Abstract
The membrane-proximal tyrosine-based sorting motif in the cytoplasmic domain of the human immunodeficiency virus type 1 Env glycoprotein is important for endocytosis from the plasma membrane, basolateral targeting of viral budding in polarized epithelial cells, and polarized budding from a localized region of the lymphocyte plasma membrane. To study the role of the Env sorting motif (Y712XXL) in infectivity, the incorporation of Env into virions, and viral entry, we disrupted the motif with a tyrosine-to-alanine substitution. To investigate the relationship between the Env sorting motif and the enhancement of infectivity by Nef, the EnvY712A substitution was made in both Nef-positive and Nef-negative backgrounds. In spreading infections, including those using primary lymphocytes, the growth of the Y712A mutant was as impaired as Nef-negative virus, and the EnvY712A/Δ-Nef combination mutant was almost completely defective. In single-round infections using CD4-positive HeLa cells, the EnvY712A mutation impaired infectivity, and Nef retained the ability to enhance the infectivity in the context of EnvY712A. EnvY712 and Nef were required for the optimal infectivity of virions produced from either HEK293T or MT4 cells, but these sequences were required for the optimal incorporation of Env only when virions were produced from MT4 cells. Despite the wild-type levels of Env in viruses produced from 293T cells, the entry of the EnvY712A and Δ-Nef mutants into target cells was impaired. We conclude that the membrane-proximal tyrosine-based sorting motif of gp41 Env is, like Nef, important for optimal viral infectivity and, in the case of MT4 T cells, virion incorporation of Env. Nef does not require the Y712XXL motif to enhance viral infectivity. The finding that EnvY712 and Nef each affect the efficiency of viral entry independently of the Env content of virions suggests that both viral proteins are involved in trafficking events that influence morphogenesis to produce maximally fusogenic virus.
The envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) binds specific receptors on the surface of a target cell and mediates fusion of the viral membrane with the cell plasma membrane. Env is a trimeric complex consisting of three heterodimers composed of a surface subunit (SU; gp120) and a transmembrane subunit (TM; gp41). Although not required for the release of progeny particles from an infected cell, the presence of Env on the viral membrane is required for mediating entry into target cells. As few as 7 to 14 Env trimers may be incorporated into each virus particle, but the optimal number of trimers per particle is unknown (10).
The cytoplasmic tail of the Env transmembrane subunit contains sequences that affect the trafficking and targeting to the plasma membrane as well as the steady-state level of Env on the surface of infected cells (3, 4, 5, 7, 14, 20, 22, 23, 27, 29, 35). Env and other viral proteins such as Gag and Gag-Pol precursors gather at the plasma membrane prior to budding from the surface of the cell. Although not required for viral budding, Env is important for the specific location of release from the plasma membrane (14, 20, 22, 23). The release of HIV from polarized epithelial cells occurs from the basolateral surface and is dependent on the membrane-proximal tyrosine-based sorting signal (Y712XXL) in the cytoplasmic tail of gp41 (22, 23). In lymphocytes, a primary target of HIV-1 infection in vivo, the tyrosine-based motif is required for the polarized release of both HIV and simian immunodeficiency virus (SIV) virions from a localized, cap-like region of the plasma membrane overlying the uropod (14, 20). In addition to polarized budding, the membrane-proximal tyrosine mediates the endocytosis of Env from the plasma membrane through interactions with adaptor protein (AP) complexes of the cellular sorting machinery, in particular the clathrin adaptor AP-2 (3, 5, 27, 29). Other determinants within the gp41 cytoplasmic tail, including a leucine-based sorting motif that binds the clathrin adaptor AP-1, have also been found to be important in determining the subcellular localization of Env (4, 7, 35).
Mutation of the membrane-proximal tyrosine-based sorting motif of gp41 affects the incorporation of Env into virions as well as the ability of the virus to establish new infections (infectivity). Paradoxically, mutation of tyrosine 712 to serine or cysteine reportedly yields virions of increased infectivity relative to the wild type (8, 33). Although this could result from the increased expression of Env at the cell surface that occurs without an intact tyrosine-based endocytosis motif, an increase in the incorporation of Env into such mutants has not been observed consistently. A twofold increase in virion incorporation relative to wild type was reported for the Y712S mutant, whereas a twofold relative decrease was reported in the incorporation of Y712C Env. Further examination of the impact of the Y712XXL sequence on viral infectivity and the incorporation of Env into virions seems necessary to understand how this motif affects viral morphogenesis.
C-terminal truncations in gp41 that disrupt the membrane-proximal tyrosine motif affect both viral infectivity and the incorporation of Env into virions in a cell-type-dependent manner (2, 18, 26, 34). In general, a direct correlation between viral infectivity and Env incorporation has been observed (2, 26). However, the extent to which infectivity and Env incorporation differed from those of the wild type depended on the cell type used to produce the viruses. For example, gp41-truncated virus produced from lymphocytic H9 cells was markedly reduced in viral infectivity and Env incorporation, whereas virus produced from lymphocytic M8166 cells was only slightly reduced in infectivity and incorporated wild-type levels of Env (2). Similarly, gp41-truncated virus produced from CEM cells was reduced 10-fold in both infectivity and Env incorporation, whereas the same mutant produced from MT4 cells retained 60% of wild-type infectivity and the incorporation of Env was reduced only threefold (26). Despite these cell-type-dependent differences, these results clearly demonstrate that the cytoplasmic tail of gp41 is involved in interactions that enable efficient incorporation into budding virions, a critical step in the production of infectious virus.
The Nef protein is another viral protein that is necessary for the production of maximally infectious HIV-1, but the mechanism of Nef-mediated enhancement of infectivity is poorly understood. One possibility is that the presence of CD4 in proximity to the location of viral budding has an inhibitory effect on the incorporation of Env and the production of infectious virus (13, 21). Consequently, the down-regulation of CD4 by Nef may contribute to the enhancement of viral infectivity. However, the observation that Nef enhances the infectivity of viruses produced from cells that do not express CD4 indicates that CD4 down-regulation is not necessary for this effect (1, 24).
Nef plays a well-known role in the alteration of intracellular trafficking, primarily via its interaction with vesicular coat proteins such as the AP complexes (6, 19). Consequently, Nef might influence the trafficking of Env to optimize viral infectivity. Indeed, several lines of evidence suggest a relationship between Nef and Env. The ability of Nef to enhance the cytoplasmic delivery of HIV-1 cores led to the suggestion that Nef may influence the function of Env to enhance viral entry (30). In an intervirion fusion assay, Nef enhanced viral infectivity when present during the production of Env-containing particles but not when present during the production of functional HIV-1 cores; these data led to the suggestion that Nef does not enhance infectivity by modifying the virus core but, instead, may influence the function of Env (36). Lastly, the F12 allele of HIV-1 Nef inhibits viral release from infected cells by a mechanism that requires the cytoplasmic domains of both gp41 and CD4 (28). Together, these data suggest the possibility that Nef plays a role in enhancing Env function. Here, we hypothesized that if Nef were to influence Env, then it would likely do so via an effect on trafficking mediated by an AP binding motif such as the membrane-proximal YXXL sequence.
