Abstract
Microbial motility frequently depends on flagella or type IV pili. Using recently developed archaeal genetic tools, archaeal flagella and its assembly machinery have been identified. Archaeal flagella are functionally similar to bacterial flagella and their assembly systems are homologous with type IV pili assembly systems of Gram-negative bacteria. Therefore elucidating their biochemistry may result in insights in both archaea and bacteria. FlaI, a critical cytoplasmic component of the archaeal flagella assembly system in Sulfolobus acidocaldarius, is a member of the type II/IV secretion system ATPase superfamily, and is proposed to be bi-functional in driving flagella assembly and movement. In the present study we show that purified FlaI is a Mn2+ -dependent ATPase that binds MANT-ATP [2′-/3′-O-(N’-methylanthraniloyl)adenosine-5′-O-triphosphate] with a high affinity and hydrolyses ATP in a co-operative manner. FlaI has an optimum pH and temperature of 6.5 and 75 °C for ATP hydrolysis. Remarkably, archaeal, but not bacterial, lipids stimulated the ATPase activity of FlaI 3–4-fold. Analytical gel filtration indicated that FlaI undergoes nucleotide-dependent oligomerization. Furthermore, SAXS (small-angle X-ray scattering) analysis revealed an ATP-dependent hexamerization of FlaI in solution. The results of the present study report the first detailed biochemical analyses of the motor protein of an archaeal flagellum.
Keywords: ATPase superfamily, lipid activation, secretion superfamily ATPase, Sulfolobus acidocaldarius, type II/IV secretion ATPase, type IV pilus
INTRODUCTION
Motility is vitally important for most prokaryotes, and various mechanisms utilized by bacteria and archaea for motility have been described [1]. In bacteria, the two most commonly used mechanisms, movement via flagellar rotation as well as twitching using the extension and retraction of T4P (type IV pili) have been studied extensively [2,3]. This has led to the identification and characterization of many components required for the assembly and function of these systems. Very little, however, is known about the assembly of archaeal surface appendages such as flagella and pili, although a diverse variety have been identified [4,5]. In Halobacterium salinarum flagella have been reported to be involved in swimming motility [6–8] and shown to be important for swarming on solid surfaces in methanogens and Sulfolobus solfataricus [7,8]. Recent evidence further revealed that flagella in S. solfataricus are involved not only in surface attachment, but also in late phases of bio-film maturation [9–11].
Remarkably, the structural and genetic information available indicated that the archaeal flagella are more similar to bacterial T4P than to the bacterial flagella [4,12–14]. Genetic analysis in different flagellated archaea has been used to identify a genetic locus that encodes an AFAS (archaeal flagella assembly system) [15]. Within different archaea, this genetic locus contains 7–13 genes. Usually it contains several flagellin genes (flaA and/or flaB genes) followed by a conserved set of flagella-related protein genes (flaC–flaJ) [16]. The genetic locus in Sulfolobales, however, contains, apart from the conserved set of flagella-related protein genes, only one gene encoding a flagellin. Several components of the archaeal flagella assembly machinery show homology with proteins involved in T4P assembly, T2S (type II secretion) and T4S (type IV secretion) systems of Gram-negative bacteria [4,7,17–21]. T2S systems in bacteria play a role in the secretion of toxins and enzymes, e.g. the cholera toxin of Vibrio cholerae [22], whereas T4S systems are used for the transport of DNA to either bacterial or eukaryal host cells, as exemplified by Agrobacterium tumefaciens [23]. T4P are involved in a variety of functions, such as twitching motility and cell adhesion which are essential for the pathogenicity of several harmful bacteria [14]. All three systems have significant structural and functional similarities pointing to a common origin [18]. Interestingly, the subunits that are found to be similar in those systems are also present in the AFAS [24]. These components include pre-flagellin peptidases (e.g. PibD of S. solfataricus) [4], FlaJ, which has homology with the polytopic membrane protein found in T4P systems (e.g. PilG of Neisseria) [17,18,25] and FlaI, which has homology with T2S ATPases and T4P assembly ATPases (e.g. PilB, GspE, EpsE, HP0525) [18,19,26–28]. Three-dimensional reconstructions of electron microscopic images of archaeal flagella and of T4P have revealed a considerable structural homology with T4P [13,20,21,29,30]. These collective results showed that the archaeal flagella and T4P systems show strong similarities and might be evolutionarily related.
A central component found in both systems is the cytoplasmic ATPase that provides the energy. These ATPases belong to the ‘secretion superfamily ATPase’ or ‘T2S/T4S ATPase’ family [18]. Proteins of this superfamily function in multiple macromolecular transport systems in bacteria and archaea, such as T2S, T4S and T4P assembly, and in archaeal flagella assembly systems [14,18,26,27,31]. Members of this superfamily contain characteristic conserved motifs such as Walker A, Walker B and P-loop motifs [GX4GK(S/T)], which are involved in nucleotide binding. They also contain the aspartate box motif involved in divalent cation binding and the histidine box motif possibly involved in stabilization of nucleotide binding [32].
Based on their sequence similarities, secretion ATPases were grouped into different clusters, including clusters for ATPases from T2S systems, T4P systems, and a cluster for archaeal ATPases [18]. Although both T2S and T4S are powered by secretion ATPases, the basic mechanism is different for these secretion systems. In the T2S system, the ATPase GspE is involved in assembly of the ‘pseudopilin’ filament called pseudopilus. In comparison, T4S system ATPases may function as protein translocators by interacting with the secreted protein already folded in the cytoplasm. In T4P assembly systems, two ATPases, PilB and PilT, are present and function in assembly and disassembly of the pilus respectively [33–35]. Interestingly, the amino acid sequence of FlaI resembles bacterial T2S- and T4P-type ATPases, suggesting a common evolutionary root for all of these secretion ATPases.
The archaeal flagellum is able to move clockwise and counterclockwise, which results in a switch in swimming direction [36]. The question arises how a T4P would be able to move in this fashion, as normally twitching motility is conferred by filament assembly and disassembly, and not by any rotational movement. Because FlaI is the only ATPase identified in archaeal flagella operons, it is most likely to be involved in two functions: first in the assembly process and secondly in the movement leading to motility. Therefore it is important to understand how this protein can perform both functions. The specific role of the secretion ATPase in archaeal flagella assembly systems has not previously been addressed. It has been shown for Methanococcus maripaludis and Halobacterium salinarum that FlaI-deletion mutants are impaired in assembly of their flagella [16,19,37], but neither have been biochemically characterized. The relationship between ATP binding and hydrolysis, and flagellum assembly and rotation remains unknown. In the present study, the secretion ATPase FlaI from S. acidocaldarius, a thermoacidophilic crenarchaeon that grows optimally at 70–80 °C and pH 2–3, is biochemically and structurally characterized.
MATERIALS AND METHODS
Construction of a FlaI overexpression vector
To enable expression of non-tagged FlaI or FlaI with an N-terminal histidine tag, the flaI gene (saci_1173) was amplified by PCR from genomic DNA of S. acidocaldarius DSM639 using primer pairs FlaI713/FlaI714 or FlaI728/FlaI729, and was cloned as NdeI/BamHI or EcoRI/HindII fragments into plasmid pETDuet-1 (Novagen), yielding plasmids pSVA251 and pSVA263 respectively. For expression analysis, the respective vectors were transformed to Escherichia coli C43 cells containing the RILChl-plasmid (Stratagene) [38].
