Abstract
Investigating escape mechanisms of human immunodeficiency virus type 1 (HIV-1) from cytotoxic T lymphocytes (CTLs) is essential for understanding the pathogenesis of HIV-1 infection and developing effective vaccines. To study the processing and presentation of known CTL epitopes, we prepared Epstein-Barr virus-transformed B cells that endogenously express the gag gene of six field isolates by adopting an env/nef-deletion HIV-1 vector pseudotyped with vesicular stomatitis virus G protein and then tested them for the recognition by Gag epitope-specific CTL lines or clones. We observed that two field variants, SLFNTVAVL and SVYNTVATL, of an A*0201-restricted Gag CTL epitope SLYNTVATL, and three field variants, KYRLKHLVW, QYRLKHIVW, and RYRLKHLVW, of an A24-restricted Gag CTL epitope KYKLKHIVW escaped from being killed by the CTL lines, despite the fact that they were recognized when the synthetic peptides corresponding to these variant sequences were exogenously loaded onto the target cells. Thus, their escape is likely due to the changes that occur during the processing and presentation of epitopes in the infected cells. Mutations responsible for this mode of escape were located within the epitope regions rather than the flanking regions, and such mutations did not influence the virus replication. The results suggest that the impaired antigen processing and presentation often occur in HIV-1 field isolates and thus are one of the major mechanisms that enable HIV-1 to escape from CTL recognition. We emphasize the importance of testing HIV-1 variants in an endogenous expression system.
Accumulated evidence has indicated a critical role of cytotoxic T lymphocytes (CTLs) in controlling human immunodeficiency virus (HIV) replication during acute and chronic infection (16). Eliciting HIV type 1 (HIV-1)-specific CTLs has been thought to be crucial for effective HIV/AIDS vaccines (15). However, despite the presence of CTLs, the majority of HIV-1-infected cases eventually progress to AIDS, probably as a consequence of the emergence of escape mutants from CTLs (8, 20). Among immunized monkeys, which developed strong cellular immune responses against HIV-1, eventual vaccine failure occurs by viral escape from CTLs (2). Thus, investigating the mechanisms of CTL failure to control the virus is essential to understanding the pathogenesis of HIV-1 infection and to develop HIV/AIDS vaccines.
The high rate of HIV-1 replication in vivo indicates that HIV-1 has tremendous ability to mutate swiftly (9, 30) and to make a dynamic adaptation to host-immune environments (3, 14, 18, 21, 31). Several mutations have been described in CTL epitopes in HIV-1-infected individuals, which result in either a lack of binding to the MHC class I molecule or nonrecognition by T-cell receptor (TCR) (3, 8, 12, 20, 21). Consequently, the virus escapes from CTL recognition. There are other mutations that do not lead to either escape effects (12); very little is known about the influence of these mutations on CTL recognition. CTL antigens are processed and presented on the cell surface in a very complex manner. Peptides are cleaved from endogenously synthesized proteins by proteasome in the cytoplasm and transported into the endoplasmic reticulum by the transporter of antigen presentation. Amino-terminal extended peptides are trimmed to the right size of peptides by aminopeptidases, which exist in both the cytoplasm and the endoplasmic reticulum (23). These steps have various degrees of substrate sequence specificity (17). The generated peptides should have sufficient affinity to bind to a major histocompatibility complex (MHC) class I molecule in the presence of various other peptides derived from host proteins and to maintain the stability of peptide-MHC complexes until they are presented on the cell surface (28). Thus, it is plausible that some amino acid substitutions in the epitope and its flanking regions have a significant influence on antigen processing and presentation. In the present study, we hypothesized that such mutations often enable HIV-1 to escape from CTL recognition.
Conventionally, the intracellular HIV-1 antigen processing and presentation has been studied with recombinant vaccinia viruses expressing an HIV-1 gene (3, 4, 11, 20, 26). Several studies have addressed this issue in the context of HIV-infected T cells (4, 29, 32, 33). Most studies, however, have only evaluated a single or a few laboratory-established strains. The CTL recognition of HIV-1 clinical isolates has been evaluated, in most cases, by exogenously applying synthetic variant peptides to the cell surface to replace MHC-bound peptides (8, 12, 20, 21). Very little is known about how the antigenic products of HIV-1 clinical isolates are processed and presented in the infected cells. To address this issue, we prepared CTL target cells that endogenously express the gag gene derived from HIV-1 clinical isolates by adopting an env/nef-deletion HIV-1-based vector pseudotyped with vesicular stomatitis virus protein G (VSV-G) proteins. Here, we show evidence that HIV-1 escapes from CTL recognition often via the impairment of antigen processing and presentation.
MATERIALS AND METHODS
Subjects.
Peripheral blood mononuclear cells (PBMC) were collected from five HIV-1-infected individuals from the HIV clinic affiliated with the Institute of Medical Science, University of Tokyo. Two individuals (IMS1 and IMS2) had no therapy; one individual (IMS6) was off drugs but had received treatment (zidovudine alone) 2 years prior to blood sampling; two individuals (IMS4 and IMS7) had received therapy (zidovudine-lamivudine-indinavir and stavudine-lamivudine-nelfinavir, respectively) but for less than 3 months. CD4 count, viral load, and HLA type of the recruited individuals are shown in Table 1. HLA class I typing was initially performed by serology. Subtyping of HLA-A2 was done by a PCR-sequence-specific primer method (Dynal Classic SSP HLA-A2; Dynal A.S., Oslo, Norway).
TABLE 1.