To study the role of the Y712XXL sorting motif of Env in infectivity and virion incorporation, we disrupted the motif by mutating tyrosine 712 to alanine. To investigate the possibility that Nef acts through the Y712XXL motif of Env, the EnvY712A substitution was made in both Nef-positive and Nef-negative backgrounds. Our results indicate that the membrane-proximal tyrosine-based sorting motif of Env is important for the virion incorporation of Env in certain cell types. However, regardless of the producer cell type and even in the absence of an effect on virion incorporation, EnvY712 is required for optimal viral infectivity. The effects of Nef and EnvY712 on viral infectivity are additive, suggesting that EnvY712 is not required for the enhancement of viral infectivity by Nef. Despite this lack of interdependence, both Nef and the gp41 YXXL motif appear to optimize infectivity by a CD4- and Env-incorporation-independent mechanism that is at least in part due to enhanced viral entry into target cells.
MATERIALS AND METHODS
Cells.
P4.R5 cells were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) (P4.R5 MAGI), from Nathaniel Landau (9) and maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (FBS) (Sigma, St. Louis, Mo.), penicillin (100 U/ml)-streptomycin (100 μg/ml) (pen/strep) (Gibco), 2 mM l-glutamine (Gibco), and puromycin (1 μg/ml). HeLa-CD4 cells (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH [HeLa CD4 clone 1022 from Bruce Chesebro]) were maintained in DMEM supplemented with 10% FBS, pen/strep, l-glutamine, and G418 (200 μg/ml) (11). HEK293T (a gift from Nathaniel Landau) and HEK293 cells were maintained in complete medium consisting of minimum essential medium (E-MEM; Quality Biological, Inc., Gaithersburg, Md.) supplemented with 10% FBS, pen/strep, and l-glutamine. MT4 T cells and CEM T cells were maintained in RPMI 1640 medium (BioWhittaker, Walkersville, Md.) supplemented with 10% FBS, pen/strep, and l-glutamine. Peripheral blood mononuclear cells (PBMCs) were isolated from healthy human donors by centrifugation of whole blood through Histopaque-1077 separation medium according to the manufacturer's directions (Accupsin tubes; Sigma). PBMCs were maintained in RPMI 1640 medium supplemented with 10% FBS, pen/strep, and l-glutamine.
Proviral constructs.
A plasmid encoding the complete proviral sequence of HIV-1 clone NL4-3 (pNL4-3) was used for the production of virus. Variants of pNL4-3 were constructed as follows. Nef-negative virus was made by the introduction of two premature termination codons in nef as previously described (12). The Env tyrosine 712 mutant was made by PCR site-directed mutagenesis of env from a plasmid containing the NheI-BamHI fragment of NL4-3 env. Primers flanking the HindIII and BamHI restriction sites of env and mutagenic primers in between the HindIII and BamHI sites were used in a two-round overlap PCR strategy that generated a 458-bp product containing a 4-bp change (ATAT to CGCC). In addition to changing the tyrosine 712 codon (TAT) to alanine (GCC), the mutagenesis created a new restriction site for NarI that aided in screening for the mutation. The PCR product was cut with HindIII and BamHI and ligated into the larger NheI-BamHI fragment of env. Sequence analysis verified the intended mutation and excluded mutations in the surrounding sequences. In preparation for introduction of the mutation into full-length provirus, the NheI-BamHI fragment was cut out of both wild-type pNL4-3 and pNLDSNef (nef double-stop) plasmids. Finally, the mutagenized NheI-BamHI fragment was ligated into both pNL4-3 and pNLDSNef to create the plasmids pNLEnvY712A and pDSEnvY712A. The EnvY712C and EnvY712F proviral constructs were provided by Eric Hunter (33).
Virus production and cell lysate preparation.
Wild-type and mutant viruses were produced by transient transfection of cell lines with proviral plasmids. Viruses produced for infection of T cells and for single-round infectivity assays were made by calcium phosphate transfection of HEK293 cells using the CellPhect reagents (Amersham-Pharmacia, Piscataway, N.J.) and the manufacturer's instructions. One million cells were transfected with 3 μg of plasmid DNA in six-well tissue culture plates. Virus-containing supernatants were collected 3 days later, centrifuged at low speed to pellet cells and cellular debris, filtered (pore size, 0.2 μm), and then frozen at −80°C. For certain single-round infectivity assays, MT4 cells were transfected using Lipofectin (Invitrogen, Carlsbad, Calif.), 4 μg of plasmid DNA, and 2 million cells in suspension. Virus-containing supernatants were collected 3 days later, centrifuged at low speed to pellet cells and cellular debris, filtered (pore size, 0.2 μm), and then frozen at −80°C. For viral entry assays and single-round infectivity assays, HEK293T cells were transiently transfected using Lipofectamine 2000 (Invitrogen), 20 μg of plasmid DNA, and 2 million cells plated in 100-mm-diameter tissue culture dishes. Virus-containing supernatants were collected 2 days later, centrifuged at low speed to pellet cells and cellular debris, filtered (pore size, 0.2 μm), and then frozen at −80°C. Prior to freezing, all virus-containing supernatants were sampled for detection of the p24 capsid protein by enzyme-linked immunosorbent assay (ELISA) (Perkin-Elmer, Boston, Mass.).
Lysates of MT4 cells were prepared at the time of virus collection. Pelleted cells were washed with phosphate-buffered saline (PBS), pelleted again, and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer containing dithiothreitol. After a sample was taken for p24 analysis, the cell lysate was frozen at −20°C for analysis by Western blot as described below.
Immunofluorescence.
CD4-positive HeLa cells (clone 1022) were transiently transfected with proviral plasmids using FuGENE 6 (Roche Applied Science, Indianapolis, Ind.). One million cells in a six-well tissue culture plate were transfected with 2 μg of plasmid DNA according to the manufacturer's instructions. Cells were replated onto glass coverslips 1 day later and then stained for gp41 Env the following day. To stain gp41, adhered cells were washed with PBS, fixed with 3% paraformaldehyde, washed with PBS, permeabilized with 0.1% NP-40, washed with PBS, blocked with 3% bovine serum albumin-PBS, incubated with mouse anti-gp41 (Advanced Biotechnologies, Columbia, Md.), washed three times with PBS, incubated with Rhodamine-X-conjugated secondary antibody, washed with PBS, and finally mounted onto glass slides. Slides were analyzed with a Zeiss Axioskop fluorescent microscope and magnified (×400), and images were processed using Adobe Photoshop.
Spreading infection assays.
Viral stocks produced as described above from HEK293 cells were normalized by p24 capsid protein concentration. For infection of CEM and MT4 cells, one million cells were pelleted in duplicate and resuspended in 1 ml of complete RPMI 1640 medium containing 5, 50, or 250 ng of virus. After incubating at 37°C for 6 h, cells were washed and resuspended in 4 ml of medium and then incubated at 37°C. Samples of the culture were taken every 2 to 3 days to quantify the concentration of p24 capsid protein by ELISA. After p24 sampling, the cell suspension was split 1:4 and returned to incubation at 37°C. Results of p24 analyses are presented on a logarithmic scale.
Freshly isolated PBMCs were stimulated with phytohemagglutinin for 3 days. One million cells were pelleted in duplicate and resuspended in 1 ml of RPMI 1640 medium containing 50 ng of virus and interleukin-2 (IL-2) (20 U/ml). After incubating at 37°C for 6 h, cells were washed and resuspended in 4 ml of medium supplemented with IL-2 (20 U/ml) and then incubated at 37°C. Samples of medium were taken over time for analysis of viral output by p24 ELISA.
P4 infectivity assay.