Site-directed mutagenesis of flaI
Mutagenesis of flaI was performed by overlap PCR on vector pSVA263. Mutants were created by changing Lys268 within the predicted Walker A motif, Asp290 within the aspartate box and Glu336 within the Walker B motif to alanine residues. The resultant plasmids were named pSVA265 (K268A), pSVA307 (D290A), pSVA308 (E336A) and pSVA314 (K268A/E336A). Sequences of the oligonucleotides used for mutagenesis are listed in Table 1.
Table 1.
Primers and plasmids used for assays in the present study
| Ampr, ampicillin resistant; Carr, chloramphenicol resistant. | ||
|---|---|---|
| Bacterial strains, primers and plasmids | Relevant characteristics | Source or reference |
| Strains | ||
| E . coli | ||
| DH5α | l2 f80d/lacZDM15 D(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK2 mK1) supE44 thi-1 gyrA relA1 | Gibco |
| Bl21(DE3)-RIL | B F-ompT hsdS(rB–mB–) dcm + Tetr E. coli gal λ (DE3) endA Hte [argU ileY leuW Camr] | Stratagene |
| Walker cells-RIL (C43) | F-ompT gal hsdSB (rB-mB-) dcm lon _DE3 endA Hte [argU ileY leuW Camr] | [38] |
| S. acidocaldarius | ||
| DSM639 | Hyperthermo-acidophilic crenarchaeon which grows optimally at 80°C and pH 2–3 | DSMZ |
| Primers | ||
| FlaI713 | 5′-GGGGGCATATGAGCTTCGTGGAAGAC-3′; forward primer for FlaI containing an NdeI restriction site (underlined) | The present study |
| FlaI714 | 5′-GGGGGCTCGAGTCAAATATTACTGACATATTTTATC-3′; reverse primer for FlaI containing a XhoI restriction site (underlined) | The present study |
| FlaI728 | 5′- CCCCCGAATTCGATGAGCTTCGTGGAAGAC-3′; forward primer for FlaI containing an EcoRI restriction site (underlined) | The present study |
| FlaI729 | 5′-CCCCCAAGCTTTCAAATATTACTGACATATTTTATC-3′; reverse primer for FlaI containing a HindIII restriction site (underlined) | The present study |
| FlaI742 | 5′-ATCTGGAGCGACAACTAC-3′; overlapping internal forward primer for the K268A mutation in FlaI | The present study |
| FlaI743 | 5′-AATGTAGTTGTCGCTCCAG-3′; overlapping internal reverse primer for the K268A mutation in FlaI | The present study |
| FlaI782 | 5′-TATTGAGGCGACTCCAG-3′; overlapping internal forward primer for the D290A mutation in FlaI | The present study |
| FlaI783 | 5′-TCTGGAGTCGCCTCAATA-3′; overlapping internal reverse primer for the D290A mutation in FlaI | The present study |
| FlaI784 | 5′-ACTTGTGGGAGCAATTAGAG-3′; overlapping internal forward primer for the E336A mutation in FlaI | The present study |
| FlaI785 | 5′-CTTTGTCTCTAATTGCTCCCACAAG-3′; overlapping internal reverse primer for the E336A mutation in FlaI | The present study |
| Plasmids | ||
| pETDuet-1 | Ampr, Carr, expression plasmid containing replicon ColE1 (pBR322) and two MCS (MCS1 and MCS2) | Novagen |
| pSVA251 | Ampr, Carr, pETDuet-1 carrying untagged FlaI in MCS2 using restriction sites NdeI-XhoI | The present study |
| pSVA263 | Ampr, Carr, pETDuet-1 carrying N-terminal His-tagged FlaI in MCS1 using restriction sites EcoRI-HindIII | The present study |
| pSVA265 | Ampr, Carr, pETDuet-1 carrying N-terminal His-tagged FlaIK268A in MCS1 using restriction sites EcoRI-HindIII | The present study |
| pSVA307 | Ampr, Carr, pETDuet-1 carrying N-terminal His-tagged FlaID290A in MCS1 using restriction sites EcoRI-HindIII | The present study |
| pSVA308 | Ampr, Carr, pETDuet-1 carrying N-terminal His-tagged FlaIE336A in MCS1 using restriction sites EcoRI-HindIII | The present study |
| pSVA314 | Ampr, Carr, pETDuet-1 carrying N-terminal His-tagged FlaIK268A/E336A in MCS1 using restriction sites EcoRI-HindIII | The present study |
Expression of recombinant proteins in E. coli
A total volume of 10 ml of an overnight culture of E. coli C43 cells containing the RIL plasmid [38] and one of the respective overexpression plasmids was used to inoculate 1 litre of Luria–Bertani medium containing ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). Cells were grown at 37 °C until a D600 of 0.6 was reached, at which time 1 mM IPTG (isopropyl β-D-thiogalactopyranoside) was added. The temperature was reduced to 16 °C, and growth was continued overnight to reduce inclusion body formation. The cells were collected by centrifugation, resuspended in lysis buffer [50 mM Hepes-NaOH (pH 7.5), 150 mM NaCl and 10 % glycerol] containing the complete EDTA-free protease inhibitor cocktail (1 tablet/50 ml of lysate; Roche), frozen in liquid nitrogen and stored at −80°C.
Purification of recombinant proteins
Before purification, frozen resuspended cell pellets were thawed on ice. Cells were lysed by sonication with a Sonoplus HD3100 sonicator (Bandelin Sonorex Biotechnique) using probe HD3100. Cell debris was removed by centrifugation at 6000 g for 15 min followed by centrifugation at 50 000 rev./min for 30 min at 4°C in a Optima™ MAX-XP ultracentrifuge (rotor MLA 55; Beckman Coulter) to remove the membranes. The supernatant was incubated at 70°C for 20 min in a water bath. The sample was cooled down on ice and precipitated proteins were removed by centrifugation at 50000 rev./min for 30 min at 4°C in a Optima™ MAX-XP ultracentrifuge (rotor MLA 55; Beckman Coulter). For purification of histidine-tagged proteins, the supernatant was applied to a Ni2+ -affinity column (Native IMAC) on the Profinia™ protein purification system (Bio-Rad Laboratories). Bound protein was eluted in lysis buffer containing 500 mM imidazole. The eluted fraction was dialysed overnight at 4°C against lysis buffer. The dialysed fraction was loaded on to a Superdex 200 10/300 gel-filtration column on an ÄKTA purifier system (GE Healthcare), equilibrated with lysis buffer (flow rate of 0.5 ml/min). The untagged FlaI was expressed, sonicated and heat treated, as described above. The protein was then precipitated at room temperature (25°C) with 20% (w/v) ammonium sulfate for 30 min. The precipitated protein was re-suspended in buffer Blue A [50 mM Tris/HCl (pH 8.0), 200 mM NaCl and 2 mM EDTA] and loaded on to a 5 ml HiTrap BLUE column in the ÄKTA purifier system (GE Healthcare). The column was washed with buffer Blue A and the bound protein was eluted with buffer Blue B [50 mM Tris/HCl (pH 8.0), 1 M NaCl and 2 mM EDTA]. The eluted protein was further purified on a 5 ml HiTrap PHENYL column. The protein was eluted using a gradient from buffer Blue B to buffer PHENYL [50 mM Tris/HCl (pH 8.0) and 2 mM EDTA]. The eluted protein was then concentrated (<2 ml) using an Amicon concentrator system (Millipore), and loaded on to a Superose 6 10/300 size-exclusion column pre-equilibrated with PHENYL buffer and eluted with 50 mM Hepes-NaOH buffer (pH 7.5) containing 150 mM NaCl.
