Abstract
Heme and chlorophyll accumulate to high levels in legume root nodules and in photosynthetic tissues, respectively, and they are both derived from the universal tetrapyrrole precursor δ-aminolevulinic acid (ALA). The first committed step in ALA and tetrapyrrole synthesis is catalyzed by glutamyl-tRNA reductase (GTR) in plants. A soybean (Glycine max) root-nodule cDNA encoding GTR was isolated by complementation of an Escherichia coli GTR-defective mutant for restoration of ALA prototrophy. Gtr mRNA was very low in uninfected roots but accumulated to high levels in root nodules. The induction of Gtr mRNA in developing nodules was subsequent to that of the gene Enod2 (early nodule) and coincided with leghemoglobin mRNA accumulation. Genomic analysis revealed two Gtr genes, Gtr1 and a 3′ portion of Gtr2, which were isolated from the soybean genome. RNase-protection analysis using probes specific to Gtr1 and Gtr2 showed that both genes were expressed, but Gtr1 mRNA accumulated to significantly higher levels. In addition, the qualitative patterns of expression of Gtr1 and Gtr2 were similar to each other and to total Gtr mRNA in leaves and nodules of mature plants and etiolated plantlets. The data indicate that Gtr1 is universal for tetrapyrrole synthesis and that a Gtr gene specific for a tissue or tetrapyrrole is unlikely. We suggest that ALA synthesis in specialized root nodules involves an altered spatial expression of genes that are otherwise induced strongly only in photosynthetic tissues of uninfected plants.
Soybean (Glycine max) and numerous other legumes can establish a symbiosis with rhizobia, resulting in the formation of root nodules comprising specialized plant and bacterial cells (for review, see Mylona et al., 1995). Rhizobia reduce atmospheric nitrogen to ammonia within nodules, which is assimilated by the plant host to fulfill its nutritional nitrogen requirement. The high energy requirement for nitrogen fixation necessitates efficient respiration by the prokaryote within the microaerobic milieu of the nodule. The plant host synthesizes a nodule-specific hemoglobin (leghemoglobin) that serves to facilitate oxygen diffusion to the bacterial endosymbiont and to buffer the free oxygen concentration at a low tension (for review, see Appleby, 1992). Both of these functions require that the hemoglobin concentration be high, and, indeed, it exceeds 1 mm in soybean nodules (Appleby, 1984) and is the predominant plant protein in that organ. Once thought to be confined to legume nodules, hemoglobins are found throughout the plant kingdom, and leghemoglobin likely represents a specialization of a general plant phenomenon (for review, see Hardison, 1996). A gene encoding a nonsymbiotic hemoglobin has been identified in soybean and other legumes (Andersson et al., 1996); therefore, expression in nodules involves the specific activation of a subset of genes within a gene family. Leghemoglobin genes may have arisen from gene duplication, followed by specialization (Andersson et al., 1996).
Hemes and chlorophyll are tetrapyrroles synthesized from common precursors; chlorophyll is quantitatively the major tetrapyrrole in plants, with heme and other tetrapyrroles being present in minor amounts. Legume root nodules represent an exception, in which heme is synthesized in high quantity in the absence of chlorophyll, thus requiring the activity of enzymes not normally expressed highly in nonphotosynthetic tissues. Heme is synthesized from the universal tetrapyrrole precursor ALA by seven successive enzymatic steps; chlorophyll formation diverges after the synthesis of protoporphyrin, the immediate heme precursor (for review, see O'Brian, 1996). Biochemical and genetic evidence shows that soybean heme biosynthesis genes are strongly induced in root nodules (Sangwan and O'Brian, 1991, 1992, 1993; Madsen et al., 1993; Kaczor et al., 1994; Frustaci et al., 1995; Santana et al., 1998), and immunohistochemical studies demonstrate that induction is concentrated in infected nodule cells (Santana et al., 1998).