The P4.R5 cell line contains the β-galactosidase indicator under the control of the HIV-1 long terminal repeat (LTR). Cells were plated at a density of 2 × 104 cells per well in a 48-well tissue culture plate. On the following day, cells were infected in duplicate with 100 μl of several dilutions of virus normalized by p24 capsid protein. After a 2-h incubation at 37°C, the volume was increased to 500 μl by adding 400 μl of complete medium. After a 2-day incubation at 37°C, cells were fixed with 1% formaldehyde-0.2% glutaraldehyde in PBS. After washing twice with PBS, cells were stained in an X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution for 4 to 8 h and finally washed again with PBS. Infectivity was assessed by counting the blue-stained, HIV-infected foci.
Syncytium formation infectivity assay.
CD4-positive HeLa cells (clone 1022) were plated at a density of 3.5 × 104 cells per well in a 24-well tissue culture plate. On the following day, cells were infected in duplicate with 200 μl of several dilutions of virus normalized by p24 capsid protein. After a 2-h incubation at 37°C, the volume was increased to 1 ml with complete medium. Following a 3-day incubation at 37°C, cells were fixed with 100% methanol and stained with 0.5% crystal violet to visualize nuclei. Infectivity was assessed by counting syncytia.
Virus pelleting.
Viruses produced as described above from HEK293T and MT4 cells were used for determination of the incorporation of Env into virions. Viral stocks were thawed and then centrifuged at 23,500 × g for 2 h at 4°C. The viral pellets were resuspended in a small volume of reducing SDS-PAGE sample buffer, and an aliquot was taken for analysis of p24 antigen by ELISA. Resuspended pellets were frozen at −20°C for Western blot analysis as described below.
Western blot.
Cell lysates and virus pellets were analyzed by Western blot for gp120 and p24. Cell lysate and viral pellet samples were previously prepared in SDS-PAGE reducing sample buffer and stored at −20°C as described above. Based on the p24 ELISA data, equal amounts of p24 per sample were prepared. Samples were boiled for 10 min, electrophoresed in a precast 12% Tris-HCl SDS-polyacrylamide gel (Bio-Rad, Hercules, Calif.), and then electroblotted to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 4% nonfat dry milk, washed with PBS-Tween, incubated with primary antibody, washed, incubated with a horseradish peroxidase-conjugated secondary antibody, washed, developed with enhanced chemiluminescent substrate (ECL) (Amersham-Pharmacia), and exposed to X-ray film. Initially the membrane was probed for gp120 using sheep antiserum to HIV-1 gp120 (a gift from Juan Lama, originally obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH [CN 288, lot 1 DV-012]). After development of the blot, antibodies were stripped from the blot by extensive washing in Western stripping buffer (62.5 mM Tris-HCl, 2% SDS, 100 mM β-mercaptoethanol) followed by PBS-Tween. The Western blot procedure was then repeated using a primary antibody against HIV-1 p24 (clone 7A8.1; Chemicon International, Temecula, Calif.). To validate the use of Western blot for comparing quantitative differences in gp120, a standard curve using recombinant gp120 (HIV-1 IIIB gp120; obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) was also assayed by Western blot. Between 50 and 150 ng of recombinant gp120 in 25-ng increments was loaded onto a 12% Tris-HCl SDS-polyacrylamide gel alongside the pelleted virions and Western blotted as above. The film images of bands were scanned and analyzed using ImageJ (version 1.29t) software (Wayne Rasband, National Institutes of Health).
Viral entry assay.
Viral entry was detected using a flow cytometric assay as described by Münk et al. (25). The assay measures a change in the fluorescence emission from the compound CCF2/AM following cleavage by the enzyme β-lactamase (BlaM). BlaM-containing virions were produced by cotransfection of 293T cells with 10 μg of a proviral plasmid and 10 μg of pMM310, a vector encoding a β-lactamase-Vpr fusion protein (a gift from Nathaniel Landau, originally from Michael Miller, Merck Research Laboratories). Virus-containing supernatants were processed as described above and then pelleted at 23,500 × g for 2 h. Viral pellets were resuspended in 1 ml of complete DMEM for use in the entry assay or 100 μl of phosphate buffer (100 mM) for use in the β-lactamase activity assay described below. After normalization by p24 ELISA, 250 ng of BlaM-Vpr viruses was used to infect P4.R5 cells plated at a density of 2.5 × 105 cells per well in six-well tissue culture plates. After 5 h, cells were washed with PBS and incubated with 2 μM CCF2/AM (GeneBLAzer Loading kit; Aurora Biosciences, San Diego, Calif.)-1% probenecid-25 mM HEPES in 1 ml of serum-free DMEM at 25°C. After 16 to 18 h, cells were washed, trypsinized, fixed with 1% paraformaldehyde, and analyzed on a MoFlo flow cytometer (Dako Cytomation, Fort Collins, Colo.) with a UV laser and a 485-nm long-pass dichroic filter with excitation at 351 to 364 nm. Uncleaved substrate was detected as green fluorescence using a 530/40-nm band-pass filter, and cleaved substrate was detected as blue fluorescence using a 450-nm band-pass filter. AMD3100 (Sigma) at a final concentration of 100 nM was used as a control to block viral entry.
β-Lactamase activity assay.
BlaM-Vpr-containing viruses produced as described above and resuspended in 100 μl of phosphate buffer were normalized by p24 capsid protein concentration. Samples were prepared by diluting 200 ng of each virus into phosphate buffer plus 10% HIV antigen specimen diluent containing Triton X-100 (Abbott Laboratories, Abbott Park, Ill.) to a final volume of 90 μl. To visualize β-lactamase activity, 10 μl of 1 mM nitrocefin (Calbiochem, San Diego, Calif.) was added, and the solution was incubated in the dark at room temperature for 22 h. A color change from yellow to red was detected in a microplate spectrophotometer using a 490-nm filter. Nitrocefin diluted into phosphate buffer-10% specimen diluent was measured as background, and 20 μl of an ampicillin-resistant bacterial culture was used as a positive control.
RESULTS
Construction and expression of the Y712A envelope glycoprotein mutant.
To investigate the role of the membrane-proximal tyrosine-based sorting signal in the cytoplasmic domain of the TM subunit of the HIV-1 envelope glycoprotein (gp41), we constructed a mutant encoding alanine instead of tyrosine at residue 712 in env within the proviral clone pNL4-3 (Fig. 1). To study the relationship between Nef and the Env Y712XXL sorting motif, this tyrosine-to-alanine substitution was also made in an NL4-3-based construct containing two premature termination codons in nef (Δ-Nef). T lymphoblastoid cells (MT4) or fibroblastoid cells (HEK293 and HEK293T) were transfected with the four different proviral plasmids (wild-type NL4-3, Δ-Nef, EnvY712A, and EnvY712A/Δ-Nef) to verify expression and to produce virus. No significant differences between the mutants were detected in the transient production of p24 antigen from transfected 293 cells (data not shown), indicating the absence of any unanticipated defects in the RNA sequence, such as altered use of the rev splice acceptor site downstream of the Y712 codon. To characterize specifically the expression of the EnvY712A glycoprotein, lysates of transfected MT4 cells were analyzed by Western blotting (Fig. 2A). After staining for gp120, the membrane was stripped and reprobed for p24 to exclude differences in transfection or loading (Fig. 2A). The Y712A substitution in Env did not impair the expression of gp160 (the gp120/gp41 precursor) in the transfected cells at steady state (Fig. 2A). We were unable to assess the relative efficiency of the processing of Y712A gp160, because processed gp120 was undetectable in these cell lysates.
FIG. 1.