Protein purity was monitored by reducing SDS/PAGE. Gels were stained with Biosafe (Bio-Rad Laboratories) protein stain. The protein concentration was determined using the Bio-Rad protein assay dye-dependent reagent (Bio-Rad Laboratories).
ATPase assay
ATPase activities were measured using the Malachite Green assay, as described previously [39]. Briefly, purified protein (2 μg) was pre-heated for 2 min in 100 μl of assay buffer [50 mM Hepes (pH 7.5), 150 mM NaCl and 5 mM MgCl2]. Reactions were initiated by the addition of 1 mM ATP, and incubated for 5 min at 70 °C. The reaction was stopped by freezing in liquid nitrogen. The amount of Pi released was determined using the Malachite Green assay. The data were corrected for non-enzymatic ATP hydrolysis. To determine the activity of FlaI as a function of temperature, pH, nucleotides and divalent cations, the assay was performed at different temperatures, in different buffers [50 mM citrate (pH 3.0–5.5), Mes (pH 6.0–6.5), Hepes (pH 7.0–7.5) or Tris/HCl (pH 8.0–9.5)], with different NTPs (5 mM of ATP, GTP, CTP or UTP), and in the absence of or with different divalent cations (5 mM Na-EDTA, 5 mM MnCl2 or 5 mM CaCl2,) respectively. The ATPase activity was determined as a function of the ATP concentration, and the resulting data were fitted to the Hill equation:
to obtain the Vmax (a), the dissociation constant, K0.5 (b), and the Hill coefficient (n). To analyse the effect of tetraether lipids on ATP hydrolysis by FlaI, total lipids of S. acidocaldarius were extracted, as described previously [40]. The ATPase activity of FlaI (1 μM purified protein) was measured as described above in the presence of different concentrations of isolated lipids (40, 80, 100, 200, 400 and 800 μg).
MANT-ATP [2′-/3′-O-(N’-methylanthraniloyl)adenosine-5′-O-triphosphate]-binding assay
Binding of MANT-ATP was followed by FRET (fluorescence resonance energy transfer) measurements using a temperature-controlled ISS PC1 spectrofluorometer. The excitation wavelength was set to 285 nm and the emission spectrum was recorded from 400 to 500 nm. The slit-widths for the excitation and the emission beam were set to 1 nm. MANT-ATP-binding affinities were determined by titrating 0.1 μM protein with increasing concentration of MANT-ATP (0.125–25 μM) at 25 °C in binding buffer [50 mM Hepes (pH 7.5), 150 mM NaCl and 5 mM MnCl2]. Fluorescence spectra were corrected for the fluorescence of unbound MANT-ATP, and the resulting fluorescence at 450 nm was fitted to the Hill equation:
to obtain the ΔFmax (a), the dissociation constant, K0.5 (b), and the Hill coefficient (n).
Liposome pull-down assay
The liposome pull-down assay was performed as described previously [41]. Briefly, isolated and dried tetraether lipids from S. acidocaldarius were hydrated with binding buffer and vigorously vortex-mixed to ensure dispersion of the various lipids in a Sonoplus HD3100 sonicator (Bandelin Sonorex Biotechnique) using probe HD3100. Liposomes were collected from the supernatant after centrifugation (2500 g for 5 min) and used in subsequent assays. To perform experiments with E. coli polar lipids, a liquid form of polar lipid mixture was purchased from Avanti Polar Lipids. The polar lipids were dried in a vacuum desiccator for approximately 5 h. Dried lipids were hydrated with binding buffer and treated in a similar manner as tetraether lipids to generate differentially sized liposomes for the pull-down assay.
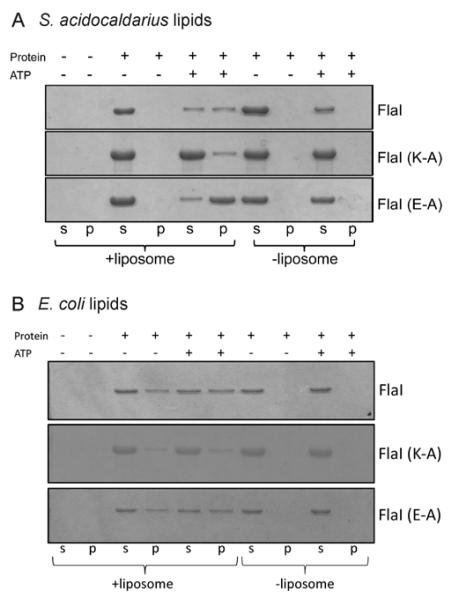
The pull-down assay was performed separately with 1 μM His6-tagged FlaI, FlaIK268A or FlaIE336A purified protein. The proteins were pre-incubated with 1 mM ATP at 25°C for 30 min in binding buffer in a total volume of 100 μl. Liposomes were added to a final lipid concentration of 80 μg/ml and reactions were incubated at room temperature for 1 h. By centrifugation at 40000 rev./min in a Beckman TLA100.3 rotor in an Optima™ MAX-XP ultracentrifuge (Beckman Coulter) at 4°C for 30 min, the lipid vesicles were separated from the soluble fraction. The supernatant and pellet were analysed on SDS/PAGE (11% gel) and visualized with Coomassie Blue.
Analytical gel filtration
Purified FlaI was concentrated to 1.0 mg/ml in 50 mM Hepes-NaOH buffer (pH 7.5) and 150 mM NaCl by centrifugation in an Amicon filter (10 kDa cut-off; Millipore). The sample was then pre-incubated at 50°C for 10 min followed by incubation at 4°C for 6 h with either 1 mM p[NH]ppA (adenosine 5′-[β,γ - imido]triphosphate) or ADP in the presence of 10 mM MgCl2. The sample was loaded on to a Superdex 200 10/300 column on the ÄKTA purifier system as described above. All of the fractions were analysed on SDS/PAGE and immunoblotted using anti-histidine antibodies (Abcam).
SAXS (small-angle X-ray scattering) analyses
SAXS data were collected at the ALS beamline 12.3.1 (SIBYLS, Lawrence Berkeley National Laboratory, Berkeley, CA, U.S.A.) [42]. The scattering vector is defined as q = 4πsinθ/λ,where 2θ is the scattering angle. Prior to SAXS data collection, wt (wild-type) FlaI or FlaIE336A were applied to a Superose 6 size-exclusion column in 20 mM Tris/HCl (pH 7.0) and 200 mM NaCl, or 20 mM Tris/HCl (pH 7.0), 200 mM NaCl, 1 mM ATP and 2.5 mM MgCl2 respectively. Fractions from the column were analysed directly, and after 2-fold and 4-fold concentration of the sample using Corning Spin-X concentrators. All experiments were performed at 20°C, and data were processed as described previously [43]. The data acquired at short- and long-time exposure (1 and 10 s) were merged for calculations using the entire scattering spectrum. The experimental SAXS data for different protein concentrations were investigated for aggregation using Guinier plots [44]. The program GNOM [45] was used to compute the pair distance distribution functions, P(r). This approach also provided the maximum dimension of the macromolecule, Dmax. The overall shapes were restored from the experimental data using the program GASBOR and averaging of 15 independent structures using the program DAMAVER [46]. Molecular graphics images were produced using the UCSF (University of California, San Francisco) Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the UCSF [supported by NIH (National Institutes of Health) grant P41 RR-01081 to Pettersen et al., 2004]. Theoretical scattering curves from crystal structures were calculated using FoXS [47].