Characteristics of five HIV-1-infected donorsa
| Donor | HLA type
|
CD4 count/μl | Virus load (copies/ ml) | No. of isolated clones | ||
|---|---|---|---|---|---|---|
| A | B | Cw | ||||
| IMS1 | *0201/2402 | 52/75 | 3 | 286 | <400 | 3 |
| IMS2 | *0201/31 | 27/5101 | 2 | 797 | <400 | 2 |
| IMS4 | *0207/2402 | 46/52 | 1 | 448 | <400 | 2 |
| IMS6 | 2402/26 | 7/5101 | 7 | 368 | 3.6 × 105 | 3 |
| IMS7 | 1/− | 37/− | 6 | 544 | 1.3 × 103 | 3 |
HLA alleles, CD4 count, viral load, and the number of isolated clones from each donor's sample are shown.
Isolation and cloning of full-length gag.
Full-length gag was amplified from proviral DNA extracted from the PBMC by nested PCR with Pfu DNA polymerase (Strategene, La Jolla, Calif.) and oligonucleotides specific for HIV-1 long terminal repeat (LTR) and reverse transcription (RT) regions. Four oligonucleotides were mixed as outer primers: the sense primers 1U5AS-S (5′-ACTCTGGTADCTAGAGATCCCTCA-3′; the position in HXB2 being 578 to 601) and TAR-2 (5′-TGAGCCTGGGAGCTCTCTGGCT-3′; 478-499) and the antisense primers RT7A-A (5′-TATGTTGAYAGGTGTAGGTC-3′; 2485 to 2504) and RT18A-A (5′-CTACYARTACTGTACCTATAG-3′; 2464 to 2484). Two oligonucleotides were used as inner primers: the sense primer TPBS1-S (5′-AAAATCTCTAGCAGTGGCGCCCGAACAGG-3′; the position in HXB2 being 622 to 650) and the antisense inner primer PRO6A (5′-ACTGTATCATCTGCTCCTGTRTCTAA-3′; 2322 to 2347). The thermocycling conditions were 95°C for 45 s, 50°C for 45 s, and 72°C for 210 s (30 cycles) and 72°C for 7 min for both primary and secondary PCR. The PCR products were purified by using spin columns (QIAquick PCR purification kit; Qiagen, Santa Clarita, Calif.) and cloned into PT7Blue3 vector by using a commercial cloning kit (Perfectly Blunt cloning kit; Novagen, Dedham, Mass.). Two to three clones for each individual were sequenced by an automated sequencer (ABI Prism 377 automated DNA sequencer; Perkin-Elmer, Norwalk, Conn.) with BigDye terminators (PE Applied Biosystems, Foster City, Calif.). The sequences of gag clones that were used in the present study are available under GenBank Accession numbers as follows: AB074049 (IMS1-28), AB074050 (IMS1-29), AB074052 (IMS2-5), AB074058 (IMS4-24), AB074061 (IMS6-34), and AB074064 (IMS7-11).
Construction of HIV-1 vector.
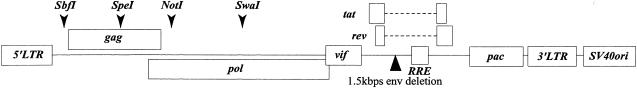
The design of HIV-1 vector, pCTLpac, is shown in Fig. 1. The backbone of the vector is derived from an infectious molecular clone, HXB2Ecogpt (22), which lacks the function of vpr, vpu, and nef genes. We deleted a 1.5-kb portion from the env-coding region but kept the function of Rev responsive element, Tat, and Rev. The nef gene was replaced with the puromycin N-acetyltransferase (pac) gene (pPUR; BD Biosciences Clontech, Palo Alto, Calif.) by using XhoI and ClaI sites where the ClaI site was introduced by site-directed mutagenesis. SbfI and SwaI sites were introduced by site-directed mutagenesis in the upstream of the gag (nucleotide 788) and in the pol (nucleotide 3717), respectively. The fragment from SpeI in the gag (nucleotide 1507) to the SwaI was then replaced with that of a previously published vector, pHXB2cv (25), which has a NotI site but lacks an SbfI site in the pol gene. Consequently, the final construct carries the single SbfI site (nucleotide 788) and the NotI site (nucleotide 2275) that corresponds to the 10th codon of protease. These sites were used for incorporating the gag clones derived from clinical isolates into the pCTLpac vector. We confirmed that the expected variant sequences were inserted in the vector by sequencing.
FIG. 1.
Structure of pCTLpac. A 1.5-kbp portion of env was deleted (▴). Puromycin N-acetyltransferase gene (pac) was inserted in the nef region. The locations of restriction enzyme sites are indicated (▾). RRE, Rev responsive element.
Generation of VSV-G pseudotype virus.
Subconfluent COS7 cells in 25-cm2 T flasks (Becton Dickinson, Lincoln Park, N.J.) were cotransfected with 4 μg of pCTLpac and 2 μg of pVSVG (BD Biosciences Clontech), which expresses VSV-G protein, by lipofection (FuGENE6; Roche Molecular Biochemicals, Mannheim, Germany) and then incubated for 48 to 60 h. The supernatant, which contains pseudotype viruses carrying the HIV-1 vector with VSV-G envelope proteins, was harvested, filtered through a 0.45-μm (pore-size) Millex filter (Millipore, Bedford, Mass.), and used as pseudotype virus stocks, some of which were stored at −80°C before use. The amount of p24 antigen in the stocks was measured by p24 antigen capture enzyme-linked immunosorbent assay (ELISA; RETRO-TEK; Zeptometrix Corp., Buffalo, N.Y.). The range of the p24 antigen yield was 40 to 100 ng/ml.
Preparation of target cells by using VSV-G pseudotyped HIV-1 vector.
Epstein-Barr virus-transformed B-lymphocyte lines (B-LCLs) were infected with pseudotype virus stocks for 6 h at 37°C. The medium was then replaced with fresh RPMI 1640 (Sigma-Aldrich, St. Louis, Mo.) supplemented with 10% fetal bovine serum (R10; HyClone, Logan, Utah), and the cells were incubated for an additional 36 h. Subsequently, 0.5 μg of puromycin (BD Biosciences Clontech)/ml was added to the R10 medium to select transduced cells. The culture was maintained until the number of transduced cells became sufficient for CTL experiments. When 106 B-LCLs were infected with 1 ml of pseudotype virus stocks, the transduction efficiency was 20 to 30%. Usually, more than 107 transduced cells were generated within 2 weeks and used as CTL target cells.