Schematic representation of the HIV-1 envelope precursor glycoprotein (gp160). The surface subunit, gp120, and transmembrane subunit, gp41, are indicated. The gp41 subunit consists of an extracellular domain, transmembrane domain (TM), and a cytoplasmic domain. Membrane-proximal amino acids of the cytoplasmic domain of HIV-1 clone NL4-3 are shown beneath the schematic, with the membrane-proximal tyrosine-based sorting motif underlined. The tyrosine at residue 712 (in boldface type) was replaced with alanine by PCR site-directed mutagenesis.
FIG. 2.
Expression of Env in MT4 and HeLa cells. (A) Western blot of MT4 cell lysates for gp160 and p24. MT4 cells were transfected with wild-type and mutant proviruses and pelleted 3 days later. Cells were washed in PBS and then resuspended in reducing SDS-PAGE sample buffer. Equal amounts of p24 capsid per lane were run on an SDS-polyacrylamide gel, transferred to a PVDF membrane, probed sequentially with sheep antiserum to HIV-1 gp120 (DV-012) and mouse anti-p24 (7A8.1), and developed with a chemiluminescent substrate. Bands corresponding to gp160 and p24 are indicated. (B) Immunofluorescence of HeLa-CD4 cells. Cells were transfected with wild-type NL4-3 or EnvY712A provirus, fixed 2 days later, permeabilized, and stained with mouse anti-gp41 followed by a Rhodamine-X-conjugated secondary antibody. Slides were analyzed with a Zeiss Axioskop fluorescent microscope (magnification, ×400), and images were processed using Adobe Photoshop.
Next, we tested the fusogenicity of EnvY712A by assessing its ability to induce the formation of syncytia. HeLa-CD4 cells were transfected with the wild type or the EnvY712A mutant plasmids and stained by immunofluorescence for gp41. No qualitative morphological differences were observed in the syncytia formed as a result of the fusion of neighboring cells with cells expressing wild type or EnvY712A (Fig. 2B). The distribution of gp41 within these syncytia was also indistinguishable. Similarly, HeLa-CD4 cells infected with wild-type or EnvY712A virus formed syncytia that were morphologically indistinguishable (data not shown). Together, these data indicated that the EnvY712A mutant is expressed normally and retains the capacity to mediate membrane fusion.
The growth rates of the EnvY712A and Δ-Nef mutants are attenuated in CEM T cells and primary PBMCs.
To characterize the phenotype of the EnvY712A mutant in spreading infections, primary human PBMCs and cells of two T lymphoblastoid lines were used for growth rate experiments. The inocula for these infections were produced by transfection of HEK293 cells with proviral plasmids containing either wild-type, Δ-Nef, EnvY712A, or EnvY712A/Δ-Nef sequences and were normalized to contain equal amounts of p24 capsid antigen.
The three mutants grew poorly relative to the wild type in the PBMC cultures (Fig. 3A). While wild-type virus yielded almost 30,000 pg of p24 antigen/ml after 15 days of culture, both the EnvY712A and Δ-Nef mutants replicated to a reduced extent, reaching about 4,500 pg of p24/ml by day 15, while the double mutant EnvY712A/Δ-Nef yielded a maximal concentration of only 300 pg/ml. Overall, the single mutants, Δ-Nef and EnvY712A, were very similarly attenuated, replicating approximately 1 order of magnitude less well than wild type, while the double mutant, EnvY712A/Δ-Nef, displayed an additive phenotype, replicating a further order of magnitude less well.
FIG. 3.
Growth of viruses in PBMC and CEM and MT4 cells. Viruses were produced by transfection of HEK293 cells. One million PBMC or CEM or MT4 cells were infected in duplicate with viruses normalized by p24 capsid protein concentration. Samples of the cultures were taken over time and analyzed for p24 protein concentration by ELISA. CEM and MT4 cells were split 1:4 after every culture sampling. Each experiment was performed at least twice, each in duplicate, with similar results in both the hierarchy and extent of replication. (A) Primary PBMCs were stimulated with phytohemagglutinin for 3 days prior to infection with virus containing 50 ng of p24 capsid, supplemented with IL-2. (B) CEM T cells were infected with 250 ng of p24 capsid. (C) MT4 T cells were infected with 5 ng of p24 capsid. Error bars indicate standard deviations of duplicate infections.
The growth of these viruses was also tested in two T-cell lines, CEM and MT4. MT4 cells were tested because these cells are reportedly permissive for the replication of mutants encoding C-terminal truncations of HIV Env (26, 34). In the CEM cultures, wild-type virus yielded 2.5 × 106 pg of p24/ml by day 13 (Fig. 3B). Both the Δ-Nef and the EnvY712A mutants replicated approximately 3 orders of magnitude less well, yielding only 1,300 and 2,100 pg of p24/ml. The EnvY712A/Δ-Nef double mutant was unable to establish an infection in CEM cells under these conditions. Although the maximal yields were different, the hierarchy of replication in CEM cells matched that observed using PBMCs. In contrast to PBMCs and CEM cells, MT4 cells were extremely permissive for the growth of these mutants; cells infected with 50 ng of p24 of each of the four viruses showed equivalently rapid rates of viral growth (data not shown). To reveal subtle differences in growth rate, these infections were repeated using 1/10 the viral inoculum. Infection of MT4 cells with 5 ng of p24 antigen also showed no major defects in viral growth (Fig. 3C), but at day 2, viral production showed a hierarchy identical to that of the PBMC and CEM growth curves: 16,900 pg/ml for wild type, ∼2,400 pg/ml for Δ-Nef and EnvY712A, and 800 pg/ml for EnvY712A/Δ-Nef. This deficiency of viral production by the mutants early after infection was overcome by day 4. Thus, MT4 cells were ultimately as permissive for the growth of the Y712A Env mutant as for viruses encoding C-terminal truncations in gp41 (26, 34).
Taken together, these data on viral growth rate in spreading infections indicated that the EnvY712A mutant was as attenuated as Δ-Nef; the data also indicated that the effects of the Y712A and Nef-negative mutations were additive.
EnvY712 is required for optimal infectivity, but the positive effect of Nef on infectivity persists in the context of the Y712A mutation.
We investigated the role of the Y712XXL motif of Env in virion infectivity and the possibility that Nef enhances infectivity via this motif. The four viruses described above were produced from either HEK293T cells or MT4 cells, and their infectivities were measured using a single-round infectious-center assay in which CD4-positive HeLa cells that contain an LTR-β-galactosidase indicator are the targets (P4 cells). The infectivity data were expressed as a ratio of the number of blue cells per nanogram of p24 in the inoculum and were normalized to a wild-type value of 100%. The infectivity of the EnvY712A mutant was reduced relative to the wild type in a cell-type-dependent manner, sevenfold using 293T-produced virus and twofold using MT4-produced virus (Fig. 4A). Nef-defective virus showed a characteristic reduction in infectivity and a similar cell type dependence. Using 293T-produced virus, Δ-Nef exhibited a 20-fold reduction in infectivity relative to wild type, whereas the infectivity of MT4-produced Δ-Nef was reduced fourfold (Fig. 4A). Even in the presence of the Y712A mutation, a difference in viral infectivity between Nef-positive and Nef-negative virus was usually observed. The double mutant EnvY712A/Δ-Nef was reduced in infectivity 14-fold compared to EnvY712A when produced from 293T cells, while MT4-produced EnvY712A/Δ-Nef virus was reduced threefold compared to EnvY712A (Fig. 4A). Together, these data indicated that Env Y712 is important for optimal infectivity. The data also indicated that Nef is able to enhance viral infectivity in the absence of the EnvY712XXL motif.