RESULTS
Expression, purification and biochemical characterization of FlaI
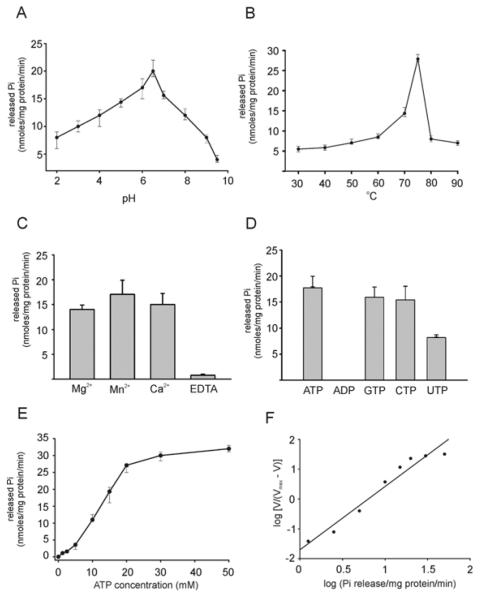
Both histidine-tagged and wt FlaI were expressed in E. coli and purified to homogeneity. Purified FlaI was analysed by SDS/PAGE and found to be over 99% pure. FlaI was tested for the ability to hydrolyse ATP under different conditions. The optimal conditions for ATP hydrolysis by FlaI were determined to be pH 6.5 and 75°C (Figures 1A and 1B), which are close to the cytoplasmic pH and the optimal growth temperature of S. acidocaldarius. Mn2+ was the preferred divalent cation for ATP hydrolysis (Figure 1C), as was observed for other secretion ATPases [26]. Considerable activity was, however, also detected in the presence of Ca2+ and Mg2+ respectively (Figure 1C). FlaI hydrolysed ATP with the highest rate, but it was also able to hydrolyse GTP, CTP and UTP (Figure 1D), as seen for other ATPases. No conversion of ADP into AMP was detected (Figure 1D).
Figure 1. ATPase activity of FlaI under various conditions.
Total Pi released upon ATP (NTP) hydrolysis was measured using the Malachite Green assay. (A) Influence of pH on FlaI ATPase activity. (B) Influence of temperature on FlaI ATPase activity. (C) Preference for optimal divalent metal ion as a cofactor in FlaI ATPase activity. The final concentration of protein used in the assays was 1 μM and the final concentration of metal ions was 5mM. (D) Analysis of optimal substrate for hydrolysis. NTPs were added to a final concentration of 5 mM. (E) FlaI ATPase activity as a function of the ATP concentration measured at 70°C with 1 μM FlaI. (F) The same FlaI ATPase activity plotted according to the Hill equation (Hill equation = 2.0). All experiments were performed in triplicate and S.D. values are indicated.
To determine the concentration-dependency of ATP hydrolysis, the ATP hydrolysis rate was determined as a function of the ATP concentration (Figures 1E and 1F). The fit of the data with the Hill equation resulted in a Vmax of 32 nmol of Pi/mg of protein per min, a K0.5 of 10 mM and a Hill coefficient of 2.0. The Hill coefficient demonstrates that ATP hydrolysis is co-operative, suggesting that FlaI oligomerizes during ATP hydrolysis. FlaI had a very low turnover rate (1 ATP/min), consistent with observations on other secretion ATPases [26]. Most of these ATPases are part of large macromolecular assemblies and other protein components of these complexes, for example, the membrane protein EpsL in T2S system, stimulate the nucleotide-hydrolysing activity of the ATPases [48,49]. Furthermore, lipids can enhance the ATPase activity of these proteins [48].
To assess the influence of archaeal tetraether lipids on the FlaI ATPase activity, the specific activity of the enzyme was measured in the presence of increasing concentrations of S. acidocaldarius tetraether lipids (Figure 2). Remarkably, the tetraether lipids increased the ATPase activity 3–4-fold, whereas the addition of an E. coli total lipid extract only led to a 1.5-fold stimulation of the ATPase activity. This is in contrast with EpsE, whose activity was only activated in the presence of both lipids and the cytoplasmic loops of the corresponding inner membrane protein, and not by lipids alone [48].
Figure 2. Effect of tetraether lipids from S. acidocaldarius and total lipids from E. coli on FlaI ATPase activity.
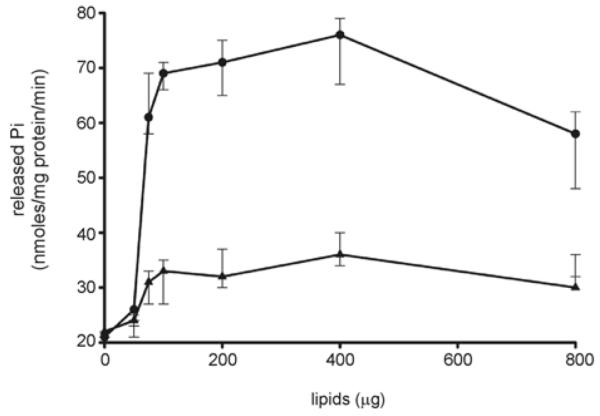
ATP hydrolysis was measured at 70°C with 1 μM purified FlaI in the presence of different concentrations (40–800 μg) of isolated tetraether-linked lipids from S. acidocaldarius, and total lipids from E. coli against respective controls in which an equivalent amount of respective lipids were present in the assay mixture without any protein, and treated in a similar manner as for the enzymatic reaction described in the Materials and methods section. All experiments were performed in triplicate and S.D. values are indicated.
Active-site mutants of FlaI
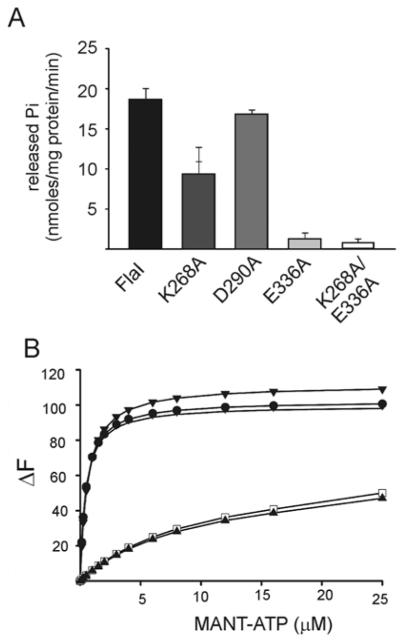
Secretion ATPases contain key conserved motifs for the Walker A and B motifs and the aspartate box. Alignment of S. acidocaldarius FlaI with GspE, i.e. a secretion ATPase of Archaeoglobus fulgidus [27], led to the identification of these motifs in FlaI (Supplementary Figure S1 at http://www.BiochemJ.org/bj/437/bj4370043add.htm). To determine the effects of mutations in these motifs, the lysine residue of the Walker A (K268A), the glutamate residue of the Walker B (E336A), and the aspartate residue of the aspartate box (D290A) were mutated. A double mutant of the Walker A and B mutations, FlaIK268A/E336A, was also constructed. These mutants were expressed and purified, and the expression levels and final purity of these proteins were equal to wt FlaI (results not shown). The different mutants were tested for their ability to hydrolyse ATP (Figure 3A), and for their ability to bind the fluorescent ATP analogue MANT-ATP (Figure 3B). Remarkably, mutation of the lysine residue of the Walker A motif to an alanine residue only resulted in a reduction of approximately 50% of ATP hydrolysis compared with wt FlaI, whereas mutagenesis of the aspartate residue of the aspartate box had no effect on ATP hydrolysis. By substituting Glu336 of the Walker B motif with an alanine residue, ATPase activity was reduced up to 90%. Analysis of the crystal structure of GspE from A. fulgidus (PDB code 2OAP) revealed that the glutamate residue (Glu336 in FlaI) in the Walker B motif is involved in the interaction with Mg-ATP and also is highly conserved among secretion superfamily ATPase members from various archaeal species (Supplementary Figure S1) [27]. Also for the Myxococcus xanthus PilT, which is the disassembly ATPase of the T4P assembly system, mutagenesis of the conserved glutamate residue in the Walker B motif to alanine resulted in loss of ATPase activity [34]. Almost no ATP hydrolysis (2–3%) was detected for the FlaIK268A/E336A double mutant.