To standardize the expression level of Gag protein in target cells, we quantified the amount of extracellular p24 antigen that 106 cells per ml of target cells had produced in 24 h. The supernatant was harvested before (supernatant A) and after (supernatant B) the 24 h of culture for the measurement of p24 antigen by p24 antigen capture ELISA (Zeptometrix Corp.). The level of p24 antigen production was defined by the difference in the concentration of p24 antigen between supernatants A and B. If the target cells produced p24 antigen that was >1 ng/ml in 24 h, they were used for CTL experiments, since the specific percent lysis did not significantly differ among target cells producing Gag protein above this level (data not shown). We also investigated the level and pattern of protein expression of gag variants by Western blot analysis, as previously described (25).
51Cr release experiments with HLA-class I-mismatched target cells in parallel with HLA-class I matched target cells of different donors confirmed that these target cells were recognized by CTLs in an HLA-restricted manner (data not shown). Repeated experiments showed that specific lysis of blank controls was equivalent to that of cells expressing gag variants that are known to escape either from TCR recognition or MHC binding. Some examples appear in the Results section below: specific lysis against IMS2-5 (Fig. 3a), IMS4-24 (Fig. 3c), IMS6-34 (Fig. 3e), and HXB2-wild (Fig. 5b). Thus, we regarded the blank control as a negative control.
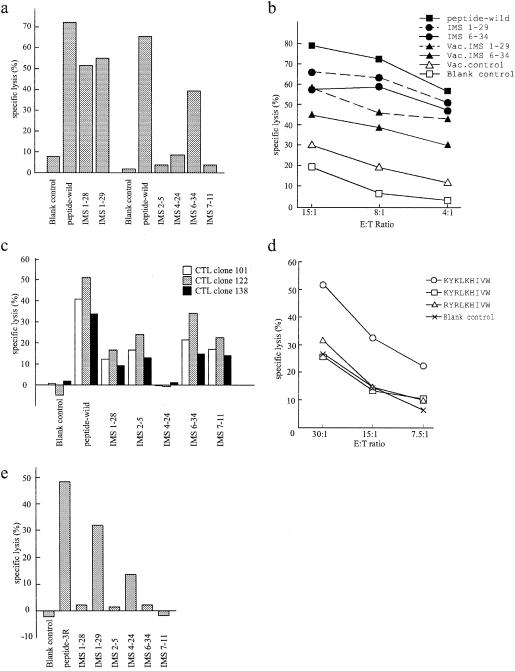
FIG. 3.
(a) Specific lysis of A*0201-matched B-LCLs (HLA-A*0201/− and HLA-B*5101/−) producing Gag proteins of clinical isolates. Peptide target cells were pulsed with the A*0201 wild-type peptide, SLYNTVATL (10 μM). A*0201-restricted SLYNTVATL -specific CTL lines were induced from a single donor (IMS1). The E:T ratio was 10:1. This experiment was repeated, with a different B-LCLs (HLA-A*0201/31 and HLA-B27/*5101), giving the same pattern of recognition (data not shown). (b) Specific lysis of A*0201-matched B-LCLs (HLA-A*0201/− and HLA-B*5101/−) expressing gag clones of two clinical isolates with the VSV-G-pseudotyped HIV-1 vector versus recombinant vaccinia viruses. Recombinant vaccinia virus expressing the human CD4 gene was used as a vaccinia virus control (1). The effector and peptide target cells were prepared as described for panel a. (c) Specific lysis of B*5101-matched B-LCLs (HLA-A*0201/− and HLA-B*5101/−) producing the Gag proteins of five clones. Three B*5101-restricted NANPDCKTI -specific CTL clones were used as effector cells at an E:T ratio of 2:1 (23). The peptide target was pulsed with the B51 wild-type peptide NANPDCKI (1μM). (d) Specific lysis of A24-matched B-LCLs (HLA-A24/− and HLA-B46/52) pulsed with the peptides KYKLKHIVW, KYRLKHIVW, and RYRLKHIVW at 10 μM. A24-restricted, KYKLKHIVW -specific CTL lines were induced from one A24-positive donor. (e) Specific lysis of A24-matched B-LCLs (HLA-A24/− and HLA-B46/52) producing variant Gag proteins. A24-restricted KYRLKHIVW (3R)-specific CTL lines were induced from another A24-positive donor. The peptide target was pulsed with 3R mutant type peptide (10 μM). The E:T ratio was 20:1. The lysis of target cells without any peptide pulsing is shown as a blank control.
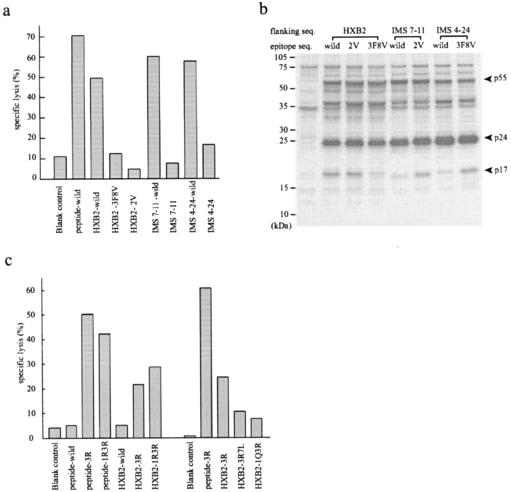
FIG. 5.
