Abstract
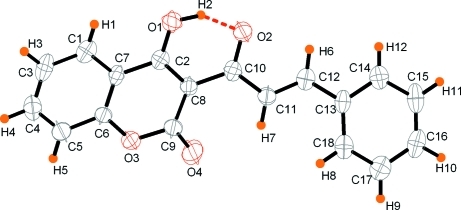
In the title molecule, C18H12O4, the phenyl ring is twisted by 23.2 (1)° from the mean plane of the chromene system. In the crystal, weak intermolecular C—H⋯O hydrogen bonds link molecules into zigzag chains extending in the [010] direction. An intramolecular O—H⋯O hydrogen bond is also present.
Related literature
For related structures, see: Traven et al. (2000 ▶); Sun & Cui (2008 ▶); Mechi et al. (2009 ▶); Hamdi et al. (2010 ▶); Asad et al. (2010 ▶). For the synthesis of coumarin chalcones, see: Claisen & Claparede (1881 ▶).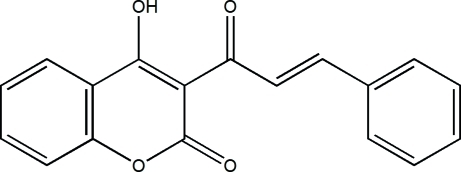
Experimental
Crystal data
C18H12O4
M r = 292.28
Monoclinic,
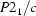
a = 11.8040 (5) Å
b = 3.8860 (5) Å
c = 29.7190 (5) Å
β = 97.164 (5)°
V = 1352.58 (18) Å3
Z = 4
Mo Kα radiation
μ = 0.10 mm−1
T = 296 K
0.3 × 0.14 × 0.06 mm
Data collection
Bruker SMART CCD area-detector diffractometer
Absorption correction: numerical (SADABS; Bruker, 2003 ▶) T min = 0.861, T max = 0.865
11154 measured reflections
2983 independent reflections
1404 reflections with I > 2σ(I)
R int = 0.070
Refinement
R[F 2 > 2σ(F 2)] = 0.069
wR(F 2) = 0.300
S = 1.04
2983 reflections
203 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.54 e Å−3
Δρmin = −0.69 e Å−3
Data collection: SMART (Bruker, 2003 ▶); cell refinement: SAINT (Bruker, 2003 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 1999 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811029801/cv5125sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811029801/cv5125Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811029801/cv5125Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C5—H5⋯O3i | 0.93 | 2.57 | 3.350 (5) | 142 |
| O1—H2⋯O2 | 0.99 (7) | 1.51 (7) | 2.413 (4) | 149 (6) |
Symmetry code: (i) 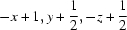 .
.
Acknowledgments
Professor A. Ben Salah is acknowledged for his contribution to the X-ray diffraction data collection at the Laboratory of Materials Science and the Environment, University of Sfax, Tunisia.
supplementary crystallographic information
Comment
In continuation of our structural and biological studies of coumarin derivatives (Mechi et al., 2009; Hamdi et al., 2010), we present the crystal structure of the title compound (I) - a new chalcone of the coumarin.
In (I) (Fig. 1), all bond lengths and angles are normal and correspond to those observed in related structures (Mechi et al., 2009; Asad et al., 2010). The presence of α, β-unsaturated ketone is indicated by the short O2–C10 and C11–C12 bond lengths of 1.289 (5)Å and 1.327 (5) Å, respectively, and the O2–C10–C11 and C10–C11–C12 bond angles of 118.2 (4) ° and 121.8 (4) °, respectively. The structure exhibits intramolecular hydrogen bonding between the hydroxyl oxygen and the ketonic oxygen in the coumarin group (Table 1). The chromen-2-one is twisted out of the plane of the phenyl ring (C13– C14) at 23.2 (1) °. The linkage between the coumarin system and phenyl ring is quite conjugated with bond lengths C10–C11 = 1.457 (5) Å, C11–C12 = 1.327 (5) Å, and C12–C13 = 1.440 (5) Å, suggesting that all non-hydrogen atoms between the electron-donors and acceptors are highly conjugated, leading to a π-bridge for the charge transfer from phenyl ring to coumarin system. Similar geometry has been observed in coumarin chalcone analogues (Mechi et al., 2009; Sun & Cui, 2008). Consequently, the C10–O2 = 1.289 (5) Å is elongated as compared with its mean value found in 3-acetyl-4hydroxycoumarin (1.253 Å) (Traven et al., 2000) owing to the localization of the hydroxyl hydrogen (H2) between the O2 ketonic oxygen and the hydroxyl oxygen O1. The O1–H2 distance (0.99 (7) Å) in (I) is shorter than that in related compounds - C18H10O7 (1.22 (7) Å) (Mechi et al., 2009) and C18H10Cl2O4 (1.27 (2) Å) (Asad et al., 2010). It should be noted that the C9–O4 bond length (1.198 (5) Å) is less than that (1.210 Å) observed in 3-acetyl-4hydroxycoumarin (Traven et al., 2000). It was concluded that it was a substantial difference for stabilizing the H atom of the hydroxyl group when we changed the nature of the substituted R group (from H to Cl and to OCH3).