Figure 3. Characterization of ATP-binding and hydrolysis motifs in FlaI.
(A) Comparative ATP-hydrolysing activity of Walker A (FlaIK268A), aspartate box (FlaID290A), Walker B (FlaIE336A), and Walker A and B double mutant (FlaIK268A/E336A) motifs against the wt FlaI protein (black). (B) Analysis of the binding of MANT-ATP by wt FlaI and the mutants as monitored by FRET. The change in fluorescence (ΔF) upon MANT-ATP binding is shown as a function of the concentration of MANT-ATP. Fluorescence spectra were corrected for the fluorescence of unbound MANT-ATP. The Kd values were calculated for each of the mutants and wt FlaI. Closed circle, FlaI; closed triangle, FlaIK268A/E336A; open square, FlaIK268A; vertical stripe, FlaID290A; closed diamonds, FlaIE336A.
The MANT-ATP-binding assay demonstrated that wt FlaI bound MANT-ATP with a Kd of 0.46 μM, a fit of the data with the Hill equation resulted in a Hill coefficient of 0.9, demonstrating that ATP binding, contrary to ATP hydrolysis, was not co-operative. Remarkably, MANT-ATP binding to both the Walker B motif (Kd = 0.58 μM) and aspartate box mutant (Kd = 0.43 μM) was comparable with wt FlaI, whereas binding to the Walker A mutant was strongly reduced (Kd = 9.6 μM). Thus our results have shown that mutagenesis of the aspartate box of FlaI does not have any effect on ATP binding and hydrolysis, whereas mutation of the Walker B motif does not affect ATP binding, but heavily abolishes ATP hydrolysis. Notably, mutagenesis of the Walker A motif results in a 20-fold lower binding affinity, but only a 2-fold reduction of ATPase hydrolysis activity. Presumably, the Walker A box substitution, i.e. K268A in FlaI, indirectly reduces ATPase activity by interfering with ATP binding. The Walker B box substitution on the other hand, i.e. E336A in FlaI, abolishes ATPase activity by directly interfering with the ATP hydrolysis, although ATP binding seems normal in this mutant (E336A). Furthermore, the results suggest no role of the conserved Asp290 in the aspartate box motif in ATP binding and hydrolysis.
Oligomerization of FlaI
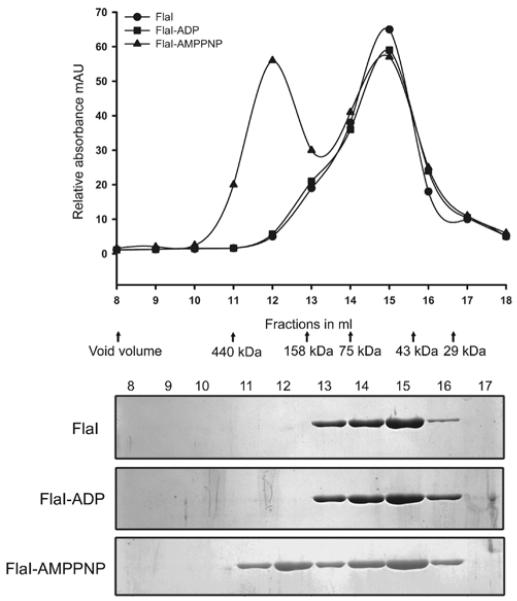
Members of the secretion ATPase superfamily can oligomerize into higher oligomeric structures [27,50,51]. To assess the oligomeric state of purified FlaI (monomer molecular mass of 55 kDa), histidine-tagged FlaI was analysed by size-exclusion chromatography. Owing to the presence of a small shoulder in the chromatogram, small amounts of a dimeric FlaI seem to be present (Figure 4, fraction 13). To determine whether FlaI could form higher oligomeric structures, the effect of nucleotide binding on FlaI oligomerization was studied using the non-hydrolysable ATP analogue p[NH]ppA. After pre-incubation with or without p[NH]ppA and Mg2+ , purified FlaI was analysed by size-exclusion chromatography. Remarkably, after incubation with p[NH]ppA, FlaI was eluted at estimated molecular masses of 330 kDa and 55 kDa respectively. This shows that FlaI assembles in an oligomeric structure in a nucleotide-dependent manner in the presence of non-hydrolysable ATP analogues. To ascertain that the same phenomenon occurs in FlaI mutants, FlaI with a mutation in either the Walker A motif (FlaIK268A) or the Walker B motif (FlaIE336A), which showed a reduced (~50%) and very low (~10%) ATPase activity respectively, were also analysed. After incubation with ATP, a higher oligomeric form was observed only for the FlaI protein containing a mutation in the Walker B (FlaIE336A), but not for the FlaI protein with a mutation in the Walker A motif (FlaIK268A) (Supplementary Figure S2 at http://www.BiochemJ.org/bj/437/bj4370043add.htm). This demonstrated that FlaI protein with a mutation in the Walker B motif (FlaIE336A) forms a stable oligomer after ATP binding. Therefore wt FlaI and FlaI protein with a mutation in the Walker B motif (FlaIE336A) oligomerize upon ATP binding. However, the oligomerization potential of the Walker B motif mutant (FlaIE336A) seems higher compared with wt FlaI, suggesting a more stable complex formation with ATP by the Walker B mutant (FlaIE336A) (Supplementary Figure S2). As expected, incubation of either the wt FlaI, or FlaI with a mutation in either the Walker A motif (FlaIK268A) or the Walker B motif (FlaIE336A) with ADP did not result in oligomerization of FlaI (results not shown).
Figure 4. Size-exclusion chromatography of FlaI, FlaI with ADP, and FlaI with p[NH]ppA.
FlaI, or FlaI in the presence of ADP or p[NH]ppA were analysed on an analytical size-exclusion column (Superdex 200). The elution fractions were analysed on SDS/PAGE and stained with Coomassie Blue. Arrows indicate the position of the marker proteins used to calibrate the column. Prior to analytical gel filtration, the purified FlaI (1 μM) was pre-incubated at 50°C for 10 min followed by an incubation at 4°C for 6 h with either 1 mM p[NH]ppA or ADP, and 10 mM MgCl2. The molecular mass standards used were: ribonuclease A, 13.7 kDa; carboxy anhydrase, 29 kDa; ovalbumin, 43 kDa; conalbumin, 75 kDa; aldolase, 158 kDa; and ferritin, 440 kDa. AMPPNP, p[NH]ppA.
