Abstract
In the title compound, C18H23NO4S, the dihedral angle between the two aromatic rings is 29.14 (7)°. The S atom has a distorted tetrahedral geometry [106.15 (9)–119.54 (10)°]. The crystal structure exhibits weak C—H⋯O and π–π interactions.
Related literature
For the biological activity of sulfonamide derivatives, see: Chumakov et al. (2006 ▶); Kremer et al. (2006 ▶). For related structures, see: Khan et al. (2010 ▶); Sharif et al. (2010 ▶).
Experimental
Crystal data
C18H23NO4S
M r = 349.43
Monoclinic,
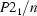
a = 5.7814 (4) Å
b = 13.9861 (12) Å
c = 21.9791 (18) Å
β = 92.949 (4)°
V = 1774.9 (2) Å3
Z = 4
Mo Kα radiation
μ = 0.20 mm−1
T = 295 K
0.30 × 0.24 × 0.20 mm
Data collection
Bruker Kappa APEXII diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.942, T max = 0.960
18724 measured reflections
3343 independent reflections
2614 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.040
wR(F 2) = 0.110
S = 1.04
3343 reflections
221 parameters
H-atom parameters constrained
Δρmax = 0.19 e Å−3
Δρmin = −0.30 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811029746/bt5588sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811029746/bt5588Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811029746/bt5588Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C12—H12⋯O1i | 0.93 | 2.54 | 3.452 (2) | 166 |
| C18—H18B⋯O2ii | 0.96 | 2.38 | 3.302 (3) | 160 |
Symmetry codes: (i) 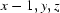 ; (ii)
; (ii)  .
.
Acknowledgments
The authors wish to acknowledge SAIF, IIT, Madras, for the data collection.
supplementary crystallographic information
Comment
Sulfonamide derivatives are extensively used in medicine as they possess a wide range of medicinal, pharmacological and antimicrobial properties (Chumakov et al., 2006, Kremer et al., 2006). We report the crystal structure of the titled compound (I) (Fig. 1).
In the title compound (I), the geometric pararameters are similar with the reported similar structures (Khan et al., 2010; Sharif et al., 2010). The S atom of the title molecule has a distorted tetrahedral geometry, with S1—O1 = 1.4210 (15), S1—O2 = 1.4195 (15), S1—N1 = 1.6391 (17), S1—C1 = 1.7538 (19) Å, O1—S1—O2 = 119.54 (10), O1—S1—N1 = 106.83 (9), O1—S1—C7 = 108.64 (9), O2—S1—N1 = 106.32 (9), O2—S1—C7 = 108.57 (9) and N1—S1—C7 = 106.15 (9)°.
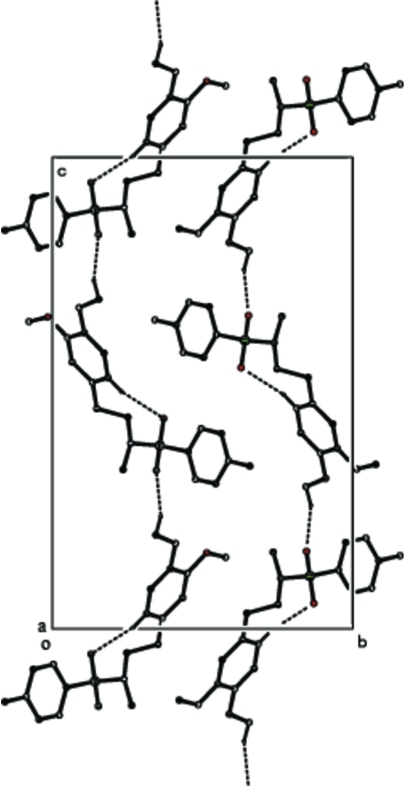
The dihedral angle between the two rings C1—C6 and C11—C16 is 29.14 (7)°. The crystal structure exhibits weak C—H···O (Fig.2 and Table 1) and π–π [Cg1···Cg2 (-x,2 - y,-z) distance of 5.2909 (13)Å and Cg2···Cg2 (-x,1 - y,-z) distance of 4.7146 (12) Å; Cg1 and Cg2 are the centroids of the rings C1—C6 and C11—C16, respectively] interactions.