In the crystal structure, weak intermolecular C– H···O hydrogen bonds (Table 1) link molecules into zigzag chains extended in [010].
Experimental
The new chalcone (I) was synthesized using the Claisen Schmidt reaction (Claisen & Claparede, 1881), by the condensation of 3-acetyl-4hydroxycoumarin (1g, 4.9 mmol) and aromatic benzaldehyde (6.4 mmol, 0.5 ml) in chloroform (5 ml) in the presence of one drop of piperidine. The mixture was refluxed in a water bath for 2 h. After cooling at room temperature, a yellow solid was obtained in good yield, filtered, washed with ethanol, and dried in air. Yellow block-shaped single crystals of the title compound, suitable for X-ray structure determination, were recrystalized by slow evaporation of dichloromethane (CH2Cl2) at room temperature after several days. Yield: 1.1 g (80%). mp= 499K. IR: ν 3468 (OH), 1690(s) (>C=O), 1578 (C= C), 1272(s) (sym) (C-O-C); 1HNMR: δppm: 7.4-8.1 (m, 10H, Ar-H+ Hethyl), 16.1(s,1H,OH). 13C NMR (ppm): 192.6(CO); 181.56 (C2); 160.22 (C9); 100.8 (C8), 116.32-147.368 (C arom); 124.39 (Cethyl1), 154.79 (Cethyl2),
Refinement
H2 atom was located on a difference map and refined isotropically. The remaining H atoms were positioned geometrically (C–H 0.93 Å) and refined using a riding model, with Uiso(H) = 1.2 Ueq(C).
Figures
Fig. 1.
The molecular structure of (I) showing 50% probability displacement ellipsoids and the atomic numbering. Dashed line denotes hydrogen bond.
Crystal data
| C18H12O4 | F(000) = 608 |
| Mr = 292.28 | Dx = 1.435 Mg m−3 |
| Monoclinic, P21/c | Melting point: 489 K |
| Hall symbol: -P2ybc | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.8040 (5) Å | Cell parameters from 203 reflections |
| b = 3.8860 (5) Å | µ = 0.10 mm−1 |
| c = 29.7190 (5) Å | T = 296 K |
| β = 97.164 (5)° | Plate, yellow |
| V = 1352.58 (18) Å3 | 0.3 × 0.14 × 0.06 mm |
| Z = 4 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2983 independent reflections |
| Radiation source: fine-focus sealed tube | 1404 reflections with I > 2σ(I) |
| graphite | Rint = 0.070 |
| φ and ω scans | θmax = 27.2°, θmin = 1.4° |
| Absorption correction: numerical (SADABS; Bruker, 2003) | h = −15→14 |
| Tmin = 0.861, Tmax = 0.865 | k = −4→4 |
| 11154 measured reflections | l = −38→37 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.069 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.300 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.1616P)2] where P = (Fo2 + 2Fc2)/3 |
| 2983 reflections | (Δ/σ)max < 0.001 |