FIG. 4.
Infectivity of HEK293T- and MT4-produced virus. (A) Viruses were produced by transfection of either 293T or MT4 cells as indicated underneath the x axis. Virus-containing supernatants normalized by p24 capsid were added to P4.R5 indicator cells that contain β-galactosidase under control of the HIV-1 LTR. Following 2 days of incubation, cells were stained with X-Gal and blue foci were counted. Infectivity data are expressed as a ratio of the number of blue cells per ng of p24 inoculum normalized to wild type. The 293T data shown are the averages of duplicates, and the MT4 data shown are the averages between two independent assays. (B) Infectivity of 293-produced virus compared in two distinct assays as indicated underneath the x axis. The P4 assay was performed as described in panel A. For the syncytium induction assay, HeLa-CD4 cells (clone 1022) were infected with viruses normalized by p24 capsid concentration and incubated for 3 days. Cells were stained with crystal violet, and the syncytia were counted. Data shown are the averages of duplicates. (C) Infectivity of various 293T-produced EnvY712 mutants tested in the P4 infectivity assay as above. Data shown are the averages of duplicates. Error bars indicate standard deviation.
To evaluate quantitatively the ability of mutants to induce fusion in target cells, we measured the infectivity of these viruses using an infectious-center assay in which the readout is syncytium formation and compared these data to those obtained using the β-galactosidase indicator cells (Fig. 4B). The relative infectivities of the mutants determined using these two assays were very similar. These data indicated that for these 293-produced viruses, the infectivity defects are unlikely to be due to a decreased fusogenicity of Env per se, at least as measured by the ability to induce fusion of target cells.
The observation that the EnvY712A mutant is less infectious than wild type was in potential conflict with published data regarding this residue, although substitutions other than alanine have previously been studied (8, 33). To reconcile these differences, the previously described NL4-3-based proviral constructs Env Y712C and Y712F were used to produce virus from 293T cells and tested alongside the EnvY712A mutant using the P4 infectivity assay. Although increases in infectivity of more than fourfold for Y712C and threefold for Y712F were previously reported, our results showed a reduction in the infectivity of Y712C and a slight increase in the infectivity of Y712F compared to wild type (Fig. 4C). To exclude inadvertent differences in these mutants, the parental wild-type constructs (pNL4-3) from both source laboratories were compared and were not significantly different (data not shown). Our data indicated that the conservative Y712F mutation does not impair infectivity, a result compatible with the ability of phenylalanine to replace tyrosine in YXXL AP-binding motifs (32). In contrast, the nonconservative Y712C mutation was, like Y712A, detrimental to infectivity.
Role of EnvY712A and Nef in the incorporation of gp120 into virions.
To investigate the possibility that the EnvY712A mutant was reduced in viral growth rate and infectivity due to the amount of envelope glycoprotein complexes incorporated into the viral membrane, we isolated virions and performed Western blotting for gp120. Recently published data indicate that gp120 and gp41 persist in virions in equimolar amounts after freeze-thawing and centrifugation, procedures that preceded our analysis (10). To validate the use of the Western blot protocol for quantifying Env, a standard curve using recombinant gp120 was generated. A plot of nanograms of recombinant gp120 versus band density showed a linear dose response and was sensitive to differences as small as 25% in the amount of input gp120 (Fig. 5A).
FIG. 5.
Env incorporation in MT4-produced virus. (A) Plot of band density versus nanograms of recombinant gp120. Known amounts of recombinant gp120 were loaded onto an SDS-polyacrylamide gel, transferred to a PVDF membrane, probed with sheep antiserum to HIV-1 gp120, and developed with a chemiluminescent substrate. Bands were analyzed using ImageJ (version 1.29t) software (Wayne Rasband, National Institutes of Health). (Inset: Western blot image of recombinant gp120.) (B) Western blot of MT4-produced virus. Virus-containing supernatants produced from transfected MT4 cells were filtered through 0.2-μm-pore-size filters and centrifuged at 23,500 × g for 2 h at 4°C. Viral pellets were then resuspended in a small volume of reducing SDS-PAGE sample buffer. Viral pellet suspensions were normalized for p24 concentration and electrophoresed and electroblotted as described in Fig. 2A. Bands corresponding to gp120 and p24 are indicated. (C) Relative amounts of gp120 incorporation. gp120 and p24 bands were scanned and analyzed using ImageJ (version 1.29t) software. Band densities are expressed as a ratio of gp120 to p24 and are an average of two independent experiments. Error bars indicate standard deviation.
Viruses produced by transfection of MT4 cells were pelleted, and equal amounts of p24 were loaded onto an SDS-polyacrylamide gel and probed for gp120. After staining for gp120, the membrane was stripped and probed again for p24 to exclude loading differences on the gel. Figure 5B shows such a Western blot probed for gp120 and p24. While the p24 level band intensities appeared indistinguishable, the band for gp120 of wild-type virions was more intense than that of Δ-Nef, EnvY712A, or EnvY712A/Δ-Nef virions. The lysates of the virus producer cells for this experiment were presented in Fig. 2A and showed no defect in gp160 expression that would account for these differences in the incorporation of gp120 into virions. This experiment was repeated using independently produced virions, and the bands were analyzed by densitometry. To normalize for any variations in p24, the data were expressed as a ratio of band intensity for gp120 to that of p24. The combined data from these two experiments using MT4-produced virions showed that the EnvY712A mutation resulted in a gp120 band intensity that was approximately 50% of wild type (Fig. 5C), a result which correlates with a 25% reduction in the absolute amount of gp120 as extrapolated from the standard curve of Fig. 5A. Nef-negative virions also showed a reduction, albeit even less dramatic, confirming the suggestion that Nef can affect the incorporation of Env into virions, at least in CD4-positive cells (21). Interestingly, the presence or absence of Nef in EnvY712A mutant viruses had no effect on Env incorporation, suggesting that Nef does not affect the incorporation of Env in the absence of an intact membrane-proximal tyrosine motif in gp41.
Using the same methods, the incorporation of Env into viruses produced from HEK293T cells was also analyzed. In contrast to the difference seen in the MT4-produced viruses, no significant difference was seen in the incorporation of Env into any of the mutant virions when produced by 293T cells (Fig. 6). Unlike MT4-produced virus, virions produced from 293T cells contained gp160 as well as gp120, and no apparent difference in the gp160/gp120 ratio was observed, indicating no processing defect caused by the Y712A mutation. Strikingly, these data indicated that the effects of both Nef and EnvY712 on the infectivity of 293T-produced virus are independent of Env incorporation.
FIG. 6.
Envelope incorporation in HEK293T-produced virus. Virus-containing supernatants produced from transfected HEK293T cells were pelleted and analyzed as described for Fig. 5B. Bands corresponding to gp160, gp120, and p24 are indicated.
Role of EnvY712A and Nef in viral entry.