Experimental
2-(3,4-dimethoxyphenyl)-N-methyl ethanamine (51 mmol) was dissolved in dichloromethane (20 ml) in a round bottom flask. To this, added triethylamine (10.2 mmol) with stirring for 5 minutes. Then 4-methylbenzene-1-sulfonyl chloride (51 mmol) was added into the reaction mass and heated to 50 °C for 6 hrs. After cooling the reaction mixture to the normal temperature, it was added to water (20 ml). The aqueous layer was separated. The ethyl acetate layer was washed twice with 10% sodium chloride solution. The organic layer was dried over 2 g of anhydrous sodium sulfate and filtered. The filtrate was evaporated under vacuum to isolate the crude compound. Recrystallization of the compound using ethyl acetate and hexane mixture yielded the diffraction quality crystals.
Refinement
All H atoms were positioned geometrically with C—H = 0.93–0.97 Å and allowed to ride on their parent atoms, with Uiso(H) = 1.2 or 1.5Ueq(C).
Figures
Fig. 1.
The molecular structure of (I), with atom labels and 30% probability displacement ellipsoids for non-H atoms.
Fig. 2.
The packing of (I), viewed down the a axis. H-bonds are shown as dashed lines; H atoms not involved in hydrogen bonding have been omitted.
Crystal data
| C18H23NO4S | F(000) = 744 |
| Mr = 349.43 | Dx = 1.308 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 6145 reflections |
| a = 5.7814 (4) Å | θ = 2.4–25.5° |
| b = 13.9861 (12) Å | µ = 0.20 mm−1 |
| c = 21.9791 (18) Å | T = 295 K |
| β = 92.949 (4)° | Block, colourless |
| V = 1774.9 (2) Å3 | 0.30 × 0.24 × 0.20 mm |
| Z = 4 |
Data collection
| Bruker Kappa APEXII diffractometer | 3343 independent reflections |
| Radiation source: fine-focus sealed tube | 2614 reflections with I > 2σ(I) |
| graphite | Rint = 0.036 |
| ω and φ scans | θmax = 25.8°, θmin = 2.4° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | h = −4→7 |
| Tmin = 0.942, Tmax = 0.960 | k = −16→17 |
| 18724 measured reflections | l = −26→26 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.110 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0471P)2 + 0.6472P] where P = (Fo2 + 2Fc2)/3 |
| 3343 reflections | (Δ/σ)max < 0.001 |
| 221 parameters | Δρmax = 0.19 e Å−3 |
| 0 restraints | Δρmin = −0.30 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.58829 (8) | 0.85799 (4) | 0.11159 (2) | 0.04660 (17) | |
| O1 | 0.6833 (3) | 0.87108 (11) | 0.05383 (7) | 0.0609 (4) | |
| O2 | 0.7388 (3) | 0.84655 (12) | 0.16418 (7) | 0.0663 (5) | |
| O3 | 0.0375 (3) | 0.48564 (11) | −0.16121 (7) | 0.0607 (4) | |
| O4 | −0.3215 (2) | 0.58964 (12) | −0.18094 (6) | 0.0612 (4) | |