| 203 parameters | Δρmax = 0.54 e Å−3 |
| 0 restraints | Δρmin = −0.69 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.0403 (2) | 0.2994 (9) | 0.14926 (11) | 0.0562 (9) | |
| O2 | 0.0632 (2) | 0.0415 (9) | 0.07711 (9) | 0.0559 (9) | |
| C1 | 0.1298 (4) | 0.5245 (10) | 0.23674 (14) | 0.0449 (11) | |
| H1 | 0.0526 | 0.5667 | 0.2281 | 0.054* | |
| C2 | 0.1481 (3) | 0.2566 (10) | 0.16162 (13) | 0.0372 (9) | |
| C10 | 0.1719 (3) | 0.0080 (10) | 0.08756 (13) | 0.0403 (10) | |
| C3 | 0.1795 (4) | 0.6154 (11) | 0.27952 (14) | 0.0531 (12) | |
| H3 | 0.1357 | 0.7162 | 0.2999 | 0.064* | |
| C14 | 0.1811 (4) | −0.5082 (11) | −0.06124 (13) | 0.0473 (11) | |
| H12 | 0.1031 | −0.5413 | −0.0610 | 0.057* | |
| C12 | 0.1891 (4) | −0.2364 (10) | 0.01330 (13) | 0.0418 (10) | |
| H6 | 0.1098 | −0.2428 | 0.0093 | 0.050* | |
| C7 | 0.1950 (3) | 0.3694 (9) | 0.20651 (12) | 0.0362 (9) | |
| C8 | 0.2206 (3) | 0.1046 (10) | 0.13289 (12) | 0.0363 (9) | |
| C15 | 0.2328 (4) | −0.6146 (11) | −0.09835 (14) | 0.0546 (13) | |
| H11 | 0.1895 | −0.7214 | −0.1227 | 0.065* | |
| C11 | 0.2385 (4) | −0.1217 (10) | 0.05304 (13) | 0.0421 (10) | |
| H7 | 0.3177 | −0.1241 | 0.0590 | 0.050* | |
| C4 | 0.2948 (4) | 0.5566 (11) | 0.29212 (14) | 0.0532 (12) | |
| H4 | 0.3282 | 0.6208 | 0.3209 | 0.064* | |
| C6 | 0.3095 (3) | 0.3138 (10) | 0.22020 (12) | 0.0379 (9) | |
| C9 | 0.3401 (4) | 0.0457 (11) | 0.14931 (13) | 0.0445 (10) | |
| C16 | 0.3483 (4) | −0.5624 (11) | −0.09925 (14) | 0.0536 (12) | |
| H10 | 0.3824 | −0.6308 | −0.1244 | 0.064* | |
| C5 | 0.3595 (4) | 0.4068 (11) | 0.26304 (14) | 0.0507 (11) | |
| H5 | 0.4368 | 0.3669 | 0.2718 | 0.061* | |
| O3 | 0.3791 (2) | 0.1608 (8) | 0.19242 (9) | 0.0485 (8) | |
| C17 | 0.4132 (4) | −0.4082 (12) | −0.06275 (15) | 0.0538 (12) | |
| H9 | 0.4912 | −0.3759 | −0.0630 | 0.065* | |
| O4 | 0.4098 (3) | −0.0973 (11) | 0.12993 (11) | 0.0753 (12) | |
| C18 | 0.3610 (4) | −0.3021 (11) | −0.02576 (13) | 0.0471 (11) | |
| H8 | 0.4046 | −0.1954 | −0.0015 | 0.057* | |
| C13 | 0.2453 (4) | −0.3519 (10) | −0.02429 (13) | 0.0386 (9) | |
| H2 | 0.022 (6) | 0.181 (17) | 0.120 (2) | 0.13 (2)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0409 (18) | 0.085 (2) | 0.0422 (18) | 0.0068 (16) | 0.0031 (14) | −0.0133 (17) |
| O2 | 0.0428 (18) | 0.083 (2) | 0.0404 (17) | 0.0022 (16) | −0.0005 (14) | −0.0144 (16) |
| C1 | 0.056 (3) | 0.040 (2) | 0.041 (2) | 0.0013 (19) | 0.017 (2) | 0.0005 (18) |
| C2 | 0.040 (2) | 0.038 (2) | 0.034 (2) | −0.0007 (17) | 0.0080 (17) | 0.0014 (17) |