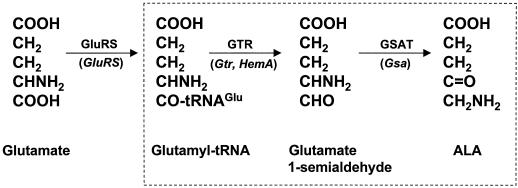
ALA is synthesized from Glu in plants by a three-step mechanism called the C5 pathway (Fig. 1); the latter two steps are committed to ALA synthesis and are catalyzed by GTR and GSAT, respectively (for review, see Beale and Weinstein, 1990; Jahn et al., 1991). Plant cDNA or genes encoding GTR (Gtr, also called HemA) and GSAT (Gsa) have been identified in several plant species (Grimm, 1990; Sangwan and O'Brian, 1993; Hofgen et al., 1994; Ilag et al., 1994; Frustaci et al., 1995; Wenzlau and Berry-Lowe, 1995; Bougri and Grimm, 1996; Kumar et al., 1996; Tanaka et al., 1996). Two genes for each enzyme have been described, and some genes are reported to be specific to a tissue, tetrapyrrole, or light regimen (Bougri and Grimm, 1996; Kumar et al., 1996; Tanaka et al., 1996). However, soybean Gsa1 is highly expressed in both leaves and nodules and contains a cis-acting element in its promoter that binds to a nuclear factor found in both tissues. (Frustaci et al., 1995). In this study we isolated soybean Gtr1 and characterized the genetic basis of GTR expression in root nodules.
Figure 1.
C5 pathway for ALA synthesis. The committed steps for ALA synthesis catalyzed by GTR and GSAT are boxed. Glutamyl-tRNA synthetase (GluRS) and glutamyl-tRNAGlu also participate in protein synthesis. The gene designations in plants are shown in parentheses below the arrows.
MATERIALS AND METHODS
Bacteria and Plants
Escherichia coli strain EV149 is an ALA auxotroph caused by a mutation in the hemA gene encoding GTR (Verkamp et al., 1993; provided by Dr. D. Söll, Yale University, New Haven, CT). It was grown in Luria broth or M9 medium (Ausubel et al., 1994) containing 50 μg mL−1 ALA and also with 50 to 100 μg mL−1 ampicillin when harboring cDNA library clones. Bradyrhizobium japonicum strain I110 was the soybean symbiont used in the present work and was cultured in glycerol-salts-yeast extract medium (Frustaci et al., 1991). We used soybean (Glycine max) cv Essex, an inbred isoline (Lorenzen et al., 1995), in the present work. Plants were either inoculated with B. japonicum or uninoculated and grown in a growth chamber under a 16-h light/8-h dark regimen at 25°C. Etiolated soybean plants were grown in total darkness for 10 d, and either left in the dark or exposed to direct light to green for the final 24 h before the leaves were harvested for RNA isolation.
Isolation of cDNA and Genomic DNA Encoding GTR
Soybean nodule and leaf cDNA expression libraries in pUC18 were gifts from Dr. M.L. Kahn (Washington State University, Pullman) and were constructed as described previously (Udvardi and Kahn, 1991). Each library was used to transform E. coli strain EV149, and cells were plated on M9 medium containing 100 μg mL−1 ampicillin in the absence of ALA. Prototrophic colonies were cultured, plasmids were isolated, and the DNA was then used to retransform strain EV149 to confirm that prototrophy was conferred by the plasmid rather than by a spontaneous genomic event. The clones were initially compared by analysis of restriction digests, and the DNA sequences of both strands of selected clones were determined.
Genomic DNA encoding Gtr1 was obtained by PCR using primers that delimited the GTR-encoding leaf cDNA clone and EcoRI-digested genomic DNA as the template. The resulting 3.6-kb DNA was sequenced, and introns were identified by comparing the genomic and cDNA sequences. A portion of Gtr2 was obtained by PCR using primers delimiting the 3′ end 610 bp from the unique EcoRV site to the end of the cloned region of Gtr1. The template was genomic DNA enriched for a 2-kb fragment that hybridized to Gtr cDNA in Southern analysis. EcoRV-digested genomic DNA was size-fractionated using a 1 to 5 m NaCl gradient, as described previously (O'Brian and Maier, 1987). Fractions of 0.5 mL were analyzed by Southern blotting to determine those enriched for either the 1- or 2-kb homologous fragment. Gtr2 was found in the 2-kb fraction, whereas Gtr1 was found in the 1-kb fraction. Errors in PCR were ruled out as the basis for differences in DNA sequence between the respective portions of Gtr1 and Gtr2 by sequencing DNA from three independent PCR reactions. In addition, RNase-protection analysis revealed differences in Gtr transcripts based on sequence variations (see below). Sequence analysis was carried out using Genetics Computer Group (Madison, WI) software (Devereaux et al., 1984).