(a) Specific lysis of A*0201-matched B-LCLs (HLA-A*0201/− and HLA-B*5101/−) that endogenously express chimeric gag clones bearing the variant CTL epitopes SLFNTVAVL and SVYNTVATL in the frame of HXB2 gag (HXB2-3F8V and HXB2-2V, respectively) or bearing the wild-type epitope in the frame of IMS7-11 and IMS4-24 Gag (IMS7-11-wild and IMS4-24-wild, respectively). A*0201-restricted SLYNTVATL CTL lines were induced from the same donor as for Fig. 3. Specific lysis of target cells expressing HXB2, IMS7-11, or IMS4-24 gag clones and being pulsed with the A*0201 wild-type peptide (10 μM) is shown in parallel. The E:T ratio was 20:1. (b) Levels and patterns of HIV-1 protein expression in target cells used in the experiments described for panel a. The Western blot was reacted with the serum from an HIV-1-infected individual. (c) Specific lysis of A24-matched B-LCLs (HLA A24/− and HLA-B46/52 or HLA-A24/26 and HLA-B51/52) that express gag clones with various point mutations. Point mutations were inserted into the A24-restricted CTL epitope region in the frame of wild-type HXB2 Gag (HXB2-wild): amino acid substitutions of Lys to Arg at position 30 (HXB2-3R) with Lys to Arg at position 28 (HXB2-1R3R), Ile to Lue at position 34 (HXB2-3R7L), or Lys to Gln at position 28 (HXB2-1Q3R). Peptide target cells were pulsed with either the KYRLKHIVW (3R) or the RYRLKHIVW (1R3R) mutant peptide at 10 μM. The effector cells were A24-restricted 3R mutant-specific CTL lines from the same donor as in the Fig. 3e experiment. The E:T ratio was 20:1.
Preparation of target cells by using recombinant vaccinia viruses.
Recombinant vaccinia viruses used in the experiment shown in Fig. 3b were made as previously described (10). HLA-matched B-LCLs were infected with recombinant vaccinia viruses at a multiplicity of infection of 3:1 overnight before being tested in a 51Cr release assay.
Effector cells.
Peptide-specific CTL lines were induced from PBMC of HIV-1-infected donors. Half of the PBMC were stimulated with phytohemagglutinin (2 μg/ml) for 24 h and then pulsed with corresponding peptides at 100 μM for 1 h and irradiated before being added to the other half of the PBMC. A total of 3 × 105 cells in each well of a 96-well U-bottom plate, with at least 10 wells for each sample, were cultured in R10; 10% Lymphocult T (Biotest, Dreieich, Germany) was added to the medium on day 3 of culture. The CTL lines were maintained by adding fresh R10 medium containing 10% Lymphocult T every 3 to 4 days and splitting the well accordingly. Assays were performed on day 14 to 28 of culture.
Synthetic peptides.
Peptides were manufactured at the Takara Shuzo Co., Ltd. (Shiga, Japan). The purity of peptides was >99% as determined by high-pressure liquid chromatography, and the identity of peptides was confirmed by matrix-assisted laser desorption ionization-mass spectrometry. Lyophilized peptides were dissolved in dimethyl sulfoxide and diluted in phosphate-buffered saline to make a stock concentration (2 mM). Further dilution was made in RPMI 1640 to make working concentrations of 200 μM for the induction of CTLs and of 20 μM for the preparation of target cells.
51Cr release assay.
In 96-well U-bottom plates, target cells were divided into aliquots at 5,000 per well. Effector cells were added to target cells at different effector/target (E:T) ratios. The amount of 51Cr release in the culture supernatants was quantified after 6 h of incubation, and the percent specific lysis was determined by using the following formula: [(E − M)/(D − M)] ×100, where E is the experimental 51Cr release, M is the 51Cr released in the presence of culture medium (which ranged between 15 and 25% of total release), and D is the total 51Cr released in the presence of 5% Triton X-100 detergent. The results were regarded as positive when recognition of the HIV target was >10% above the control. The SD50 is the peptide concentration giving 50% of maximal specific lysis of target cells pulsed with 10 μM synthetic peptide (28).
Replication kinetics assay.
Subconfluent 293T cells in Falcon 25-cm2 T flasks (Becton Dickinson) were transfected by lipofection (Roche Molecular Biochemicals), with 2 μg of HXB2cv replication-competent HIV-1 plasmids, in which various mutations were introduced. After 60 h of culture, the supernatant was harvested, filtered through a 0.45-μm-pore-size filter, and used as mutant virus stocks. Two million Jurkat cells or eight million H9 cells were infected with an equivalent of 40 ng of p24 antigen of mutant viruses in 2 ml of R10 for 1 h. Cells were washed three times with 10 ml of R10, resuspended with 5 ml of R10, and cultured in a 12.5-cm2 T flask at 37°C in 5% CO2. Every 2 or 3 days, 1.5 ml of supernatant was harvested and replaced with fresh R10. The concentration of p24 was measured by using a p24 ELISA kit (Zeptometrix Corp.).
This study was approved by the Ethics Committee of the University of Tokyo.
RESULTS
Full-length gag clones of field isolates.
We used 6 of 13 full-length gag clones that were isolated from the five infected individuals (Fig. 2). All of the clones did not have any stop codons. In the present study, we focused on the processing and presentation of three CTL epitopes: the HLA-A*0201-restricted epitope SLYNTVATL, the A24-restricted epitope KYKLKHIVW in p17 matrix protein (MA), and the HLA-B*5101-restricted epitope NANPDCKTI in p24 capsid protein (CA) (11, 26, 27). Amino acid sequences within the three epitope regions and the N- and C-terminal 15-amino-acid residues flanking each epitope were analyzed; the six clones were selected to maximize the diversity of amino acid sequences in the epitopes and its flanking regions.
FIG. 2.
Sequence variation in three CTL epitopes and their flanking regions. The amino acid sequences of six gag clones are shown. The reference sequence is derived from HXB2, and the differences are indicated. The numbering is done according to the HIV sequence database, Los Alamos National Laboratory, Los Alamos, N.Mex. The CTL epitope regions are boxed.