| N1 | 0.4284 (3) | 0.76123 (11) | 0.10662 (7) | 0.0430 (4) | |
| C1 | 0.3980 (3) | 0.95272 (13) | 0.12431 (8) | 0.0425 (4) | |
| C2 | 0.3502 (4) | 0.97724 (15) | 0.18308 (9) | 0.0537 (5) | |
| H2 | 0.4308 | 0.9487 | 0.2160 | 0.064* | |
| C3 | 0.1823 (4) | 1.04423 (16) | 0.19236 (11) | 0.0628 (6) | |
| H3 | 0.1501 | 1.0610 | 0.2320 | 0.075* | |
| C4 | 0.0602 (4) | 1.08733 (15) | 0.14425 (11) | 0.0597 (6) | |
| C5 | 0.1148 (4) | 1.06328 (16) | 0.08611 (11) | 0.0615 (6) | |
| H5 | 0.0368 | 1.0929 | 0.0532 | 0.074* | |
| C6 | 0.2822 (4) | 0.99631 (15) | 0.07548 (9) | 0.0522 (5) | |
| H6 | 0.3167 | 0.9807 | 0.0358 | 0.063* | |
| C7 | −0.1298 (5) | 1.15787 (19) | 0.15501 (15) | 0.0872 (9) | |
| H7A | −0.0652 | 1.2209 | 0.1591 | 0.131* | |
| H7B | −0.2038 | 1.1412 | 0.1916 | 0.131* | |
| H7C | −0.2416 | 1.1565 | 0.1212 | 0.131* | |
| C8 | 0.3156 (4) | 0.73564 (17) | 0.16243 (10) | 0.0587 (6) | |
| H8A | 0.1778 | 0.7732 | 0.1656 | 0.088* | |
| H8B | 0.4194 | 0.7478 | 0.1971 | 0.088* | |
| H8C | 0.2755 | 0.6690 | 0.1613 | 0.088* | |
| C9 | 0.2771 (4) | 0.75162 (14) | 0.05110 (9) | 0.0492 (5) | |
| H9A | 0.3440 | 0.7863 | 0.0181 | 0.059* | |
| H9B | 0.1272 | 0.7796 | 0.0580 | 0.059* | |
| C10 | 0.2458 (4) | 0.64822 (14) | 0.03316 (9) | 0.0497 (5) | |
| H10A | 0.3961 | 0.6211 | 0.0256 | 0.060* | |
| H10B | 0.1841 | 0.6135 | 0.0669 | 0.060* | |
| C11 | 0.0868 (3) | 0.63423 (13) | −0.02258 (8) | 0.0408 (4) | |
| C12 | −0.1148 (3) | 0.68548 (15) | −0.03237 (9) | 0.0489 (5) | |
| H12 | −0.1572 | 0.7298 | −0.0034 | 0.059* | |
| C13 | −0.2558 (3) | 0.67224 (15) | −0.08459 (9) | 0.0498 (5) | |
| H13 | −0.3904 | 0.7082 | −0.0905 | 0.060* | |
| C14 | −0.1991 (3) | 0.60677 (14) | −0.12757 (8) | 0.0420 (4) | |
| C15 | 0.0004 (3) | 0.55133 (13) | −0.11718 (8) | 0.0416 (4) | |
| C16 | 0.1404 (3) | 0.56603 (13) | −0.06575 (9) | 0.0431 (5) | |
| H16 | 0.2742 | 0.5296 | −0.0596 | 0.052* | |
| C17 | 0.2289 (4) | 0.42275 (18) | −0.15152 (13) | 0.0742 (7) | |
| H17A | 0.3703 | 0.4588 | −0.1508 | 0.111* | |
| H17B | 0.2287 | 0.3767 | −0.1839 | 0.111* | |
| H17C | 0.2169 | 0.3903 | −0.1133 | 0.111* | |
| C18 | −0.4992 (4) | 0.65515 (19) | −0.19872 (11) | 0.0662 (7) | |
| H18A | −0.6191 | 0.6526 | −0.1701 | 0.099* | |
| H18B | −0.5626 | 0.6386 | −0.2386 | 0.099* | |
| H18C | −0.4363 | 0.7186 | −0.1995 | 0.099* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0386 (3) | 0.0522 (3) | 0.0483 (3) | 0.0061 (2) | −0.0042 (2) | −0.0112 (2) |
| O1 | 0.0549 (9) | 0.0663 (10) | 0.0630 (10) | 0.0020 (7) | 0.0177 (7) | −0.0123 (8) |
