) | 0.0281 (17) | 0.0258 (17) | 0.0230 (17) |
| C7 | 0.060 (2) | 0.062 (2) | 0.057 (2) | 0.0295 (17) | 0.0209 (16) | 0.0093 (17) |
| C8 | 0.0478 (17) | 0.0402 (17) | 0.063 (2) | 0.0208 (14) | 0.0067 (15) | 0.0011 (15) |
| C9 | 0.062 (2) | 0.0343 (16) | 0.064 (2) | 0.0220 (15) | 0.0160 (17) | 0.0118 (15) |
| C10 | 0.0442 (16) | 0.0380 (16) | 0.0522 (18) | 0.0220 (14) | 0.0138 (14) | 0.0040 (14) |
| C11 | 0.0411 (16) | 0.0410 (16) | 0.0460 (17) | 0.0220 (13) | 0.0142 (13) | 0.0049 (13) |
| C12 | 0.0458 (18) | 0.0543 (19) | 0.058 (2) | 0.0212 (15) | 0.0230 (16) | 0.0138 (16) |
| C13 | 0.066 (2) | 0.077 (2) | 0.056 (2) | 0.0363 (19) | 0.0247 (18) | 0.0244 (18) |
| C14 | 0.056 (2) | 0.084 (3) | 0.055 (2) | 0.037 (2) | 0.0141 (17) | 0.0121 (19) |
| C15 | 0.0462 (18) | 0.056 (2) | 0.057 (2) | 0.0235 (15) | 0.0120 (16) | −0.0021 (16) |
Geometric parameters (Å, °)
| N1—C9 | 1.353 (3) | C4—C5 | 1.382 (4) |
| N1—C11 | 1.385 (3) | C5—C6 | 1.373 (4) |
| N1—C1 | 1.445 (3) | C5—H5 | 0.9300 |
| N2—C9 | 1.308 (3) | C6—C7 | 1.389 (4) |
| N2—C10 | 1.389 (3) | C6—H6 | 0.9300 |
| N3—C2 | 1.356 (3) | C7—C8 | 1.364 (4) |
| N3—C4 | 1.383 (3) | C7—H7 | 0.9300 |
| N3—C1 | 1.450 (3) | C8—H8 | 0.9300 |
| N4—C2 | 1.312 (4) | C9—H9 | 0.9300 |
| N4—C3 | 1.393 (3) | C10—C11 | 1.390 (4) |
| O1—H1E | 0.8499 | C10—C15 | 1.392 (4) |
| O1—H1F | 0.8500 | C11—C12 | 1.386 (4) |
| O2—H2C | 0.8500 | C12—C13 | 1.371 (4) |
| O2—H2D | 0.8500 | C12—H12 | 0.9300 |
| C1—H1A | 0.9700 | C13—C14 | 1.384 (4) |
| C1—H1B | 0.9700 | C13—H13 | 0.9300 |
| C2—H2 | 0.9300 | C14—C15 | 1.364 (4) |
| C3—C8 | 1.384 (4) | C14—H14 | 0.9300 |
| C3—C4 | 1.400 (3) | C15—H15 | 0.9300 |
| C9—N1—C11 | 106.2 (2) | C5—C6—H6 | 119.3 |
| C9—N1—C1 | 126.7 (2) | C7—C6—H6 | 119.3 |
| C11—N1—C1 | 127.1 (2) | C8—C7—C6 | 122.0 (3) |
| C9—N2—C10 | 103.8 (2) | C8—C7—H7 | 119.0 |
| C2—N3—C4 | 106.7 (2) | C6—C7—H7 | 119.0 |
| C2—N3—C1 | 126.1 (2) | C7—C8—C3 | 117.7 (3) |
| C4—N3—C1 | 127.1 (2) | C7—C8—H8 | 121.2 |
| C2—N4—C3 | 104.4 (2) | C3—C8—H8 | 121.2 |
| H1E—O1—H1F | 109.6 | N2—C9—N1 | 114.4 (3) |
| H2C—O2—H2D | 108.3 | N2—C9—H9 | 122.8 |
| N1—C1—N3 | 112.1 (2) | N1—C9—H9 | 122.8 |
| N1—C1—H1A | 109.2 | N2—C10—C11 | 110.6 (2) |
| N3—C1—H1A | 109.2 | N2—C10—C15 | 129.4 (3) |
| N1—C1—H1B | 109.2 | C11—C10—C15 | 120.0 (3) |
| N3—C1—H1B | 109.2 | N1—C11—C12 | 133.1 (2) |
| H1A—C1—H1B | 107.9 | N1—C11—C10 | 105.0 (2) |
| N4—C2—N3 | 113.8 (3) | C12—C11—C10 | 122.0 (3) |
| N4—C2—H2 | 123.1 | C13—C12—C11 | 116.8 (3) |
| N3—C2—H2 | 123.1 | C13—C12—H12 | 121.6 |
| C8—C3—N4 | 130.0 (3) | C11—C12—H12 | 121.6 |
| C8—C3—C4 | 120.0 (3) | C12—C13—C14 | 121.8 (3) |
| N4—C3—C4 | 109.9 (2) | C12—C13—H13 | 119.1 |
| C5—C4—N3 | 132.7 (2) | C14—C13—H13 | 119.1 |
| C5—C4—C3 | 122.1 (3) | C15—C14—C13 | 121.5 (3) |
| N3—C4—C3 | 105.1 (2) | C15—C14—H14 | 119.3 |
| C6—C5—C4 | 116.7 (3) | C13—C14—H14 | 119.3 |
| C6—C5—H5 | 121.7 | C14—C15—C10 | 117.9 (3) |
| C4—C5—H5 | 121.7 | C14—C15—H15 | 121.0 |