The A*0201-restricted epitope and its flanking regions were highly variable. However, we did not observe a previously recognized variation in the flanking region, Arg (R) to Lys (K) at position 76 in our clones (4). In contrast, the B*5101-restricted epitope and its flanking regions were conserved except for clones IMS2-5 and IMS4-24. In the A24-restricted epitope and its flanking regions, variations were seen almost exclusively within the epitope region with two exceptions, a Lys (K)-to-Ser (S) mutation at position 26 (K26S) in clone IMS4-24 and an Arg (R)-to-Lys (K) at position 15 in clone IMS2-5. The Lys (K)-to-Arg (R) mutation at position 30 within the A24-restricted epitope was seen more frequently than any other sequences; none of the 13 clones had the wild-type sequence of KYKLKHIVW. We incorporated the six gag clones into the HIV-1 vector with env and nef deleted, pCTLpac (Fig. 1), to make target cells expressing gag genes of these filed isolates.
CTL recognition of target cells endogenously expressing gag genes of clinical isolates.
We generated A*0201-restricted SLYNTVATL (wild type) epitope-specific oligoclonal CTL lines from one HIV-1-infected individual (IMS1) with A*0201 and used the lines to test the killing of the six different gag clones expressed on A*0201-matched B-LCLs by a conventional 51Cr release assay. The A*0201-restricted CTLs efficiently recognized target cells expressing gag clones IMS1-29, IMS1-28, and IMS6-34, which encode either wild type or the SLYNTIATL sequence in the CTL epitope region. In contrast, the same CTLs did not recognize cells expressing gag clones IMS2-5, IMS4-24, and IMS7-11, which encode SLYNLVATL, SLFNTVAVL, and SVYNTVATL, respectively, indicating that these clones escaped from A*0201-restricted CTL recognition (Fig. 3a).
CTL recognition of IMS1-29 and IMS6-34 was also tested with recombinant vaccinia viruses expressing the gag gene of these variants in parallel with the VSV-G-pseudotyped HIV-1 vectors. The HIV-1 vector method demonstrated the CTL killing as well or slightly better than the vaccinia method did (Fig. 3b).
We used three B*5101-restricted NANPDCKTI -specific CTL clones to test the CTL recognition of five representative gag clones. The CTL clones recognized four gag clones, which convey the wild-type B*5101-restricted epitope sequence; they also recognized IMS2-5 that had a substitution in the flanking region. None of the clones recognized the IMS4-24 clone, which had the variant sequence NSNPDCKNI in the epitope region (Fig. 3c).
A24-restricted KYKLKHIVW (wild type) specific-CTL lines did not recognize synthetic peptides of the most common sequence, KYRLKHIVW (3R mutant type), and the other variant, RYRLKHIVW (Fig. 3d). These two variants were shown to bind to the A*2402 MHC class I molecule in a binding assay (data not shown). We screened eight A24-positive individuals for the presence of CTL activities against the 3R mutant epitopes and found one individual who carried CTLs recognizing the 3R mutant peptide. A24-restricted 3R mutant-reactive CTL lines were induced from this A24-positive individual and used for the remaining experiments. The 3R mutant-reactive CTL lines recognized target cells expressing IMS1-29 and IMS4-24 gag clones, both of which carry the 3R mutant sequence, but did not recognize any other target cells expressing different variants (Fig. 3e). Interestingly, IMS4-24 with Lys (K)-to-Ser (S) mutation at position 26 outside the epitope region was less well recognized than IMS1-29. We consistently observed this phenomenon in repeated experiments (data not shown).
CTL recognition of exogenously loaded variant peptides.
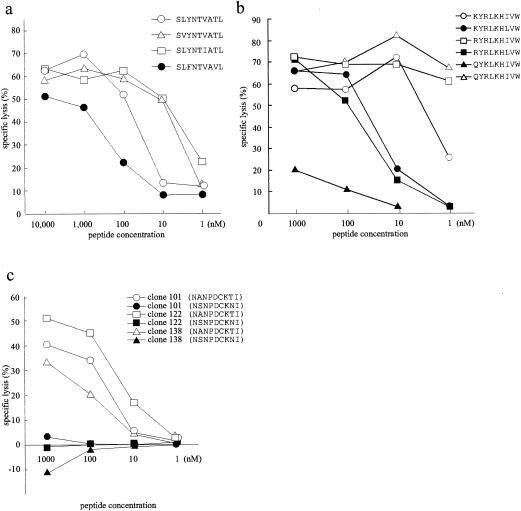
To investigate whether the above findings of escape phenomenon from CTL killing were due to either loss of peptide binding to the MHC class I molecule or to the lack of TCR recognition, we prepared synthetic peptides that represented the variant epitopes and tested them for cross-recognition of the peptides in peptide titration assays by using the same CTL lines or clones that were used in experiments described for Fig. 3. To our surprise, A*0201-restricted CTL lines recognized the peptides of two A*0201-restricted CTL epitope variants, SVYNTVATL and SLFNTVAVL, which were not recognized by the CTLs when expressed endogenously. They recognized the SLFNTVAVL peptide less efficiently, with an SD50 of >100 nM (Fig. 4a). Target cells pulsed with SLYNLVATL peptide representing clone IMS 2-5 were not cross-recognized even at a saturated concentration (10 μM) (data not shown).
FIG. 4.
Peptide titration assays. (a) Specific lysis of A*0201-matched B-LCLs pulsed with A*0201 variant peptides by A*0201-restricted CTLs at an E:T ratio of 20:1. (b) Specific lysis of A24-matched B-LCLs pulsed with 3R and its variant peptides by A24-restricted 3R mutant reactive CTLs at an E:T ratio of 20:1. (c) Specific lysis of B51-matched B-LCLs pulsed with B51 variant peptides by B51-restricted CTL clones at an E:T ratio of 2:1. The same effector and target cells were used as for Fig. 3. The percent lysis of the blank control has been subtracted.