| O2 | 0.0512 (8) | 0.0763 (11) | 0.0685 (10) | 0.0111 (8) | −0.0249 (7) | −0.0178 (8) |
| O3 | 0.0600 (9) | 0.0594 (9) | 0.0614 (9) | 0.0207 (7) | −0.0094 (7) | −0.0241 (7) |
| O4 | 0.0541 (8) | 0.0736 (11) | 0.0538 (9) | 0.0195 (7) | −0.0170 (7) | −0.0185 (8) |
| N1 | 0.0453 (9) | 0.0435 (9) | 0.0394 (9) | 0.0073 (7) | −0.0075 (7) | −0.0039 (7) |
| C1 | 0.0457 (10) | 0.0391 (10) | 0.0425 (11) | 0.0008 (8) | 0.0010 (8) | −0.0059 (8) |
| C2 | 0.0714 (14) | 0.0478 (12) | 0.0420 (12) | 0.0067 (10) | 0.0031 (10) | −0.0046 (9) |
| C3 | 0.0869 (17) | 0.0490 (13) | 0.0548 (14) | 0.0079 (12) | 0.0246 (12) | −0.0054 (11) |
| C4 | 0.0648 (14) | 0.0407 (12) | 0.0757 (16) | 0.0065 (10) | 0.0224 (12) | 0.0010 (11) |
| C5 | 0.0687 (14) | 0.0523 (13) | 0.0631 (14) | 0.0140 (11) | −0.0003 (11) | 0.0081 (11) |
| C6 | 0.0634 (13) | 0.0507 (12) | 0.0426 (11) | 0.0062 (10) | 0.0041 (9) | −0.0023 (9) |
| C7 | 0.0838 (18) | 0.0608 (16) | 0.120 (2) | 0.0226 (14) | 0.0352 (17) | 0.0052 (16) |
| C8 | 0.0647 (14) | 0.0592 (14) | 0.0520 (13) | 0.0055 (11) | 0.0017 (10) | −0.0009 (11) |
| C9 | 0.0508 (11) | 0.0448 (11) | 0.0501 (12) | 0.0040 (9) | −0.0139 (9) | −0.0035 (9) |
| C10 | 0.0555 (12) | 0.0445 (12) | 0.0480 (12) | 0.0055 (9) | −0.0088 (9) | −0.0028 (9) |
| C11 | 0.0425 (10) | 0.0379 (10) | 0.0416 (11) | 0.0011 (8) | −0.0021 (8) | −0.0006 (8) |
| C12 | 0.0471 (11) | 0.0535 (12) | 0.0459 (11) | 0.0088 (9) | 0.0014 (9) | −0.0142 (9) |
| C13 | 0.0390 (10) | 0.0563 (12) | 0.0536 (12) | 0.0140 (9) | −0.0031 (9) | −0.0107 (10) |
| C14 | 0.0378 (9) | 0.0457 (11) | 0.0420 (11) | 0.0025 (8) | −0.0031 (8) | −0.0035 (9) |
| C15 | 0.0424 (10) | 0.0368 (10) | 0.0453 (11) | 0.0031 (8) | 0.0012 (8) | −0.0068 (8) |
| C16 | 0.0407 (10) | 0.0354 (10) | 0.0525 (12) | 0.0076 (8) | −0.0036 (8) | −0.0023 (9) |
| C17 | 0.0615 (14) | 0.0631 (16) | 0.0972 (19) | 0.0222 (12) | −0.0040 (13) | −0.0318 (14) |
| C18 | 0.0567 (13) | 0.0845 (18) | 0.0556 (14) | 0.0189 (12) | −0.0147 (11) | −0.0004 (12) |
Geometric parameters (Å, °)
| S1—O2 | 1.4195 (15) | C8—H8A | 0.9600 |
| S1—O1 | 1.4210 (15) | C8—H8B | 0.9600 |
| S1—N1 | 1.6391 (17) | C8—H8C | 0.9600 |
| S1—C1 | 1.7538 (19) | C9—C10 | 1.507 (3) |
| O3—C15 | 1.359 (2) | C9—H9A | 0.9700 |
| O3—C17 | 1.421 (3) | C9—H9B | 0.9700 |
| O4—C14 | 1.360 (2) | C10—C11 | 1.506 (3) |
| O4—C18 | 1.417 (2) | C10—H10A | 0.9700 |
| N1—C8 | 1.463 (3) | C10—H10B | 0.9700 |
| N1—C9 | 1.471 (2) | C11—C12 | 1.376 (3) |
| C1—C6 | 1.378 (3) | C11—C16 | 1.392 (3) |
| C1—C2 | 1.378 (3) | C12—C13 | 1.386 (3) |