SAXS analysis of oligomerization states of FlaI
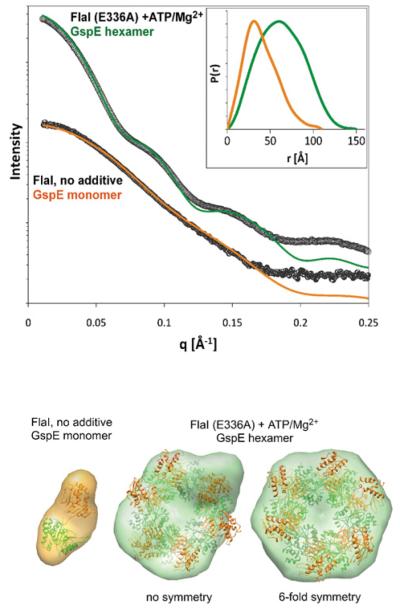
To further analyse the oligomeric structure of FlaI, SAXS was performed. SAXS is a powerful method to structurally analyse proteins in solution under physiological conditions [43]. In Figure 5, the SAXS analysis of wt FlaI is visualized in comparison with the Walker B mutant (FlaIE336A) in the presence of ATP and Mg2+ . The experimental data was compared with theoretical SAXS curves from the crystal structures of monomeric and hexameric GspE. In order to obtain a reasonable fit of the monomeric GspE to the SAXS data from monomeric FlaI, the N-terminal domain (in orange) had to be extended from the ATPase domain (green), which is an indication that there is a flexible conformation of these domains relative to one another. FlaIE336A with ATP clearly forms a hexameric ring similar to the one determined for GspE.
Figure 5. Solution structures of S. acidocaldarius FlaI with and without ATP.
(A) Experimental scattering profiles are shown for wt FlaI (lower curve) and for the Walker B mutant FlaIE336A in the presence of ATP and MgCl2 (upper curve). The theoretical scattering from A. fulgidus GspE is shown in green for a hexamer and in orange for a monomer. In the presence of ATP and MgCl2, FlaI forms a hexameric structure similar to the hexameric ring of GspE, without nucleotide this ring falls apart in what seems to be similar to the GspE monomer. Inset: distance distribution functions P(r) of the FlaI hexamer and monomer computed from experimental SAXS data are shown in green and orange respectively. The P(r) functions are normalized to unity at their maxima. (B) Average of 15 ab initio models for FlaI with and without ATP overlaid with the crystal structures of hexameric and monomeric GspE respectively. The model for hexameric FlaI was calculated without and with applied six-fold symmetry in the modelling.
Ab initio structures for both complexes agree well with the GspE models. For calculating the structure of the hexameric protein, we used both no symmetry constraint and six-fold symmetry. Even without symmetry constraints, we obtained the closed ring structure, but the conformation of the N-terminal domains (orange) seems to be different for the single subunits. The SAXS data from FlaI in monomeric and hexameric form can be analysed using the Kratky plot (Supplementary Figure S3 at http://www.BiochemJ.org/bj/437/bj4370043add.htm) to determine amounts of intrinsic flexibility. In the monomeric form of FlaI that is present in the cytoplasm, the amount of flexibility within the protein is much higher compared with the hexameric membrane-associated form. From these data, we propose that the N-terminal domains are flexible when FlaI has a cytoplasmic localization, but may adopt a more rigid conformation when the proteins attach to the membrane or membrane proteins.
Liposome-interaction studies
The structure of GspE showed an accumulation of hydrophobic patches at the interface which probably interacts with membrane-integral components. The same hydrophobicity was observed in FlaI models based on the GspE structures. Based on the 3–4-fold increase in FlaI ATPase activity upon addition of archaeal tetraether lipids, it was essential to ascertain that a direct interaction between lipids and FlaI occurs.
To this end, a liposome pull-down assay was performed in which FlaI, FlaIK268A and FlaIE336A were incubated either with S. acidocaldarius tetraether lipid liposomes (Figure 6A) or E. coli lipid liposomes (Figure 6B). In the case of the tetraether lipids, the FlaI association with the liposomes was ATP-dependent. When ATP was present, FlaIE336A associated most strongly with the tetraether liposomes, whereas FlaIK268A bound the least. The same results were observed when the same experiment was performed in the presence of p[NH]ppA (Supplementary Figure S4 at http://www.BiochemJ.org/bj/437/bj4370043add.htm). These data imply that ATP binding by FlaI is essential for its interaction with tetraether lipids. In contrast, the interaction of FlaI with the E. coli lipids was independent of the presence or absence of ATP, showing that this is an unspecific interaction that is caused by the hydrophobic nature of the upper surface of the hexameric protein.
Figure 6. Liposome pull-down assay of FlaI in the presence of S. acidocaldarius tetraether lipids and E. coli lipid extract.
The interaction of either FlaI, FlaIK268A or FlaIE336A protein was tested either with S. acidocaldarius tetraether liposomes (A) or lipids from E. coli (B) in the presence or absence of ATP. After centrifugation, proteins remaining in the supernatant (s) or those associated with liposomes in the pellet (p) were separated by SDS/PAGE.
DISCUSSION
Although archaeal flagella systems were identified some time ago, the role that the various flagellar subunits play during assembly and rotation and how they interact with each other during this process has not been elucidated [14,18]. The secretion ATPase FlaI is a central component of the flagellum assembly system. Previously it was shown that FlaI could hydrolyse ATP under in vitro conditions [26]. Genetic analysis revealed its importance in the assembly process. flaI-deletion mutants of M. maripaludis and H. salinarum were impaired in flagella assembly [16,19,37]. Furthermore, in H. salinarum it has been shown that flagellar rotation is dependent on the intercellular ATP concentration and not on the protonmotive force, as is the case in bacteria [52]. These findings implicate FlaI as an essential ATPase for flagellar assembly and/or rotation. However, to provide further insights into its roles, it is necessary to understand the molecular mechanism that FlaI employs. In the present study we have demonstrated that FlaI hydrolyses ATP and undergoes an ATP-dependent oligomerization.
Computational analysis has identified the conserved ATP-binding (Walker A), metal-binding (aspartate box), and ATP hydrolysis (Walker B) motifs in FlaI. We demonstrated the importance of each of these predicted domains in ATP binding and hydrolysis by site-directed mutagenesis. ATP-hydrolysing activity of FlaI is more efficient using Mn2+ as the co-factor rather than Mg2+ or Ca2+ . This preference for Mn2+ as a co-factor is congruent with secretion ATPases and kinases characterized from S. solfataricus [26]. Our results also revealed that the ATPase activity of FlaI displays positive co-operativity, indicating that interactions between subunits occur in an ATP-dependent manner. However, we found that MANT-ATP binding is non-co-operative, probably because ATP analogues bind the monomeric or dimeric state of FlaI under in vitro conditions. Analytical gel filtration demonstrated that, in the presence of a non-hydrolysable ATP analogue, the monomer shifted to a higher oligomer, and SAXS confirmed that the higher molecular species was a hexamer.
Importantly, archaeal tetraether lipids were found to enhance the ATPase activity of FlaI, indicating that the FlaI transition state intermediate is possibly stabilized in an ATP-bound state. The interactions of the ATPase motor with membrane lipids may therefore help regulate its actions.