Analysis of RNA
Isolation of RNA from leaves, roots, and nodules and preparation of poly(A+) RNA were carried out as described previously (Sangwan and O'Brian, 1993). RNA-blot analysis was carried out with poly(A)+ RNA under high-stringency conditions. Gtr1 and Gtr2 mRNAs were analyzed by RNase-protection analysis using a kit (Hybspeed, Ambion, Austin, TX). The protocol used took advantage of differences in the RNA sequence between the two genes by digesting unpaired nucleotides in imperfect RNA hybrids. Antisense probes of 100 and 98 bp, complementary to Gtr1 and Gtr2 mRNA, respectively, were prepared using an in vitro transcription kit (MAXIscript, Ambion) according to the manufacturer's instructions. The specificity of each antisense probe for the cognate mRNA was established by RNase-protection analysis using in vitro-synthesized complementary sense-strand RNAs.
Conditions in which each antisense RNA would form an RNase-sensitive duplex of an imperfect hybrid and a stable duplex with a perfect hybrid were as follows. Hybridizations were carried out overnight at 47°C in hybridization buffer described by Ausubel et al. (1994) rather than the buffer provided in the kit. Hybridized RNA was digested for 45 min at 30°C with 20 units of RNase T1 and 20 units of RNase A. Then, 8 × 104 cpm of probe was used in each reaction, and products were analyzed as autoradiograms of 7.5% acrylamide gels. Gtr1 and Gtr2 mRNAs were analyzed using antisense probes of almost the same size and of the same specific activity and analyzed on the same gels. Therefore, the relative amounts of each transcript in tissues could be assessed. Autoradiogram bands were quantified using an imaging densitometer (model GS-700, Bio-Rad) in the transmittance mode and the Molecular Analyst software package. Several exposures were analyzed to quantitations made in the linear region of the densitometer.
RESULTS
Isolation of Soybean cDNA Encoding GTR
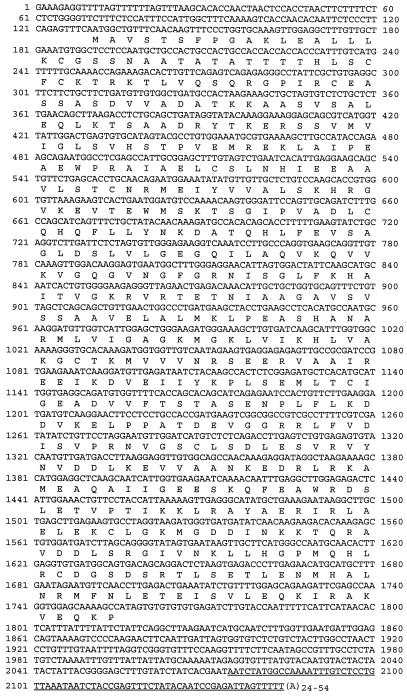
E. coli strain EV149 is defective in hemA, the gene encoding GTR, and behaves as an ALA auxotroph (Verkamp et al., 1993). To isolate soybean-nodule cDNA encoding GTR, strain EV149 was transformed en masse with a soybean-nodule cDNA expression library, and cells that were functionally complemented were selected as ampicillin-resistant, ALA-prototrophic colonies on agar medium. Eight complementing plasmids had identical restriction-enzyme patterns, and partial DNA sequencing of the 3′ ends of the clones revealed identical sequences, with variation only in the length of the polyadenylated tail. One clone, pGTRN1, was chosen for further analysis. The insert of pGTRN1 contained a 1629-bp open reading frame that encoded a peptide 542 amino acids in length beginning with a Met codon (Fig. 2). In addition, a termination codon was identified upstream of the Met codon and in the same reading frame, showing that the entire coding region was present in the cloned cDNA. This peptide was highly homologous to GTRs from other plants, with the highest identity to that from cucumber (83%; Tanaka et al., 1996). This homology, along with complementation of the E. coli hemA mutant, provides strong evidence that the cloned cDNA encodes GTR.