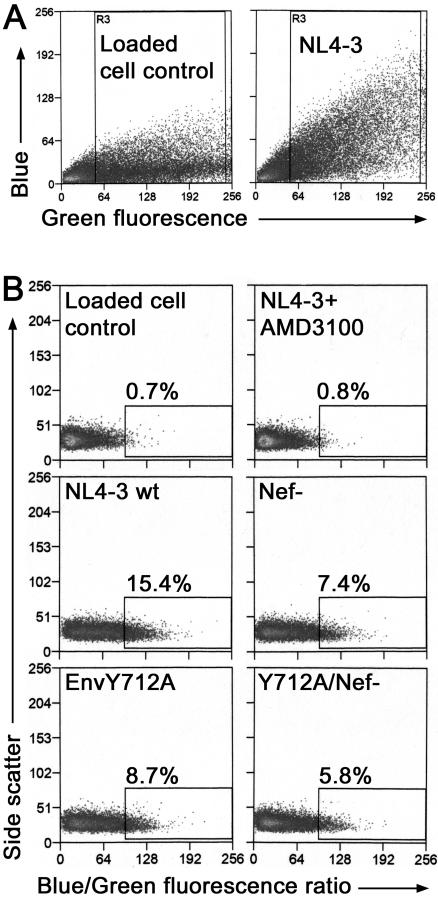
To identify the step in the virus replication cycle that is impaired by these mutations, we investigated the ability of the mutants to enter target cells. In the case of Nef, controversy exists regarding a putative positive effect on biologically relevant entry (30, 31), whereas no data exist regarding the EnvY712A mutant. To measure biologically relevant entry, virions containing a β-lactamase-Vpr fusion indicator protein (BlaM-Vpr) were produced by cotransfection of 293T cells with proviral plasmids and the BlaM-Vpr expression construct. The infectivity of pelleted and nonpelleted BlaM-Vpr-containing viruses was tested in the P4 single-cycle infectivity assay and closely matched the infectivity data shown in Fig. 4A for 293T-produced virions (data not shown). After normalization by p24 capsid concentration, equal amounts of pelleted BlaM-Vpr viruses were used to infect P4 cells (the same target cell used in the infectivity assays of Fig. 4A and C). After 5 h, the virions were removed, and the cells were incubated overnight with a fluorescent substrate, CCF2/AM, that is cleaved by β-lactamase. CCF2/AM contains two fluorochromes that undergo fluorescence resonance energy transfer, allowing for the detection of BlaM activity by flow cytometry (a shift of emission from green to blue after cleavage of the substrate and the loss of fluorescence resonance energy transfer). Uninfected cells loaded with CCF2/AM exhibited a wide range of green fluorescence intensity, with relatively few cells showing high levels of blue fluorescence (Fig. 7A, left panel). Cells infected with virus containing BlaM-Vpr shifted the population towards higher intensity blue fluorescence (Fig. 7A, right panel). For analysis, the results of this assay were plotted as a ratio of blue/green intensity per cell to account for possible differential dye loading (25). Uninfected cells loaded with CCF2/AM were used to set a gate for positive cells. During one representative experiment, uninfected cells showed a background level of 0.7% (Fig. 7B, upper-left panel). Cells infected with wild-type virus containing BlaM-Vpr resulted in 15.4% positive cells, while infection of cells in the presence of the CXCR4 antagonist AMD3100 completely blocked entry (0.8% positive cells) (Fig. 7B, left-center and upper-right panels). Δ-Nef, EnvY712A, and EnvY712A/Δ-Nef viruses each showed a reduction in BlaM-positive cells (Fig. 7B). Combined data from three independent experiments showed that all three mutant viruses exhibited an approximately 50% reduction in viral entry compared to wild type (Fig. 8A). To exclude the possibility that the indicator viruses varied in BlaM activity, the mutant virions were tested using a cell-free assay and a chromogenic substrate, nitrocefin; all preparations contained similar enzyme activity (Fig. 8B). In addition, AMD3100 had no effect on β-lactamase activity in vitro, validating the use of this compound to demonstrate the biological relevance of the entry assay (data not shown). Together, these data suggested that a defect in viral entry accounts for at least part of the infectivity defect of the 293T-produced Δ-Nef and EnvY712A mutants, despite their apparently wild-type phenotypes with respect to the virion incorporation of Env and the induction of fusion among target cells.
FIG. 7.
β-Lactamase-Vpr entry assay. Viruses were produced by cotransfection of the indicated provirus and pMM310, a β-lactamase-Vpr (BlaM-Vpr) fusion protein expression vector. Viruses were pelleted, resuspended in 1 ml of medium, normalized by p24 capsid concentration, and then used to infect P4.R5 cells for 5 h. After washing, cells were incubated with the β-lactamase-sensitive fluorochrome CCF2/AM for 16 to 18 h at 25°C. Analysis was performed by flow cytometry. Uncleaved substrate was detected as green fluorescence, and cleaved substrate was detected as blue fluorescence. (A) Plot of green fluorescence versus blue fluorescence. Loaded cell control refers to uninfected cells loaded with CCF2/AM. Box R3 encloses cells used for the ratio analysis in panel B, cells positive for CCF2/AM loading. (B) Data are expressed as a ratio of blue to green fluorescence versus side scatter and are gated using uninfected cells (loaded cell control). AMD3100 (100 nm) is a CXCR4 antagonist used to block entry into cells.
FIG. 8.
Entry assay summary and β-lactamase activity. (A) Data from three independent experiments as shown in Fig. 7 were expressed as a percentage of wild-type entry and averaged. (B) β-Lactamase activity of BlaM-Vpr-containing viruses. Pelleted viruses normalized by p24 capsid concentration were incubated with the chromogenic substrate nitrocefin for 22 h at room temperature. Samples were analyzed in a microplate spectrophotometer at 490 nm. Nitrocefin diluted into sample diluent was measured as background, and 20 μl of an ampicillin-resistant bacterial culture was used as a positive control. Error bars, standard deviations.
DISCUSSION
In this report we show that the membrane-proximal tyrosine-based sorting motif of gp41 Env (Y712XXL) is important for viral infectivity, viral entry, and, in the case of at least one T-cell line, optimal virion incorporation of envelope glycoprotein. The requirement of EnvY712 for infectivity was found in virions produced from either CD4-negative (HEK293T) or CD4-positive (MT4) cells; however, only virions produced from MT4 cells showed a reduction in the incorporation of Y712A Env. Consequently, the HEK293-producer cell data established unequivocally a role for the YXXL motif in infectivity that is independent of Env incorporation. The YXXL motif was essential for optimal viral replication in primary cultures of mononuclear cells from the peripheral blood, emphasizing its importance in a physiological context. Interestingly, these attributes of the Y712A mutant were similar both qualitatively and quantitatively with those of Nef-negative virus. Nef was required for optimal infectivity in both single-round, infectious-center assays and in spreading infections; it was required for the optimal incorporation of Env into MT4- but not HEK293-produced virions; and at least part of its effects were due to enhanced viral entry independent of the incorporation of Env into virions. However, the effects of nonsense mutations in nef were additive with those induced by the EnvY712A mutation, clearly indicating that Nef does not require the YXXL motif in Env for its effects. Nevertheless, the ability of both the EnvY712XXL and Nef sequences to enhance the entry of virions into target cells independently of the virion incorporation of Env suggests an intriguing hypothesis: both these sequences may influence trafficking events during viral morphogenesis in the producer cell such that the progeny virions are more fusogenic.