Our results thus provide the first substantial insights into archaeal FlaI function and probably preface the understanding of a common mechanism of FlaI homologues in the archaea. Generally, these data suggest a hexameric state of FlaI in its ATP-bound state, when it is involved in flagellum assembly. Collectively, these data provide the basis for a model for the mode-of-function of FlaI (Figure 7). Under in vivo conditions, FlaI hydrolyses ATP and provides energy to the AFAS. It is still a matter of contention whether the energy gained in this process is utilized for flagellar assembly by directing the translocation of structural components such as flagellins, or for the rotational movement of flagella, or even for both.
Figure 7. Model describing the sequence of events that FlaI undergoes in AFAS as inferred from the present study.
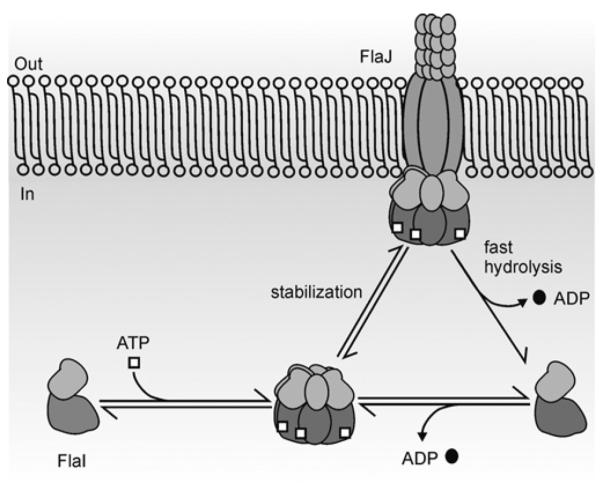
FlaI undergoes ATP-dependent oligomerization and attains a hexameric conformation. This ATP bound hexameric FlaI is possibly stabilized via interactions with other membrane components (such as FlaJ) in AFAS and also with tetraether lipids in the membrane. ATPase activity of FlaI, already associated with the macromolecular archaeal flagella assembly complex, might provide energy to drive the flagellum assembly and/or rotation.
Crystal structures of the archaeal secretion ATPase GspE, the Helicobacter pylori and E. coli hexameric T4S ATPases, as well as the hexameric PilT ATPase in the T4P assembly system of Pseudomonas aeruginosa have been shown to use N-terminal domain movement to facilitate closed ATP-bound and open ADP-bound hexameric conformations [27,28,51,53–55]. Furthermore, it has been postulated that these ATPases act as inner-membrane gated transporters for structural proteins and may thus mediate the whole cycle of assembly by executing a conformational change from closed to open upon ATP hydrolysis. It is possible that ATP hydrolysis is closely coupled to flagellin export into the growing flagellar filament. In contrast with this idea, ATP hydrolysis of FlaI might generate energy for the rotation of the flagellum, as it was shown that in H. salinarum flagellar rotation is ATP-dependent [52].
Understanding how components in AFAS function to effect proper assembly on the archaeal surface and also how they communicate to execute these functions are still at an early stage. The secretion ATPase FlaI and polytopic membrane protein FlaJ are potentially involved in formation of the core of the assembly complex, and it has been postulated that on this FlaIJ core is the main platform for flagellar assembly [12,56]. However, another incomplete ATPase, FlaH has also been postulated to function in association with the FlaIJ core to facilitate the process of assembly, disassembly or rotation. Understanding the biochemical and structural features of the FlaI protein provides a basis to understand the function that this protein fulfils in AFAS which can be transferred to homologous ATPases from other T4P systems of bacteria.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Benjamin Meyer for isolating the archaeal lipids. We thank Michal Hammel for expert SAXS data collection assistance. We thank Andrew Bozarth for critical reading of the manuscript prior to submission.
FUNDING A.G. received a Max Planck Postdoctoral fellowship and S.V.A. was supported by a VIDI grant of the Dutch Science Organization (NWO) and intramural funds of the Max Planck Society. T4P system studies by S.H. and J.A.T. are supported by the NationalInstitutes of Health [grant number AI022160] and the microbial complex SAXS studies are supported by the ENIGMA Program of the Department of Energy, Office of Biological and Environmental Research [contract number DE-AC02-05CH11231], the Lawrence Berkeley National Laboratory and by the National Institutes of Health [grant number AI022160]. The SIBYLS beamline (BL12.3.1) at the Advanced Light Source is supported by the United States Department of Energy program Integrated Diffraction Analysis Technologies (IDAT) [grant number DE-AC02-05CH11231].
Abbreviations used
- AFAS
archaeal flagella assembly system
- FRET
fluorescence resonance energy transfer
- MANT-ATP
2′-/3′-O-(N’-methylanthraniloyl)adenosine-5′-O-triphosphate
- p[NH]ppA
adenosine 5′-[β,γ-imido]triphosphate
- SAXS
small-angle X-ray scattering
- T4P
type IV pili
- T2S
type II secretion
- T4S
type IV secretion
- wt
wild-type
Footnotes
AUTHOR CONTRIBUTION Abhrajyoti Ghosh, Sophia Hartung, Chris van der Does and Sonja-Verena Albers conceived and designed the experiments. Abhrajyoti Ghosh and Sophia Hartung performed the experiments. Abhrajyoti Ghosh, Sophia Hartung, Chris van der Does and Sonja-Verena Albers analysed the data. John Tainer contributed reagents, materials and analysis tools. Abhrajyoti Ghosh, Chris van der Does and Sonja-Verena Albers wrote the paper.