Figure 2.
Nucleotide sequence and deduced product of cDNA encoding soybean GTR. The underlined nucleotides denote the sequence found in a leaf cDNA clone. The deduced protein shares 83% identity with GTR from cucumber.
The gene corresponding to the complementing clone was designated Gtr1. A single complementing clone was isolated from a leaf cDNA library using the same selection procedure described for the nodule library. The cDNA sequence was identical to pGTRN1 except that an additional 70 bp was found immediately prior to the poly(A+) tail (underlined sequence in Fig. 2). The sequence variation likely arose from differential processing of Gtr1 mRNA rather than from transcription of two genes (see below). RNase-protection analysis of leaf and nodule mRNA using an antisense probe corresponding to the 70-bp region showed that the additional sequence was not leaf specific and was present as a minor species (data not shown). The basis for this variation was not studied further.
Gtr Is Induced in Root Nodules
Hemoglobin synthesis is highly induced in root nodules, as is Glu-dependent ALA-synthesis activity and Gsa expression (Sangwan and O'Brian, 1991, 1992, 1993; Frustaci et al., 1995). To determine the expression pattern of Gtr, RNA-blot analysis was performed on poly(A+) RNA from various tissues from soybean plants using a portion of the Gtr cDNA as a probe. Gtr mRNA accumulated to very low levels in uninfected roots but was strongly expressed in root nodules to a level somewhat lower than was observed in leaves from the same plants (Fig. 3A). These observations indicate that, like Gsa (Sangwan and O'Brian, 1993; Frustaci et al., 1995; also see Fig. 3A), induction of ALA synthesis is correlated with the activation of Gtr.
Figure 3.

Northern analysis of Gtr mRNA from soybean tissues. A, Poly(A)+ RNA (approximately 5 μg) was analyzed from leaves (L), roots (R), and nodules (N) of 24-d-old plants. A single filter was probed separately with radiolabeled cDNA from Gtr, Gsa, and ubiquitin (Ubi), and the filter was stripped after each hybridization and exposure. Ubiquitin was used as a control for a constitutively expressed gene. B, Leaves from illuminated (I) or dark-treated (D) etiolated plantlets were analyzed for Gtr, Cab, and Ubi mRNA. Cab was used as a control for a light-regulated gene.
Evidence indicates that ALA synthesis is induced by light in photosynthetic tissues of some plants, at least in part because of induction of the Gtr (HemA) gene (Ilag et al., 1994; Bougri and Grimm, 1996; Tanaka et al., 1996). To assess the light requirement for soybean Gtr mRNA expression in leaves, RNA-blot analysis was carried out with poly(A+) from leaves of etiolated plants grown completely in the dark or those exposed to light for 24 h prior to harvesting (Fig. 3B). Cab (chlorophyll a/b-binding protein) was used as a control for a light-regulated gene (Chang and Walling, 1992). Gtr transcripts accumulated to high levels in etiolated leaves. Exposure to light resulted in an approximately 3-fold increase in expression (see also Fig. 6). Thus, although Gtr mRNA is modestly light induced, light is not required for expression.
Figure 6.
Analysis of Gtr1 and Gtr2 mRNA. A, Sequence comparison of a 3′ portion of Gtr1 and Gtr2 DNA. The sequence of Gtr1 is shown, with differences in Gtr2 shown below it. “X” denotes a gap in one sequence where a nucleotide is present in the other. The underlined sequence denotes the antisense riboprobe used to analyze Gtr1 (probe 1) in B and C. The same region was used as a probe for Gtr2 (probe 2), except that it contained the nucleotide additions, deletions, and substitutions noted in the figure. B, Antisense probe 1 and probe 2 are specific for Gtr1 and Gtr2 mRNA, respectively. RNA sense strand 1 (S1) and sense strand 2 (S2) are identical to portions of Gtr1 and Gtr2 mRNA, respectively, and were synthesized in vitro and used in RNase-protection assays with radiolabeled antisense probes 1 and 2. Each probe formed an RNase-resistant duplex with the perfectly complementary hybrid only. C, RNase-protection analysis of RNA from leaves (L) and nodules (N) of 24-d-old plants and from leaves of illuminated (I) and dark-treated (D) etiolated plantlets using riboprobes specific to Gtr1 or Gtr2 (probes 1 and 2, respectively). The intensities of bands in the first two rows can be directly compared. Gtr2 (long) is a longer exposure of the autoradiogram above it, which allows a better comparison of Gtr2 between tissues.