We also obtained similar discordant results in experiments of A24-restricted CTL epitope variants. A24-restricted 3R mutant-specific CTL lines recognized peptides of three variants—KYRLKHLVW, RYRLKHLVW, and QYRLKHIVW— that were not recognized by the CTLs when they were expressed endogenously. In fact, the CTLs recognized QYRLKHIVW peptide even better than the 3R mutant peptide but did not cross-recognize the QYKLKHIVW peptide (Fig. 4b).
We tested one B*5101 variant peptide, NSNPDCKNI, in a peptide titration assay. This variant was not cross-recognized by any of the CTL clones even at a high concentration (1 μM) (Fig. 4c). The two amino acid mutations in this epitope coincided with two anchor residues to the MHC biding, suggesting that the lack of recognition of this variant was likely due to loss of peptide binding.
Mutations responsible for impairing the epitope processing and presentation.
The discrepancies seen above between the CTL recognition of endogenously expressed and exogenously loaded antigen indicate that some mutations have caused the impairment of epitope processing and presentation. To locate specific variations that were responsible for the poor recognition of endogenously expressed HIV-1 gag variants, we constructed four different target vectors: an HXB2 gag sequence with A*0201-restricted epitope variations (SLFNTVAVL [HXB2-3F8V] or SVYNTVATL [HXB2-2V]) and IMS4-24- or IMS7-11-derived gag sequence with the wild-type A*0201 epitope sequence (IMS4-24-wild or IMS7-11-wild, respectively). The replacement of the variant epitope region with the wild-type epitope sequence restored CTL recognition of the escape variants, whereas replacement of the wild-type epitope with the two variant epitopes resulted in no CTL recognition of HXB2 Gag (Fig. 5a). The levels and patterns of Gag protein expression in target cells were analyzed by Western blot experiments (Fig. 5b). The expression levels of p55 Gag precursor and p24 CA did not significantly differ between the mutants and the wild type. The p17 MA band was not clear in HXB2-2V, IMS7-11-wild, and IMS4-24-wild, but the appearance of this band did not correlate with CTL killing. These results indicate that amino acid substitutions within the A*0201-restricted epitope region, rather than those in the flanking regions, have caused the inhibition of CTL recognition in our endogenous expression system.
To further investigate the effect of amino acid substitutions within the A24-restricted epitope on antigen processing and presentation, we introduced various point mutations into the wild-type HXB2 vector, pCTLpac, and tested them for the recognition by A24-restricted 3R mutant-reactive CTL lines. The A24-restricted 3R mutant-specific CTLs did not cross-recognize the wild-type peptide and the wild-type HXB2 vector but did recognize HXB2 with a 3R mutation (HXB2-1R). The substitution of Lys (K) with Arg (R) at position 28 (HXB2-1R3R) did not affect the A24-restricted 3R mutant-specific CTL recognition, but a Lys (K)-to-Gln (Q) substitution at position 28 (HXB2-1Q3R) or an Ile (I)-to-Leu (L) substitution at position 34 (HXB2-3R7L) resulted in the escape from CTL killing (Fig. 5c).
Replication kinetics of HIV-1 mutant viruses.
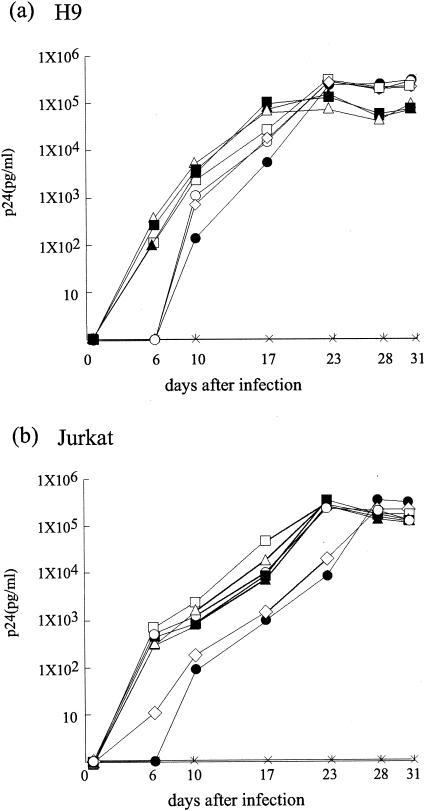
We analyzed the replication kinetics of recombinant viruses carrying mutations that have affected the epitope processing and presentation by infecting H9 or Jurkat cells. All mutants were found to replicate to equivalent levels, suggesting that these mutations do not have a significant influence on HIV-1 replication (Fig. 6).
FIG. 6.
Replication of HIV-1 clones with mutations that impaired the processing and presentation of A*0201 or A24 CTL epitopes in H9 (a) and Jurkat (b) cells. The kinetics of each recombinant virus replication were monitored as the production of p24 antigen by p24 ELISA. Symbols: ○, wild-type; □, A24-3R; ▪, A24-K26S+3R; ▵, A24-3R7L; ▴, A24-1Q3R; •, A*0201-3F8V; ⋄, A*0201-2V; ×, mock.
DISCUSSION
The present study focused on three Gag CTL epitopes restricted by three common HLA alleles in Japanese people (24). The Gag protein is most commonly targeted by CTL-inducing HIV/AIDS vaccines (15). In our endogenous expression system, three A*0201-restricted epitope variants and one B*5101-restricted epitope variant escaped from the wild-type CTL recognition, and four A24-restricted epitope variants escaped from the A24-restricted 3R mutant-reactive CTL recognition. Intriguingly, two A*0201-restricted variants and three A24-restricted variants escaped from CTL killing when the gag clones were expressed endogenously in the target cells by the HIV-1 vector, despite the fact that the synthetic variant peptides were well recognized by the CTLs when loaded onto the MHC class I molecule exogenously. The peptide titration experiments have revealed that the strength of these variant peptides' recognition was almost equivalent to that of the A*0201-restricted wild-type peptide or the A24-restricted 3R mutant peptide. The results were not likely due to differences in the pattern of Gag protein expression, as shown in the Western blot experiments. All target cells were confirmed to express a sufficient level of Gag protein by p24 antigen production. Therefore, we believe that the escape mechanism of these variants resides in the antigen processing and presentation, as has been observed in a mouse model with murine leukemia virus infection (19). The observation of such phenomenon in two epitopes restricted by different alleles implies that this finding is not unique to a particular epitope-MHC pair.