REFERENCES
- 1.Bardy SL, Jarrell KF. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 2003;50:1339–1347. doi: 10.1046/j.1365-2958.2003.03758.x. [DOI] [PubMed] [Google Scholar]
- 2.Macnab RM. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta. 2004;1694:207–217. doi: 10.1016/j.bbamcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Hansen JK, Forest KT. Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J. Mol. Microbiol. Biotechnol. 2006;11:192–207. doi: 10.1159/000094054. [DOI] [PubMed] [Google Scholar]
- 4.Albers SV, Pohlschroder M. Diversity of archaeal type IV pilin-like structures. Extremophiles. 2009;13:403–410. doi: 10.1007/s00792-009-0241-7. [DOI] [PubMed] [Google Scholar]
- 5.Pohlschroder M, Ghosh A, Tripepi M, Albers SV. Archaeal type IV pilus-like structures: evolutionarily conserved prokaryotic surface organelles. Curr. Opin. Microbiol. 2011 doi: 10.1016/j.mib.2011.03.002. doi:10.1016/j.mib.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Marwan W, Alam M, Oesterhelt D. Rotation and switching of the flagellar motor assembly in Halobacterium halobium. J. Bacteriol. 1991;173:1971–1977. doi: 10.1128/jb.173.6.1971-1977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szabo Z, Sani M, Groeneveld M, Zolghadr B, Schelert J, Albers SV, Blum P, Boekema EJ, Driessen AJ. Flagellar motility and structure in the hyperthermoacidophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 2007;189:4305–4309. doi: 10.1128/JB.00042-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewus P, Ford RM. Temperature-sensitive motility of Sulfolobus acidocaldarius influences population distribution in extreme environments. J. Bacteriol. 1999;181:4020–4025. doi: 10.1128/jb.181.13.4020-4025.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zolghadr B, Klingl A, Koerdt A, Driessen AJ, Rachel R, Albers SV. Appendage-mediated surface adherence of Sulfolobus solfataricus. J. Bacteriol. 2010;192:104–110. doi: 10.1128/JB.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koerdt A, Godeke J, Berger J, Thormann KM, Albers SV. Crenarchaeal biofilm formation under extreme conditions. PLoS One. 2010;5:e14104. doi: 10.1371/journal.pone.0014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellack A, Huber H, Rachel R, Wanner G, Wirth R. Methanocaldococcus villosus sp. nov., a heavily flagellated archaeon adhering to surfaces and forming cell-cell contacts. Int. J. Syst. Evol. Microbiol. 2010 doi: 10.1099/ijs.0.023663-0. doi 10.1099/ijs.0.023663-0. [DOI] [PubMed] [Google Scholar]
- 12.Ng SY, Chaban B, Jarrell KF. Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications. J. Mol. Microbiol. Biotechnol. 2006;11:167–191. doi: 10.1159/000094053. [DOI] [PubMed] [Google Scholar]
- 13.Trachtenberg S, Cohen-Krausz S. The archaeabacterial flagellar filament: a bacterial propeller with a pilus-like structure. J. Mol. Microbiol. Biotechnol. 2006;11:208–220. doi: 10.1159/000094055. [DOI] [PubMed] [Google Scholar]
- 14.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 15.Ng SY, Zolghadr B, Driessen AJ, Albers SV, Jarrell KF. Cell surface structures of archaea. J. Bacteriol. 2008;190:6039–6047. doi: 10.1128/JB.00546-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaban B, Ng SY, Kanbe M, Saltzman I, Nimmo G, Aizawa S, Jarrell KF. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol. Microbiol. 2007;66:596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- 17.Albers SV, Szabo Z, Driessen AJ. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 2003;185:3918–3925. doi: 10.1128/JB.185.13.3918-3925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- 19.Patenge N, Berendes A, Engelhardt H, Schuster SC, Oesterhelt D. The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum. Mol. Microbiol. 2001;41:653–663. doi: 10.1046/j.1365-2958.2001.02542.x. [DOI] [PubMed] [Google Scholar]
- 20.Cohen-Krausz S, Trachtenberg S. The structure of the archeabacterial flagellar filament of the extreme halophile Halobacterium salinarum R1M1 and its relation to eubacterial flagellar filaments and type IV pili. J. Mol. Biol. 2002;321:383–395. doi: 10.1016/s0022-2836(02)00616-2. [DOI] [PubMed] [Google Scholar]
- 21.Cohen-Krausz S, Trachtenberg S. The flagellar filament structure of the extreme acidothermophile Sulfolobus shibatae B12 suggests that archaeabacterial flagella have a unique and common symmetry and design. J. Mol. Biol. 2008;375:1113–1124. doi: 10.1016/j.jmb.2007.10.048. [DOI] [PubMed] [Google Scholar]
- 22.Mudrak B, Kuehn MJ. Specificity of the type II secretion systems of enterotoxigenic Escherichia coli and Vibrio cholerae for heat-labile enterotoxin and cholera toxin. J. Bacteriol. 2010;192:1902–1911. doi: 10.1128/JB.01542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo M, Jin S, Sun D, Hew CL, Pan SQ. Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20019–20024. doi: 10.1073/pnas.0701738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh A, Albers SV. Assembly and function of the archaeal flagellum. Biochem. Soc. Trans. 2011;39:64–69. doi: 10.1042/BST0390064. [DOI] [PubMed] [Google Scholar]
- 25.Bardy SL, Eichler J, Jarrell KF. Archaeal signal peptides: a comparative survey at the genome level. Protein Sci. 2003;12:1833–1843. doi: 10.1110/ps.03148703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albers SV, Driessen AJ. Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology. 2005;151:763–773. doi: 10.1099/mic.0.27699-0. [DOI] [PubMed] [Google Scholar]
- 27.Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 29.Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, Lloyd SJ, Shin DS, Getzoff ED, Yeager M, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 30.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Possot O, Pugsley AP. Molecular characterization of PulE, a protein required for pullulanase secretion. Mol. Microbiol. 1994;12:287–299. doi: 10.1111/j.1365-2958.1994.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 33.Chiang P, Sampaleanu LM, Ayers M, Pahuta M, Howell PL, Burrows LL. Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology. 2008;154:114–126. doi: 10.1099/mic.0.2007/011320-0. [DOI] [PubMed] [Google Scholar]
- 34.Jakovljevic V, Leonardy S, Hoppert M, Sogaard-Andersen L. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 2008;190:2411–2421. doi: 10.1128/JB.01793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner LR, Lara JC, Nunn DN, Lory S. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 1993;175:4962–4969. doi: 10.1128/jb.175.16.4962-4969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlesner M, Miller A, Streif S, Staudinger WF, Muller J, Scheffer B, Siedler F, Oesterhelt D. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol. 2009;9:56. doi: 10.1186/1471-2180-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas NA, Mueller S, Klein A, Jarrell KF. Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. Mol. Microbiol. 2002;46:879–887. doi: 10.1046/j.1365-2958.2002.03220.x. [DOI] [PubMed] [Google Scholar]
- 38.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 39.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 40.Albers SV, Konings WN, Driessen AJ. Membranes of thermophiles and other extremophiles. In: Rainey F, Oren A, editors. Methods in Microbiology. Elsevier; Amsterdam: 2006. pp. 161–171. [Google Scholar]
- 41.Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, Bresson SM, Tomchick DR, Alto NM. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469:107–111. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, II, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat. Methods. 2009;6:606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 44.Guinier A, Fournet G. Small Angle Scattering of X-rays. Wiley Interscience; New York: 1995. [Google Scholar]
- 45.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 46.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38(Suppl.):W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camberg JL, Johnson TL, Patrick M, Abendroth J, Hol WG, Sandkvist M. Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. EMBO J. 2007;26:19–27. doi: 10.1038/sj.emboj.7601481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiue SJ, Kao KM, Leu WM, Chen LY, Chan NL, Hu NT. XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. EMBO J. 2006;25:1426–1435. doi: 10.1038/sj.emboj.7601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patrick M, Korotkov KV, Hol WG, Sandkvist M. Oligomerization of EpsE coordinates residues from multiple subunits to facilitate ATPase activity. J. Biol. Chem. 2011;286:10378–10386. doi: 10.1074/jbc.M110.167031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Misic AM, Satyshur KA, Forest KT. P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor. J. Mol. Biol. 2010;400:1011–1021. doi: 10.1016/j.jmb.2010.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Streif S, Staudinger WF, Marwan W, Oesterhelt D. Flagellar rotation in the archaeon Halobacterium salinarum depends on ATP. J. Mol. Biol. 2008;384:1–8. doi: 10.1016/j.jmb.2008.08.057. [DOI] [PubMed] [Google Scholar]
- 53.Krause S, Pansegrau W, Lurz R, de la Cruz F, Lanka E. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krause S, Barcena M, Pansegrau W, Lurz R, Carazo JM, Lanka E. Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3067–3072. doi: 10.1073/pnas.050578697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 56.Albers SV, Szabo Z, Driessen AJ. Protein secretion in the Archaea: multiple paths towards a unique cell surface. Nat. Rev. Microbiol. 2006;4:537–547. doi: 10.1038/nrmicro1440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.