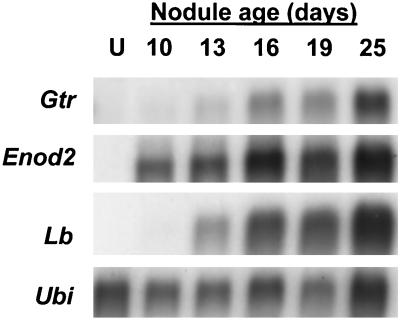
Root-nodule ontogeny is broadly divided into early and late development, with the latter stage commencing with the onset of nitrogen fixation. We compared the temporal expression of Gtr with those of the nodule-specific genes Enod2 and Lb, which are well-described markers of early and late development, respectively (for review, see Mylona et al., 1995). RNA-blot analysis showed that Enod2 mRNA was not detected in uninfected roots but was found by 10 d postinfection (Fig. 4). Gtr mRNA was weakly expressed in 13-d-old nodules and easily discerned by 16 d and, therefore, does not correspond well with early development. However, the temporal pattern of Gtr expression correlated well with that of Lb, which encodes nodule hemoglobin, and therefore Gtr is likely to be activated later in nodule development when needed for high levels of heme synthesis.
Figure 4.
Temporal expression of Gtr mRNA in developing nodules and comparison with Enod2 and Lb. Approximately 5 μg of poly(A+) RNA from uninfected roots (U) and from nodules 10, 13, 16, 19, and 25 d postinfection were loaded onto each lane. A single filter was hybridized with each radiolabeled cDNA separately, and the filter was stripped after each hybridization and exposure.
Isolation of Genomic DNA Encoding Gtr1 and Evidence for a Second Gtr Gene
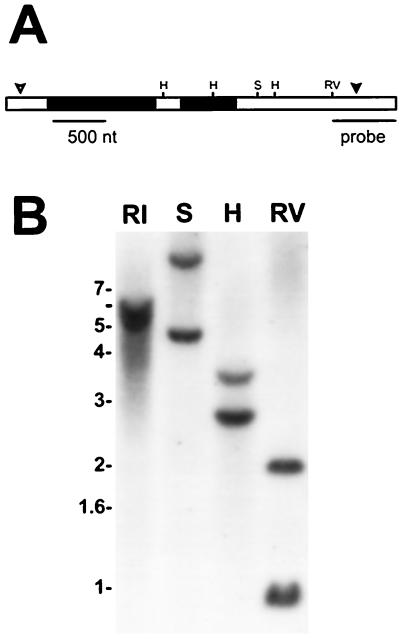
Genomic DNA was isolated by PCR using primers that delimited the cDNA sequence, and the DNA sequence was determined. The cloned gene contained two introns 1007 and 513 bp in size (Fig. 5A), which is much larger than the introns found in the Gtr1 (HemA1) gene of Arabidopsis (Ilag et al., 1994). The exon sequences were identical to the corresponding sequence in the cDNA, indicating that the mRNA from which the cDNA was synthesized is a transcript of the identified gene. To determine whether more than one Gtr gene was present in the soybean genome, Southern analysis of genomic DNA was carried out using restriction enzymes and a radiolabeled probe that would yield only one fragment if only Gtr were present (Fig. 5B). Each digested DNA sample yielded two bands of approximately equal intensity, indicating the presence of two Gtr genes that are very homologous, at least in the region corresponding to the probe.
Figure 5.
Gene structure of Gtr1 and evidence for two Gtr genes. A, Representation of Gtr1 showing three exons (white bars) and two introns (black bars). Restriction sites are shown for HindIII (H), SphI (S), and EcoRV (RV). The open and closed arrowheads denote the translation start and termination sites, respectively. The probe used for the Southern analysis in B is shown. B, Southern analysis of soybean DNA cut with EcoRI (RI), SphI (S), HindIII (H), or EcoRV (RV).