| C2—C3 | 1.372 (3) | C12—H12 | 0.9300 |
| C2—H2 | 0.9300 | C13—C14 | 1.367 (3) |
| C3—C4 | 1.380 (3) | C13—H13 | 0.9300 |
| C3—H3 | 0.9300 | C14—C15 | 1.399 (3) |
| C4—C5 | 1.374 (3) | C15—C16 | 1.372 (2) |
| C4—C7 | 1.504 (3) | C16—H16 | 0.9300 |
| C5—C6 | 1.375 (3) | C17—H17A | 0.9600 |
| C5—H5 | 0.9300 | C17—H17B | 0.9600 |
| C6—H6 | 0.9300 | C17—H17C | 0.9600 |
| C7—H7A | 0.9600 | C18—H18A | 0.9600 |
| C7—H7B | 0.9600 | C18—H18B | 0.9600 |
| C7—H7C | 0.9600 | C18—H18C | 0.9600 |
| O2—S1—O1 | 119.54 (10) | N1—C9—H9A | 109.4 |
| O2—S1—N1 | 106.32 (9) | C10—C9—H9A | 109.4 |
| O1—S1—N1 | 106.83 (9) | N1—C9—H9B | 109.4 |
| O2—S1—C1 | 108.57 (9) | C10—C9—H9B | 109.4 |
| O1—S1—C1 | 108.64 (9) | H9A—C9—H9B | 108.0 |
| N1—S1—C1 | 106.15 (9) | C11—C10—C9 | 113.42 (16) |
| C15—O3—C17 | 117.52 (16) | C11—C10—H10A | 108.9 |
| C14—O4—C18 | 117.51 (16) | C9—C10—H10A | 108.9 |
| C8—N1—C9 | 113.66 (16) | C11—C10—H10B | 108.9 |
| C8—N1—S1 | 114.86 (13) | C9—C10—H10B | 108.9 |
| C9—N1—S1 | 116.11 (13) | H10A—C10—H10B | 107.7 |
| C6—C1—C2 | 120.49 (18) | C12—C11—C16 | 117.84 (17) |
| C6—C1—S1 | 119.59 (15) | C12—C11—C10 | 122.44 (17) |
| C2—C1—S1 | 119.67 (15) | C16—C11—C10 | 119.71 (16) |
| C3—C2—C1 | 119.1 (2) | C11—C12—C13 | 121.17 (18) |
| C3—C2—H2 | 120.4 | C11—C12—H12 | 119.4 |
| C1—C2—H2 | 120.4 | C13—C12—H12 | 119.4 |
| C2—C3—C4 | 121.5 (2) | C14—C13—C12 | 120.65 (18) |
| C2—C3—H3 | 119.2 | C14—C13—H13 | 119.7 |
| C4—C3—H3 | 119.2 | C12—C13—H13 | 119.7 |
| C5—C4—C3 | 118.2 (2) | O4—C14—C13 | 125.54 (17) |
| C5—C4—C7 | 120.8 (2) | O4—C14—C15 | 115.54 (16) |
| C3—C4—C7 | 121.0 (2) | C13—C14—C15 | 118.91 (17) |
| C4—C5—C6 | 121.5 (2) | O3—C15—C16 | 125.43 (17) |
| C4—C5—H5 | 119.3 | O3—C15—C14 | 114.69 (16) |
| C6—C5—H5 | 119.3 | C16—C15—C14 | 119.88 (17) |
| C5—C6—C1 | 119.15 (19) | C15—C16—C11 | 121.46 (17) |
| C5—C6—H6 | 120.4 | C15—C16—H16 | 119.3 |
| C1—C6—H6 | 120.4 | C11—C16—H16 | 119.3 |
| C4—C7—H7A | 109.5 | O3—C17—H17A | 109.5 |
| C4—C7—H7B | 109.5 | O3—C17—H17B | 109.5 |
| H7A—C7—H7B | 109.5 | H17A—C17—H17B | 109.5 |
| C4—C7—H7C | 109.5 | O3—C17—H17C | 109.5 |
| H7A—C7—H7C | 109.5 | H17A—C17—H17C | 109.5 |
| H7B—C7—H7C | 109.5 | H17B—C17—H17C | 109.5 |
| N1—C8—H8A | 109.5 | O4—C18—H18A | 109.5 |
| N1—C8—H8B | 109.5 | O4—C18—H18B | 109.5 |
| H8A—C8—H8B | 109.5 | H18A—C18—H18B | 109.5 |
| N1—C8—H8C | 109.5 | O4—C18—H18C | 109.5 |
| H8A—C8—H8C | 109.5 | H18A—C18—H18C | 109.5 |
| H8B—C8—H8C | 109.5 | H18B—C18—H18C | 109.5 |
| N1—C9—C10 | 111.29 (16) | ||
| O2—S1—N1—C8 | 51.06 (16) | C8—N1—C9—C10 | −75.5 (2) |