Previous studies regarding the role of the EnvY712XXL motif in viral infectivity have utilized various missense mutations, including Y712S, Y712C, and Y712F (8, 33). Paradoxically, the resulting mutants were not reported as impaired in infectivity and in some cases were characterized by supranormal infectivity. Nevertheless, our results demonstrate that replacement of Y712 with alanine yields virions that are less than half as infectious as wild type in single-round infectivity assays; in some cases the infectivity was as little as 20% that of wild type (Fig. 4). To reconcile these differences, we studied the Y712F and Y712C mutants in parallel with the alanine substitution mutant. Here, the Y712C mutation, like Y712A, impaired infectivity, whereas the Y712F mutation had little effect. The differences between the results herein and those reported previously may relate to different methods for standardization of the inocula (RT activity versus p24 antigen content) or to the use of different target cells (Magi versus P4). However, our results are potentially more consistent with the putative role of the motif as an AP-binding sequence; specifically, the conservative substitution of phenylalanine for tyrosine could be predicted to preserve the function of such a motif, whereas the replacement by cysteine would not (32). In addition, our data are consistent with the strict conservation of the YXXL sequence among isolates of HIV-1, HIV-2, and SIV and the importance of the YXXL motif for the pathogenesis of disease caused by SIV in macaques (16).
What may be the basis for the reduced infectivity of viruses with mutation of the Env YXXL motif? In the case of MT4-produced virus, the defect in infectivity may be due in part to the reduction of the incorporation of Env into virions (Fig. 5C). Although the optimal number of Env trimers per virus particle is unknown, a recent study estimated that only 7 to 14 trimers are incorporated per particle (10). Based on our quantification of gp120 by Western blot and using the same estimate for Gag molecules per virion (1,200 to 2,500) (10), we estimated that there are 18 to 38 Env trimers per virus particle produced from HEK293T cells. If, indeed, there are this few Env trimers per virion, then a 25% reduction could conceivably account for the reduced infectivity of EnvY712A virus. By analogy, tyrosine-based sorting motifs within the Env cytoplasmic domain of bovine leukemia virus are important for viral entry and the incorporation of Env into virions (17). However, in contrast to the MT4-produced virus, virions produced from 293T cells did not show a difference in the incorporation of EnvY712A, but they did show a marked defect in infectivity as a result of this mutation. These results clearly indicated that the infectivity defect of this mutant does not rely on a defect in the virion incorporation of Env.
The mechanism by which the YXXL motif influences infectivity independently of the virion incorporation of Env is unclear. However, at least part of this effect appears related to the impaired efficiency of viral entry into target cells, as evidenced by the reduced delivery of β−lactamase activity to target cells by virions. These findings imply that the virions are more fusogenic when the YXXL motif is intact.
The hypothesis that the YXXL motif enhances the fusogenicity of virions seems paradoxical considering that quantitatively similar defects in the infectivity of the EnvY712A mutant were observed using two distinct infectious-center assays: one using trans-activation of the expression of β-galactosidase as the indicator (P4 assay) and the other using the morphological scoring of syncytia (Fig. 4B). The similar defects detected in these two assays suggests that the fusogenicity of Y712A Env itself is likely unimpaired by the mutation; if it were impaired, then the syncytium-formation assay would be predicted to yield a greater apparent defect than the P4 assay. One resolution of this paradox is that cell-cell fusion is less sensitive than virus-cell fusion to an impaired fusogenicity of Y712A Env. A second resolution, which we favor, is that the YXXL motif in the cytoplasmic domain of gp41 renders the virion more fusogenic via its function as an AP-binding sequence, targeting the trafficking of Env and consequently viral assembly to specific compartments such as endosomal or plasma membrane domains. In this model, the specific lipid and/or protein content of the membranes on which virions assemble is the varying determinant of fusogenicity in the experiments herein, such that optimal infectivity requires the targeting activity of the YXXL motif. Consequently, the intrinsic fusogenicity of the Y712A Env glycoprotein need not be affected.
The hypothesis that the trafficking function of the YXXL motif in Env directs viral assembly to optimal cellular membranes has the potential to resolve two problems: one is the question of why the cytoplasmic domain of gp41 contains AP-binding motifs and the other is the unclear purpose of targeting Env to specific, largely internal, cellular membranes. Several hypotheses have been proposed to explain these observations: limiting the amount of Env at the surface of an infected cell may prevent certain immune responses from detecting and killing the cell (15), directing Env to subdomains of the plasma membrane may optimize cell-to-cell transmission of the virus (14), and endocytosis may maintain an optimal steady-state level of Env at the plasma membrane prior to viral budding (15). Although any or all of these mechanisms may be operational, we favor the hypothesis that AP-binding and other protein sorting motifs in gp41, such as the YW motif that binds TIP47 (4), are required for optimal infectivity because they target viral assembly to an appropriate internal membrane compartment. This compartment is likely endosomal, given the juxtanuclear location of gp41 at steady state (Fig. 2).
This trafficking-assembly model has the potential to encompass the role of Nef in viral infectivity and fusogenicity, providing a general basis for the similar phenotypes of the Y712A and Δ-Nef mutants. Indeed, we initially hypothesized that Nef enhances the infectivity of virions by influencing the trafficking of Env, such that the YXXL motif would be required for the Nef effect. Instead, the virologic effects of the Y712A and nef-nonsense mutations, though qualitatively and quantitatively similar, were additive in every format (except the entry assay). Taken together, these results suggest that the effects of the Env YXXL and Nef sequences are not interdependent. Nevertheless, they may be mechanistically related. Nef has widespread effects on the endosomal system (19), where the majority of gp41 appears to accumulate. Furthermore, mutation of the EXXXLL AP-binding motif in Nef, like mutation of the YXXL motif in Env, impairs infectivity in a CD4-independent manner that is manifest at least in part at the level of viral entry (data not shown). Apparently, AP-binding motifs in two distinct viral proteins, Env and Nef, make similar contributions to viral infectivity and replication.
The data herein regarding Nef and viral entry may be controversial. Although Nef reportedly enhances the cytoplasmic delivery of virion cores as detected using a cell fractionation assay (30), a very recent report that used an entry assay similar to that described herein failed to reveal an entry defect in the case of Nef-negative virions (31). Notably, our data regarding the CD4-independent effect of Nef nominally indicate that Nef-negative virions enter cells with half the efficiency of the wild type, but they are only 1/20 as infectious (compare Fig. 4A, 7B, and 8A). One interpretation of these data is that a defect in entry only partly explains the defect in infectivity. However, another interpretation is that these differences in the magnitude of the defects are due to an inoculum effect; data regarding entry derive from the exposure of cells to inocula 10- to 100-fold more concentrated than those used to obtain the infectivity data. Consequently, although the defect in entry may account for only part of the Nef (and Env YXXL) infectivity effect, it remains conceivable that it may account for the entire effect.
In summary, we provide evidence that the YXXL AP-binding motif in gp41 contributes directly to the optimal infectivity and replication of HIV-1. Although the virologic functions of the YXXL motif and the HIV-1 Nef protein do not appear interdependent, the similar contributions of these sequences to replication, infectivity, and entry by a process independent of CD4 and the virion incorporation of Env suggest a common underlying mechanism. We hypothesize that this mechanism involves AP-mediated trafficking, targeting viral morphogenesis to specific, probably endosomal, membranes. Proof of this hypothesis, and an understanding of why assembly on such membranes yields more fusogenic virions, remains to be determined.
ADDENDUM IN PROOF
In support of a common mechanism underlying the virologic effects of the gp41 YXXL motif and Nef, a recent study reported the emergence of compensatory mutations in the cytoplasmic domain of gp41 during serial passage of nef-deleted SIV in rhesus monkeys (L. Alexander, P. O. Illyinskii, S. M. Lang, R. E. Means, J. Lifson, K. Mansfield, and R. C. Desrosiers, J. Virol. 77:6823-6835, 2003). Strikingly, these mutations were identical or very similar to those that emerged during passage in vivo of SIV containing a deletion in the gp41 YXXL motif, indicating a common compensatory response to the loss of Nef or YXXL sequences (16).