To further investigate whether there were two Gtr genes, DNA corresponding to the 3′ end of Gtr was amplified from size-fractionated EcoRV-digested genomic DNA enriched for either the 1- or 2-kb fragment that hybridized to the probe (Fig. 5B). DNA sequencing revealed that the 1-kb EcoRV fragment corresponded to Gtr1, whereas the 2-kb fragment contained a sequence highly related but not identical to Gtr1 (Fig. 6A), indicating the existence of a second gene tentatively named Gtr2. The isolated 3′ portion of Gtr2 was different from Gtr1 in 5.3% of the nucleotides and comprised substitutions, deletions, and additions. The high degree of relatedness of the two genes suggests a gene-duplication event.
Soybean Gtr1 Is Universal for Tetrapyrrole Synthesis
The identification of two Gtr genes raises the possibility that high expression in root nodules involves activation of a gene specific for that tissue or for heme synthesis in general. To examine the expression of the two genes, RNase-protection analysis of RNA from various tissues was carried out using probes specific to Gtr1 or Gtr2 (Fig. 6, B and C). In addition, the experiments were carried out so that the relative quantity Gtr1 and Gtr2 mRNA in each tissue could be determined by the intensity of the bands on autoradiograms (see Methods). Gtr1 mRNA was 15- to 20-fold more abundant than the Gtr2 message in leaves and nodules of 24-d-old plants and etiolated plantlets (Fig. 6C). Thus, if Gtr2 is a functional gene, it can account for only a minor portion of total Gtr expression. In addition, the qualitative patterns of expression of the two genes were similar, with the highest levels found in leaves from mature plants and light-exposed etiolated plantlets and lesser but significant levels found in nodules and leaves of dark-treated etiolated plants. Therefore, strong expression of Gtr in different tissues or light conditions cannot be explained by differential expression of Gtr1 and Gtr2. Finally, the expression pattern of each gene was similar to that for total Gtr expression, as discerned by northern analysis (Fig. 3). The data indicate that Gtr1 is expressed both in nodules for heme synthesis and in leaves, where chlorophyll is the predominant tetrapyrrole, and the findings argue against significant expression of a Gtr gene specific for a tissue, tetrapyrrole, or light condition.
DISCUSSION
We isolated a soybean Gtr gene that is strongly induced in symbiotic root nodules. Gsa1 is induced similarly (Sangwan and O'Brian, 1993; Frustaci et al., 1995), and thus ALA synthesis correlates with the activation of both committed steps of the C5 pathway at the mRNA level. Heterogeneity in Gtr mRNA could be attributed to the presence of two transcribed genes, Gtr1 and Gtr2, and the likely differential processing of Gtr1 transcripts at the 3′ end. The temporal expression of Gtr in developing nodules was correlated with that of the soybean hemoglobin gene Lb, and thus regulation of Gtr is likely to be coordinated with nodule function rather than with the early stages of nodule development.
ALA formation in symbiotic root nodules is unique among plants in that synthesis is high but none is incorporated into chlorophyll. In addition, synthesis is induced in response to parameters associated with symbiosis rather than photosynthesis, suggesting that there should be fundamental differences in regulation in these two contexts. In cucumber a Gtr (HemA) gene specific for chlorophyll and one for nonchlorophyll tetrapyrroles was proposed based on the light-dependent regulation of the former (Tanaka et al., 1996). However, soybean Gtr1 mRNA was strongly expressed in both leaves and nodules, and Gtr2 was expressed to a lesser extent in a qualitatively similar manner. In addition, Gtr1 mRNA accumulation did not require light in leaves; therefore, expression in subterranean nodules did not require a compensatory regulatory mechanism.
The data suggest that the soybean Gtr1 gene is activated in tissues where high levels of ALA are necessary for the synthesis of heme or chlorophyll, and the data argue against a Gtr gene specific for a tissue or tetrapyrrole. Analysis of the Gsa1 gene yielded essentially the same conclusion (Frustaci et al., 1995). Therefore, activation of the two committed steps of the C5 pathway during nodule development likely requires an altered spatial pattern of expression of genes normally induced strongly only in photosynthetic tissue. Evidence suggests that chlorophyll synthesis is coordinated with chloroplast development (Beator and Kloppstech, 1993). If so, then the high expression of Gtr1 in nodules and etiolated leaves indicates that ALA synthesis can be uncoupled from chloroplast development, and thus Gtr1 is likely to be affected by separate and independent signal transduction pathways.