Since all variants investigated here were derived from clinical samples and those mutations did not affect the virus replication, our observations are relevant for discussing what may be going on in HIV-infected individuals. Our results indicate that the impaired antigen processing and presentation often occurs in HIV-1 field isolates and thus is one of the major mechanisms that enable HIV-1 to escape from the CTL recognition. To understand further the significance of this escape mechanism, it is important to evaluate an accumulation of such escape variants in infected hosts in a longitudinal study or at a population level. A previous report using a vaccinia virus expression system did not reveal that any mutations in the A*0201-restricted p17 epitope of HIV-1 and its flanking region altered the processing and presentation of its variant epitope (4). However, that study did not investigate A*0201-restricted 2V and 3F8V variants, which we found affected epitope processing and presentation.
Experiments with chimeric genes, as well as point mutations, showed that escapes from epitope processing and presentation were mostly attributable to mutations within the epitope regions rather than its flanking regions. In the present study, we demonstrated that point mutations of Lys (L) to Gln (Q) at position 28 and of Ile to Leu at position 34 drastically impaired the processing and presentation of the A24-restricted CTL epitope. Moreover, the experiment with HXB2 clone carrying IMS 7-11 variant of A*0201-restricted CTL epitope indicates that a substitution of Leu (L) to Val (V) at position 78 was responsible for the impaired processing and presentation of the epitope. These mutations in the epitope region may have induced a proteasome cleavage site within the epitope (19). On the other hand, we observed that the variations in the 15 amino acids up- and downstream of the epitope did not affect CTL recognition. An exception was a Lys (L)-to-Ser (S) substitution (−2S) at position 26, which is only two amino acids adjacent to the N terminus of the A24-restricted epitope. However, this −2S substitution did not void the A24-restricted 3R mutant-reactive CTL recognition completely. One possible explanation is that the −2S substitution shifted the optimal proteasome cleavage site, resulting in the generation of a larger peptide, which has a lower affinity to the MHC class I molecule.
We have first attempted to investigate the antigen processing and presentation by the conventional recombinant vaccinia virus method for all variants before we established this VSV-G-pseudotyped HIV-1 vector method. Soon, we realized that preparing recombinant vaccinia viruses was much more laborious and time-consuming. Early experiments of comparing two methods by using the first available recombinant vacinia viruses concluded that the HIV-1 vector method demonstrated CTL killing better than did the recombinant vaccinia virus method (Fig. 3b). In the recombinant vaccinia virus expression system, the massive production of vaccinia virus proteins inevitably takes place, along with the expression of an HIV-1 gene and sometimes causes a high background lysis. The expression manner and the production ratio to non-HIV proteins may also influence antigen processing and presentation (27, 34). Thus, we thought that the antigen processing and presentation in the HIV-1 vector expression system is more physiological than the recombinant vaccinia virus expression system and that continuing vaccinia virus experiments would not be significantly beneficial to address the issue of antigen processing and presentation. Nevertheless, there remains a concern that there might be a potential difference in the antigen processing and presentation between immortalized B cells that were used here and primary CD4+ T cells (32, 33). Perhaps it is important to reevaluate the interaction of CTLs and these variants in experiments with variant HIV-1-infected T cells. Our HIV-1 vector carries neither the nef gene nor the vpu gene, which significantly affect antigen presentation by downregulating MHC class I cell surface expression (5, 13). From this point of view, one might expect that more variants would escape from the CTL recognition in the actual HIV-1 infection than what is shown in our experiments. However, we think that our system is suited to identify a specific association between a certain mutation and the escape from antigen processing and presentation. To prove the existence of this mode of escape mechanism, we may need a new system that can directly detect a trace of specific epitopes that are eluted from MHC class I molecules of HIV-1 antigen-producing cells.
Although the structure analysis of MHC class I molecules and its binding motif has facilitated the prediction of CTL epitopes from the primary amino acid sequence data of HIV-1 (6, 11, 26), it remains difficult to envisage the efficiency of epitope processing and presentation. Enormous diversity realized in HIV-1 field isolates causes a further complexity (7). Our data emphasize the importance of testing HIV-1 variants in an endogenous expression system. Detailed analysis of epitope processing and presentation among HIV-1 field isolates, particularly of non-B subtypes circulating in the vaccine trial fields, is essential, since such information allows us to forecast which virus may elude the immunity elicited by vaccines, thus providing a clue for a rational design for effective HIV/AIDS vaccines.
Acknowledgments
We thank the donors in this study for their participation, Sachiko Tateishi for assistance, and Kunito Yoshiike for critical reading of the manuscript.
This study was supported by the Japanese Ministry of Health, Labor, and Welfare and the Japan Health Science Foundation.