Acknowledgments
We thank Sherry Rostami, Heather Overton, Melissa Moore, and Nancy Keating of the UCSD Center for AIDS Research p24 Core for assaying samples; Juan Lama and María José Cortés Mateos for technical advice and the provision of gp120 antiserum; Nathaniel Landau for the gift of HEK293T cells and pMM310; Peggy O'Keefe of the VA Research Core, San Diego, Calif., for flow cytometry assistance; and Darica Smith for assistance in preparation of the manuscript.
This work was supported by grants from the National Institutes of Health (AI38201), the UCSD Center for AIDS Research (AI36214), and the Research Center for AIDS and HIV Infection of the San Diego Veterans Affairs Medical Center.
REFERENCES
- 1.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69:5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akari, H., T. Fukumori, and A. Adachi. 2000. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J. Virol. 74:4891-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blot, G., K. Janvier, S. Le Panse, R. Benarous, and C. Berlioz-Torrent. 2003. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for Env incorporation into virions and infectivity. J. Virol. 77:6931-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boge, M., S. Wyss, J. S. Bonifacino, and M. Thali. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273:15773-15778. [DOI] [PubMed] [Google Scholar]
- 6.Bresnahan, P. A., W. Yonemoto, S. S. Ferrell, D. G. R. Williams-Herman, and W. C. Greene. 1998. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr. Biol. 8:1235-1238. [DOI] [PubMed] [Google Scholar]
- 7.Bültmann, A., W. Muranyi, B. Seed, and J. Haas. 2001. Identification of two sequences in the cytoplasmic tail of the human immunodeficiency virus type 1 envelope glycoprotein that inhibit cell surface expression. J. Virol. 75:5263-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cervantes-Acosta, G., R. Lodge, G. Lemay, and E. A. Cohen. 2001. Influence of human immunodeficiency virus type 1 envelope glycoprotein YXXL endocytosis/polarization signal on viral accessory protein functions. J. Hum. Virol. 4:249-259. [PubMed] [Google Scholar]
- 9.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 10.Chertova, E., J. J. Bess, Jr., B. J. Crise, R. C. Sowder II, T. M. Schaden, J. M. Hilburn, J. A. Hoxie, R. E. Benveniste, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), Is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesebro, B., and K. Wehrly. 1988. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J. Virol. 62:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowers, M. Y., C. A. Spina, T. J. Kwoh, N. J. S. Fitch, D. D. Richman, and J. C. Guatelli. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 68:2906-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortés, M. J., F. Wong-Staal, and J. Lama. 2002. Cell surface CD4 interferes with the infectivity of HIV-1 particles released from T cells. J. Biol. Chem. 277:1770-1779. [DOI] [PubMed] [Google Scholar]
- 14.Deschambeault, J., J. P. Lalonde, G. Cervantes-Acosta, R. Lodge, E. A. Cohen, and G. Lemay. 1999. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J. Virol. 73:5010-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan, M. A., L. M. Carruth, J. F. Rowell, X. Yu, and R. F. Siliciano. 1996. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J. Virol. 70:6547-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fultz, P. N., P. J. Vance, M. J. Endres, B. Tao, J. D. Dvorin, I. C. Davis, J. D. Lifson, D. C. Montefiori, M. Marsh, M. H. Malim, and J. A. Hoxie. 2001. In vivo attenuation of simian immunodeficiency virus by disruption of a tyrosine-dependent sorting signal in the envelope glycoprotein cytoplasmic tail. J. Virol. 75:278-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inabe, K., M. Nishizawa, S. Tajima, K. Ikuta, and Y. Aida. 1999. The YXXL sequences of a transmembrane protein of bovine leukemia virus are required for viral entry and incorporation of viral envelope protein into virions. J. Virol. 73:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwatani, Y., T. Ueno, A. Nishimura, X. Zhang, T. Hattori, A. Ishimoto, M. Ito, and H. Sakai. 2001. Modification of virus infectivity by cytoplasmic tail of HIV-1 TM protein. Virus Res. 74:75-87. [DOI] [PubMed] [Google Scholar]
- 19.Janvier, K., H. Craig, D. Hitchin, R. Madrid, N. Sol-Foulon, L. Renault, J. Cherfils, D. Cassel, S. Benichou, and J. Guatelli. 2003. HIV-1 Nef stabilizes the association of adaptor protein complexes with membranes. J. Biol. Chem. 275:8725-8732. [DOI] [PubMed] [Google Scholar]
- 20.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, M. Marsh, and J. A. Hoxie. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J. Virol. 69:5217-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lama, J., A. Mangasarian, and D. Trono. 1999. Cell-surface expresion of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr. Biol. 9:622-631. [DOI] [PubMed] [Google Scholar]
- 22.Lodge, R., H. gottlinger, D. Gabuzda, E. A. Cohen, and G. Lemay. 1994. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J. Virol. 68:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodge, R., J. P. Lalonde, G. Lemay, and E. A. Cohen. 1997. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 16:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, M. D., M. T. Warmerdam, K. A. Page, M. B. Feinberg, and W. C. Greene. 1995. Expression of the human immunodeficiency virus type 1 (HIV-1) nef gene during HIV-1 production increases progeny particle infectivity independently of gp160 or viral entry. J. Virol. 69:579-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Münk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno, H., R. C. Aguilar, M. C. Fournier, S. Hennecke, P. Cosson, and J. S. Bonifacino. 1997. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238:305-315. [DOI] [PubMed] [Google Scholar]
- 28.Olivetta, E., K. Pugliese, R. Bona, P. d'Aloja, F. Ferrantelli, A. C. Santarcangelo, G. Mattia, P. Verani, and M. Federico. 2000. cis expression of the F12 human immunodeficiency virus (HIV) Nef allele transforms the highly productive NL4-3 HIV type 1 to a replication-defective strain: involvement of both Env gp41 and CD4 intracytoplasmic tails. J. Virol. 74:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowell, J. F., P. E. Stanhope, and R. F. Siliciano. 1995. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J. Immunol. 155:473-488. [PubMed] [Google Scholar]
- 30.Schaeffer, E., R. Geleziunas, and W. C. Greene. 2001. Human immunodeficiency virus type 1 Nef functions at the level of virus entry by enhancing cytoplasmic delivery of virions. J. Virol. 75:2993-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tobiume, M., J. E. Lineberger, C. A. Lundquist, M. D. Miller, and C. Aiken. 2003. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J. Virol. 77:10645-10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trowbridge, I. S., J. F. Collawn, and C. R. Hopkins. 1993. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu. Rev. Cell Biol. 9:129-161. [DOI] [PubMed] [Google Scholar]
- 33.West, J. T., S. K. Weldon, S. Wyss, X. Lin, Q. Yu, M. Thali, and E. Hunter. 2002. Mutation of the dominant endocytosis motif in human immunodeficiency virus type 1 gp41 can complement matrix mutations without increasing Env incorporation. J. Virol. 76:3338-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilk, T., T. Pfeiffer, and V. Bosch. 1992. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology 189:167-177. [DOI] [PubMed] [Google Scholar]
- 35.Wyss, S., C. Berlioz-Torrent, M. Boge, G. Blot, S. Honing, R. Benarous, and M. Thali. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor. J. Virol. 75:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, J., and C. Aiken. 2001. Nef enhances human immunodeficiency virus type 1 infectivity resulting from intervirion fusion: evidence supporting a role for Nef at the virion envelope. J. Virol. 75:5851-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]