| C10 | 0.045 (2) | 0.040 (2) | 0.037 (2) | −0.0008 (18) | 0.0070 (19) | 0.0017 (18) |
| C3 | 0.086 (4) | 0.044 (2) | 0.034 (2) | −0.002 (2) | 0.021 (2) | −0.0042 (18) |
| C14 | 0.053 (3) | 0.049 (2) | 0.038 (2) | 0.005 (2) | 0.001 (2) | 0.0004 (19) |
| C12 | 0.048 (2) | 0.044 (2) | 0.034 (2) | 0.0010 (19) | 0.0072 (19) | −0.0007 (18) |
| C7 | 0.045 (2) | 0.036 (2) | 0.028 (2) | −0.0032 (17) | 0.0103 (17) | 0.0047 (16) |
| C8 | 0.042 (2) | 0.038 (2) | 0.029 (2) | −0.0014 (17) | 0.0038 (17) | 0.0021 (16) |
| C15 | 0.084 (4) | 0.047 (3) | 0.030 (2) | 0.011 (2) | 0.001 (2) | −0.0057 (19) |
| C11 | 0.046 (2) | 0.047 (2) | 0.033 (2) | 0.0025 (19) | 0.0059 (18) | −0.0005 (18) |
| C4 | 0.076 (3) | 0.050 (3) | 0.032 (2) | −0.010 (2) | 0.000 (2) | −0.0010 (19) |
| C6 | 0.043 (2) | 0.041 (2) | 0.030 (2) | −0.0036 (18) | 0.0080 (17) | 0.0012 (17) |
| C9 | 0.043 (2) | 0.056 (3) | 0.034 (2) | 0.008 (2) | 0.0060 (19) | −0.0003 (19) |
| C16 | 0.078 (4) | 0.052 (3) | 0.034 (2) | 0.018 (2) | 0.016 (2) | 0.003 (2) |
| C5 | 0.055 (3) | 0.055 (3) | 0.041 (2) | −0.009 (2) | −0.002 (2) | 0.000 (2) |
| O3 | 0.0369 (16) | 0.072 (2) | 0.0357 (16) | 0.0048 (15) | 0.0014 (12) | −0.0042 (14) |
| C17 | 0.060 (3) | 0.056 (3) | 0.047 (3) | 0.007 (2) | 0.014 (2) | 0.003 (2) |
| O4 | 0.053 (2) | 0.123 (3) | 0.050 (2) | 0.032 (2) | 0.0068 (17) | −0.021 (2) |
| C18 | 0.060 (3) | 0.047 (2) | 0.033 (2) | 0.001 (2) | 0.005 (2) | 0.0004 (18) |
| C13 | 0.052 (2) | 0.035 (2) | 0.029 (2) | 0.0043 (18) | 0.0049 (18) | 0.0029 (16) |
Geometric parameters (Å, °)
| O1—C2 | 1.290 (5) | C7—C6 | 1.379 (5) |
| O1—H2 | 0.99 (7) | C8—C9 | 1.452 (5) |
| O2—C10 | 1.289 (5) | C15—C16 | 1.383 (7) |
| C1—C3 | 1.378 (6) | C15—H11 | 0.9300 |
| C1—C7 | 1.391 (5) | C11—H7 | 0.9300 |
| C1—H1 | 0.9300 | C4—C5 | 1.354 (6) |
| C2—C8 | 1.411 (5) | C4—H4 | 0.9300 |
| C2—C7 | 1.447 (5) | C6—O3 | 1.371 (4) |
| C10—C8 | 1.447 (5) | C6—C5 | 1.383 (5) |
| C10—C11 | 1.457 (5) | C9—O4 | 1.198 (5) |
| C3—C4 | 1.385 (7) | C9—O3 | 1.381 (5) |
| C3—H3 | 0.9300 | C16—C17 | 1.384 (6) |
| C14—C15 | 1.388 (6) | C16—H10 | 0.9300 |
| C14—C13 | 1.394 (6) | C5—H5 | 0.9300 |
| C14—H12 | 0.9300 | C17—C18 | 1.388 (6) |
| C12—C11 | 1.327 (5) | C17—H9 | 0.9300 |
| C12—C13 | 1.440 (5) | C18—C13 | 1.385 (6) |
| C12—H6 | 0.9300 | C18—H8 | 0.9300 |
| C2—O1—H2 | 107 (4) | C12—C11—C10 | 121.8 (4) |
| C3—C1—C7 | 120.0 (4) | C12—C11—H7 | 119.1 |
| C3—C1—H1 | 120.0 | C10—C11—H7 | 119.1 |
| C7—C1—H1 | 120.0 | C5—C4—C3 | 120.8 (4) |