| C5—C6—C7 | 121.4 (3) | C10—C15—H15 | 121.0 |
| C9—N1—C1—N3 | 98.4 (3) | N4—C3—C8—C7 | 177.7 (3) |
| C11—N1—C1—N3 | −80.0 (3) | C4—C3—C8—C7 | −0.9 (4) |
| C2—N3—C1—N1 | 96.9 (3) | C10—N2—C9—N1 | 0.0 (3) |
| C4—N3—C1—N1 | −79.1 (3) | C11—N1—C9—N2 | −0.2 (3) |
| C3—N4—C2—N3 | 0.1 (3) | C1—N1—C9—N2 | −178.9 (2) |
| C4—N3—C2—N4 | 0.2 (3) | C9—N2—C10—C11 | 0.3 (3) |
| C1—N3—C2—N4 | −176.5 (2) | C9—N2—C10—C15 | −178.7 (3) |
| C2—N4—C3—C8 | −179.1 (3) | C9—N1—C11—C12 | 179.9 (3) |
| C2—N4—C3—C4 | −0.4 (3) | C1—N1—C11—C12 | −1.5 (4) |
| C2—N3—C4—C5 | 177.7 (3) | C9—N1—C11—C10 | 0.4 (3) |
| C1—N3—C4—C5 | −5.6 (4) | C1—N1—C11—C10 | 179.1 (2) |
| C2—N3—C4—C3 | −0.4 (3) | N2—C10—C11—N1 | −0.5 (3) |
| C1—N3—C4—C3 | 176.2 (2) | C15—C10—C11—N1 | 178.6 (2) |
| C8—C3—C4—C5 | 1.0 (4) | N2—C10—C11—C12 | 180.0 (2) |
| N4—C3—C4—C5 | −177.9 (2) | C15—C10—C11—C12 | −0.9 (4) |
| C8—C3—C4—N3 | 179.4 (2) | N1—C11—C12—C13 | −179.8 (3) |
| N4—C3—C4—N3 | 0.5 (3) | C10—C11—C12—C13 | −0.4 (4) |
| N3—C4—C5—C6 | −177.8 (3) | C11—C12—C13—C14 | 1.2 (4) |
| C3—C4—C5—C6 | 0.1 (4) | C12—C13—C14—C15 | −0.6 (5) |
| C4—C5—C6—C7 | −1.3 (4) | C13—C14—C15—C10 | −0.8 (4) |
| C5—C6—C7—C8 | 1.4 (5) | N2—C10—C15—C14 | −179.6 (3) |
| C6—C7—C8—C3 | −0.3 (4) | C11—C10—C15—C14 | 1.5 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1E···N2i | 0.85 | 1.99 | 2.841 (3) | 178. |
| O1—H1F···O1ii | 0.85 | 2.41 | 2.911 (5) | 118. |
| O2—H2C···N4iii | 0.85 | 2.23 | 3.083 (3) | 176. |
| O2—H2D···N4iv | 0.85 | 2.12 | 2.968 (3) | 177. |
| C2—H2···O1ii | 0.93 | 2.39 | 3.299 (4) | 167 |
| C9—H9···O2i | 0.93 | 2.56 | 3.363 (4) | 145 |
| C12—H12···O1ii | 0.93 | 2.57 | 3.495 (4) | 173 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x, −y, −z+1; (iii) x+1, y+1, z+1; (iv) −x+1, −y, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: VM2104).
References
- Abrahams, B. F., Hoskins, B. F., Robson, R. & Slizys, D. A. (2002). CrystEngComm, 4, 478–482.
- Bruker (2002). SADABS, SMART and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chang, Q., Meng, X. R., Song, Y. L. & Hou, H. W. (2005). Inorg. Chim. Acta, 358, 2117–2124.
- Fan, J., Sun, W. Y., Okamura, T., Zheng, Y. Q., Sui, B., Tang, W. X. & Ueyama, N. (2004). Cryst. Growth Des. 4, 579–584.
- Jin, S. W. & Chen, W. Z. (2007). Inorg. Chim. Acta, 12, 3756–3764.
- Lavandera, J. L., Cabildo, P. & Claramunt, R. M. (1988). J. Heterocycl. Chem. 25, 771–778.
- Ma, J. F., Yang, J., Zheng, G. L., Li, L. & Liu, J. F. (2003). Inorg. Chem. 42, 7531–7534. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wen, L. L., Li, Y. Z., Lu, Z. D., Lin, J. G., Duan, C. Y. & Meng, Q. J. (2006). Cryst. Growth Des. 6, 530–537.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811027966/vm2104sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811027966/vm2104Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811027966/vm2104Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report