The accession number for the Gtr1 gene sequence reported in this paper is AF105221.
ACKNOWLEDGMENTS
We thank Dr. M.L. Kahn for soybean nodule and leaf cDNA libraries, Dr. D. Söll for E. coli strain EV149, and Drs. T. Bisseling, K. Marcker, D.P.S. Verma, and L. Walling for soybean cDNA clones.
Abbreviations:
- ALA
δ-aminolevulinic acid
- GSA
glutamate 1-semialdehyde
- GSAT
glutamate 1-semialdehyde aminotransferase
- GTR
glutamyl-tRNA reductase
Footnotes
This work was supported by the Cooperative State Research Service, U.S. Department of Agriculture, under agreement no. 95-37305-2253.
LITERATURE CITED
- Andersson CR, Jensen EO, Llewellyn DJ, Dennis ES, Peacock WJ. A new hemoglobin gene from soybean: a role for hemoglobin in all plants. Proc Natl Acad Sci USA. 1996;93:5682–5687. doi: 10.1073/pnas.93.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleby CA (1984) Plant hemoglobin properties, function and genetic origin. In PW Ludden, JE Burris, eds, Nitrogen Fixation and CO2 Metabolism. Elsevier Science Publishing, New York, pp 41–51
- Appleby CA. The origins and functions of haemoglobins in plants. Sci Prog. 1992;76:365–398. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current Protocols in Molecular Biology. Wiley Interscience, New York
- Beale SI, Weinstein JD. Tetrapyrrole metabolism in photosynthetic orgnisms. In: Dailey HA, editor. Biosynthesis of Heme and Chlorophylls. Publishing Co, New York: McGraw-Hill; 1990. pp. 287–391. [Google Scholar]
- Beator J, Kloppstech K. The circadian oscillator coordinates the synthesis of apoproteins and their pigments during chloroplast development. Plant Physiol. 1993;103:191–196. doi: 10.1104/pp.103.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougri O, Grimm B. Members of a low-copy number gene family encoding glutamyl-tRNA reductase are differentially expressed in barley. Plant J. 1996;9:867–878. doi: 10.1046/j.1365-313x.1996.9060867.x. [DOI] [PubMed] [Google Scholar]
- Chang YC, Walling LL. Spatial and temporal expression of Cab mRNAs in cotyledons of the developing soybean seedling. Planta. 1992;186:262–272. doi: 10.1007/BF00196256. [DOI] [PubMed] [Google Scholar]
- Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frustaci JM, Sangwan I, O'Brian MR. Aerobic growth and respiration of a δ-aminolevulinic acid synthase (hemA) mutant of Bradyrhizobium japonicum. J Bacteriol. 1991;173:1145–1150. doi: 10.1128/jb.173.3.1145-1150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frustaci JM, Sangwan I, O'Brian MR. gsa1 is a universal tetrapyrrole synthesis gene in soybean and is regulated by a GAGA element. J Biol Chem. 1995;270:7387–7393. doi: 10.1074/jbc.270.13.7387. [DOI] [PubMed] [Google Scholar]
- Grimm B. Primary structure of a key enzyme in plant tetrapyrrole synthesis: glutamate 1-semialdehyde aminotransferase. Proc Natl Acad Sci USA. 1990;87:4169–4173. doi: 10.1073/pnas.87.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC. A brief history of hemoglobins: plant, animal, protist and bacteria. Proc Natl Acad Sci USA. 1996;93:5675–5679. doi: 10.1073/pnas.93.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofgen R, Axelsen KB, Kannangara CG, Schuttke I, Pohlenz H-D, Willmitzer L, Grimm B, von Wettstein D. A visible marker for antisense mRNA expression in plants: inhibition of chlorophyll synthesis with a glutamate-1-semialdehyde aminotransferase antisense gene. Proc Natl Acad Sci USA. 1994;91:1726–1730. doi: 10.1073/pnas.91.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilag LL, Kumar AM, Söll D. Light regulation of chlorophyll biosynthesis at the level of 5-aminolevulinate formation in Arabidopsis. Plant Cell. 1994;6:265–275. doi: 10.1105/tpc.6.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn D, Verkamp E, Söll D. Glutamyl-transfer RNA: a precursor of heme and chlorophyll biosynthesis. Trends Biochem Sci. 1991;17:215–218. doi: 10.1016/0968-0004(92)90380-r. [DOI] [PubMed] [Google Scholar]