| O1—C2—C8 | 122.2 (4) | C5—C4—H4 | 119.6 |
| O1—C2—C7 | 118.3 (3) | C3—C4—H4 | 119.6 |
| C8—C2—C7 | 119.5 (4) | O3—C6—C7 | 122.0 (3) |
| O2—C10—C8 | 117.8 (4) | O3—C6—C5 | 116.7 (4) |
| O2—C10—C11 | 118.2 (4) | C7—C6—C5 | 121.4 (4) |
| C8—C10—C11 | 124.0 (4) | O4—C9—O3 | 115.3 (4) |
| C1—C3—C4 | 119.9 (4) | O4—C9—C8 | 127.5 (4) |
| C1—C3—H3 | 120.1 | O3—C9—C8 | 117.3 (3) |
| C4—C3—H3 | 120.1 | C15—C16—C17 | 120.0 (4) |
| C15—C14—C13 | 120.4 (4) | C15—C16—H10 | 120.0 |
| C15—C14—H12 | 119.8 | C17—C16—H10 | 120.0 |
| C13—C14—H12 | 119.8 | C4—C5—C6 | 119.4 (4) |
| C11—C12—C13 | 127.0 (4) | C4—C5—H5 | 120.3 |
| C11—C12—H6 | 116.5 | C6—C5—H5 | 120.3 |
| C13—C12—H6 | 116.5 | C6—O3—C9 | 122.9 (3) |
| C6—C7—C1 | 118.6 (4) | C16—C17—C18 | 119.5 (5) |
| C6—C7—C2 | 118.2 (3) | C16—C17—H9 | 120.3 |
| C1—C7—C2 | 123.2 (4) | C18—C17—H9 | 120.3 |
| C2—C8—C10 | 118.1 (4) | C13—C18—C17 | 121.4 (4) |
| C2—C8—C9 | 120.0 (3) | C13—C18—H8 | 119.3 |
| C10—C8—C9 | 121.9 (3) | C17—C18—H8 | 119.3 |
| C16—C15—C14 | 120.3 (4) | C18—C13—C14 | 118.5 (4) |
| C16—C15—H11 | 119.9 | C18—C13—C12 | 122.1 (4) |
| C14—C15—H11 | 119.9 | C14—C13—C12 | 119.3 (4) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C5—H5···O3i | 0.93 | 2.57 | 3.350 (5) | 142. |
| O1—H2···O2 | 0.99 (7) | 1.51 (7) | 2.413 (4) | 149 (6) |
Symmetry codes: (i) −x+1, y+1/2, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CV5125).
References
- Asad, M., Oo, C.-W., Osman, H., Quah, C. K. & Fun, H.-K. (2010). Acta Cryst. E66, o3022–o3023. [DOI] [PMC free article] [PubMed]
- Brandenburg, K. (1999). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2003). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Claisen, L. & Claparede, A. (1881). Ber. Dtsch. Chem. Ges. 14, 2460–2468.
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Hamdi, N., Bouabdallah, F., Romerosa, A. & Ben Hassen, R. (2010). C. R. Chim. 13, 1261–1268.
- Mechi, L., Chtiba, S., Hamdi, N. & Ben Hassen, R. (2009). Acta Cryst. E65, o1652–o1653. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sun, Y. F. & Cui, Y. P. (2008). Dyes Pigments, 78, 65–76.
- Traven, V. F., Manaev, A. V., Safronova, O. B., Chibisova, T. A., Lyssenko, K. A. & Antipin, M. Yu. (2000). Russ. J. Gen. Chem. 70, 798–808.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536811029801/cv5125sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811029801/cv5125Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811029801/cv5125Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report