| O1—S1—N1—C8 | 179.76 (14) | S1—N1—C9—C10 | 147.97 (15) |
| C1—S1—N1—C8 | −64.43 (15) | N1—C9—C10—C11 | 178.43 (16) |
| O2—S1—N1—C9 | −172.91 (13) | C9—C10—C11—C12 | −41.2 (3) |
| O1—S1—N1—C9 | −44.21 (15) | C9—C10—C11—C16 | 140.1 (2) |
| C1—S1—N1—C9 | 71.59 (15) | C16—C11—C12—C13 | −2.3 (3) |
| O2—S1—C1—C6 | 161.10 (17) | C10—C11—C12—C13 | 178.9 (2) |
| O1—S1—C1—C6 | 29.63 (19) | C11—C12—C13—C14 | 0.8 (3) |
| N1—S1—C1—C6 | −84.94 (18) | C18—O4—C14—C13 | 11.4 (3) |
| O2—S1—C1—C2 | −24.6 (2) | C18—O4—C14—C15 | −168.70 (19) |
| O1—S1—C1—C2 | −156.06 (17) | C12—C13—C14—O4 | −178.3 (2) |
| N1—S1—C1—C2 | 89.36 (18) | C12—C13—C14—C15 | 1.8 (3) |
| C6—C1—C2—C3 | 1.4 (3) | C17—O3—C15—C16 | 4.2 (3) |
| S1—C1—C2—C3 | −172.90 (17) | C17—O3—C15—C14 | −176.0 (2) |
| C1—C2—C3—C4 | 0.1 (4) | O4—C14—C15—O3 | −2.4 (3) |
| C2—C3—C4—C5 | −1.6 (4) | C13—C14—C15—O3 | 177.47 (19) |
| C2—C3—C4—C7 | 177.6 (2) | O4—C14—C15—C16 | 177.38 (18) |
| C3—C4—C5—C6 | 1.7 (4) | C13—C14—C15—C16 | −2.7 (3) |
| C7—C4—C5—C6 | −177.6 (2) | O3—C15—C16—C11 | −179.08 (18) |
| C4—C5—C6—C1 | −0.2 (3) | C14—C15—C16—C11 | 1.2 (3) |
| C2—C1—C6—C5 | −1.3 (3) | C12—C11—C16—C15 | 1.4 (3) |
| S1—C1—C6—C5 | 172.95 (17) | C10—C11—C16—C15 | −179.82 (18) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2···O2 | 0.93 | 2.59 | 2.942 (3) | 103 |
| C8—H8B···O2 | 0.96 | 2.44 | 2.895 (3) | 109 |
| C9—H9A···O1 | 0.97 | 2.39 | 2.880 (3) | 111 |
| C12—H12···O1i | 0.93 | 2.54 | 3.452 (2) | 166 |
| C18—H18B···O2ii | 0.96 | 2.38 | 3.302 (3) | 160 |
Symmetry codes: (i) x−1, y, z; (ii) x−3/2, −y+3/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5588).
References
- Bruker (2004). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Chumakov, Y. M., Tsapkov, V. I., Bocelli, G., Antonsyak, B. Y., PalomaresSa’nches, S. A., Ortiz, R. S. & Gulya, A. P. (2006). J. Struct. Chem. 47, 923–929.
- Khan, I. U., Akkurt, M., Sharif, S. & Ahmad, W. (2010). Acta Cryst. E66, o3053. [DOI] [PMC free article] [PubMed]
- Kremer, E., Facchin, G., Este’vez, E., Albore’ s, P., Baran, E. J., Ellena, J. & Torre, M. H. (2006). J. Inorg. Biochem. 100, 1167–1175. [DOI] [PubMed]
- Sharif, S., Iqbal, H., Khan, I. U., John, P. & Tiekink, E. R. T. (2010). Acta Cryst. E66, o1288. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (1996). SADABS, University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811029746/bt5588sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811029746/bt5588Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811029746/bt5588Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report