REFERENCES
- 1.Aoki, N., T. Shioda, H. Satoh, and H. Shibuta. 1991. Syncytium formation of human and non-human cells by recombinant vaccinia viruses carrying the HIV env gene and human CD4 gene. AIDS 5:871-875. [DOI] [PubMed] [Google Scholar]
- 2.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 3.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 4.Brander, C., O. O. Yang, N. G. Jones, Y. Lee, P. Goulder, R. P. Johnson, A. Trocha, D. Colbert, C. Hay, S. Buchbinder, C. C. Bergmann, H. J. Zweerink, S. Wolinsky, W. A. Blattner, S. A. Kalams, and B. D. Walker. 1999. Efficient processing of the immunodominant, HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic-T-lymphocyte epitope despite multiple variations in the epitope flanking sequences. J. Virol. 73:10191-10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 6.Falk, K., O. Rotzschke, S. Stevanovic, G. Jung, and H. G. Rammensee. 1991. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351:290-296. [DOI] [PubMed] [Google Scholar]
- 7.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 8.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 9.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 10.Hoshikawa, N., A. Kojima, A. Yasuda, E. Takayashiki, S. Masuko, J. Chiba, T. Sata, and T. Kurata. 1991. Role of the gag and pol genes of human immunodeficiency virus in the morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: an ultrastructural study. J. Gen. Virol. 72:2509-2517. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda-Moore, Y., H. Tomiyama, M. Ibe, S. Oka, K. Miwa, Y. Kaneko, and M. Takiguchi. 1998. Identification of a novel HLA-A24-restricted cytotoxic T-lymphocyte epitope derived from HIV-1 Gag protein. AIDS 12:2073-2074. [DOI] [PubMed] [Google Scholar]
- 12.Kawana, A., H. Tomiyama, M. Takiguchi, T. Shioda, T. Nakamura, and A. Iwamoto. 1999. Accumuation of specific amino acid substitutions in HLA-B35-restricted human immunodeficiency virus type 1 cytotoxic T lymphocyte epitopes. AIDS Res. Hum. Retrovir. 15:1099-1107. [DOI] [PubMed] [Google Scholar]
- 13.Kerkau, T., I. Bacik, J. R. Bennink, J. W. Yewdell, T. Hunig, A. Schimpl, and U. Schubert. 1997. The human immunodeficiency virus type 1 (HIV-1) Vpu protein interferes with an early step in the biosynthesis of major histocompatibility complex (MHC) class I molecules. J. Exp. Med. 185:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koenig, S., A. J. Conley, Y. A. Brewah, G. M. Jones, S. Leath, L. J. Boots, V. Davey, G. Pantaleo, J. F. Demarest, C. Carter, et al. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330-336. [DOI] [PubMed] [Google Scholar]
- 15.McMichael, A. J., and T. Hanke. 2003. HIV vaccines 1983-2003. Nat. Med. 9:874-880. [DOI] [PubMed] [Google Scholar]
- 16.McMichael, A. J., and S. L. Rowland-Jones. 2001. Cellular immune responses to HIV. Nature 410:980-987. [DOI] [PubMed] [Google Scholar]
- 17.Momburg, F., and G. J. Hammerling. 1998. Generation and TAP-mediated transport of peptides for major histocompatibility complex class I molecules. Adv. Immunol. 68:191-256. [DOI] [PubMed] [Google Scholar]
- 18.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 19.Ossendorp, F., M. Eggers, A. Neisig, T. Ruppert, M. Groettrup, A. Sijts, E. Mengede, P. M. Kloetzel, J. Neefjes, U. Koszinowski, and C. Melief. 1996. A single residue exchange within a viral CTL epitope alters proteasome-mediated degradation resulting in lack of antigen presentation. Immunity 5:115-124. [DOI] [PubMed] [Google Scholar]
- 20.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T-cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 21.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratner, L., A. Fisher, L. L. Jagodzinski, H. Mitsuya, R. S. Liou, R. C. Gallo, and F. Wong-Staal. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retrovir. 3:57-69. [DOI] [PubMed] [Google Scholar]
- 23.Rock, K. L., and A. L. Goldberg. 1999. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17:739-779. [DOI] [PubMed] [Google Scholar]
- 24.Saito, S., S. Ota, E. Yamada, H. Inoko, and M. Ota. 2000. Allele frequencyes and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens 56:522-529. [DOI] [PubMed] [Google Scholar]
- 25.Sugiura, W., Z. Matsuda, Y. Yokomaku, K. Hertogs, B. Larder, T. Oishi, A. Okano, T. Shiino, M. Tatsumi, M. Matsuda, H. Abumi, N. Takata, S. Shirahata, K. Yamada, H. Yoshikura, and Y. Nagai. 2002. Interference between D30N and L90M in selection and development of protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 46:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomiyama, H., T. Sakaguchi, K. Miwa, S. Oka, A. Iwamoto, Y. Kaneko, and M. Takiguchi. 1999. Identification of multiple HIV-1 CTL epitopes presented by HLA-B*5101 molecules. Hum. Immunol. 60:177-186. [DOI] [PubMed] [Google Scholar]
- 27.Tsomides, T. J., A. Aldovini, R. P. Johnson, B. D. Walker, R. A. Young, and H. N. Eisen. 1994. Naturally processed viral peptides recognized by cytotoxic T lymphocytes on cells chronically infected by human immunodeficiency virus type 1. J. Exp. Med. 180:1283-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsomides, T. J., B. D. Walker, and H. N. Eisen. 1991. An optimal viral peptide recognized by CD8+ T cells binds very tightly to the restricting class I major histocompatibility complex protein on intact cells but not to the purified class I protein. Proc. Natl. Acad. Sci. USA 88:11276-11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Baalen, C. A., M. Schutten, R. C. Huisman, P. H. Boers, R. A. Gruters, and A. D. Osterhaus. 1998. Kinetics of antiviral activity by human immunodeficiency virus type 1-specific cytotoxic T lymphocytes (CTL) and rapid selection of CTL escape virus in vitro. J. Virol. 72:6851-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 31.Wei, X., J. M. Decker, S. Wang, H Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 32.Yang, O. O., S. A. Kalams, M. Rosenzweig, A. Trocha, N. Jones, M. Koziel, B. D. Walker, and R. P. Johnson. 1996. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J. Virol. 70:5799-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, O. O., S. A. Kalams, A. Trocha, H. Cao, A. Luster, R. P. Johnson, and B. D. Walker. 1997. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. J. Virol. 71:3120-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yewdell, J. W., and J. R. Bennink. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51-88. [DOI] [PubMed] [Google Scholar]