- Kaczor CM, Smith MW, Sangwan I, O'Brian MR. Plant δ-aminolevulinic acid dehydratase. Expression in soybean root nodules and evidence for a bacterial lineage of the Alad gene. Plant Physiol. 1994;104:1411–1417. doi: 10.1104/pp.104.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AM, Csankovszki G, Söll D. A second and differentially expressed glutamyl-tRNA reductase gene from Arabidopsis thaliana. Plant Mol Biol. 1996;30:419–426. doi: 10.1007/BF00049321. [DOI] [PubMed] [Google Scholar]
- Lorenzen LL, Bourin S, Young N, Specht JE, Shoemaker RC. Soybean pedigree analysis using map-based molecular markers. I. Tracking RFLP markers in cultivars. Crop Sci. 1995;35:1326–1336. [Google Scholar]
- Madsen O, Sandal L, Sandal N, Marcker KA. A soybean coproporphyrinogen oxidase gene is highly expressed in root nodules. Plant Mol Biol. 1993;23:35–43. doi: 10.1007/BF00021417. [DOI] [PubMed] [Google Scholar]
- Mylona P, Pawlowski K, Bisseling T. Symbiotic nitrogen fixation. Plant Cell. 1995;7:869–885. doi: 10.1105/tpc.7.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brian MR. Heme synthesis in the rhizobium-legume symbiosis: a palette for bacterial and eukaryotic pigments. J Bacteriol. 1996;178:2471–2478. doi: 10.1128/jb.178.9.2471-2478.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brian MR, Maier RJ. Isolation of a cytochrome aa3 gene from Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1987;84:3219–3223. doi: 10.1073/pnas.84.10.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan I, O'Brian MR. Evidence for an inter-organismic heme biosynthetic pathway in symbiotic soybean root nodules. Science. 1991;251:1220–1222. doi: 10.1126/science.251.4998.1220. [DOI] [PubMed] [Google Scholar]
- Sangwan I, O'Brian MR. Characterization of δ-aminolevulinic acid formation in soybean root nodules. Plant Physiol. 1992;98:1074–1079. doi: 10.1104/pp.98.3.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan I, O'Brian MR. Expression of the soybean (Glycine max) glutamate 1-semialdehyde aminotransferase gene in symbiotic root nodules. Plant Physiol. 1993;102:829–834. doi: 10.1104/pp.102.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana MA, Pihakaski-Maunsbach K, Sandal N, Marcker KA, Smith AG. Evidence that the plant host synthesizes the heme moiety of leghemoglobin in root nodules. Plant Physiol. 1998;116:1259–1269. doi: 10.1104/pp.116.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Yoshida K, Nakayashiki T, Masuda T, Tsuji H, Inokuchi H, Tanaka A. Differential expression of two hemA mRNAs encoding glutamyl-tRNA reductase proteins in greening cucumber seedlings. Plant Physiol. 1996;110:1223–1230. doi: 10.1104/pp.110.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Kahn ML. Isolation and analysis of a cDNA clone that encodes an alfalfa (Medicago sativa) aspartate aminotransferase. Mol Gen Genet. 1991;231:97–105. doi: 10.1007/BF00293827. [DOI] [PubMed] [Google Scholar]
- Verkamp E, Bachman VM, Bjornsson JM, Söll D, Eggertsson G. The periplasmic dipeptide permease system transports 5-aminolevulinic acid in Escherichia coli. J Bacteriol. 1993;175:1452–1456. doi: 10.1128/jb.175.5.1452-1456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzlau JM, Berry-Lowe SL. Nucleotide sequence of a gene encoding glutamate 1-semialdehyde aminotransferase ( U10278) from Arabidopsis thaliana Columbia. Plant Physiol. 1995;108:1342. [Google Scholar]