Abstract
A single protein, termed Gag, is responsible for retrovirus particle assembly. After the assembled virion is released from the cell, Gag is cleaved at several sites by the viral protease (PR). The cleavages catalyzed by PR bring about a wide variety of physical changes in the particle, collectively termed maturation, and convert the particle into an infectious virion. In murine leukemia virus (MLV) maturation, Gag is cleaved at three sites, resulting in formation of the matrix (MA), p12, capsid (CA), and nucleocapsid (NC) proteins. We introduced mutations into MLV that inhibited cleavage at individual sites in Gag. All mutants had lost the intensely staining ring characteristic of immature particles; thus, no single cleavage event is required for this feature of maturation. Mutant virions in which MA was not cleaved from p12 were still infectious, with a specific infectivity only ∼10-fold below that of the wild type. Particles in which p12 and CA could not be separated from each other were noninfectious and lacked a well-delineated core despite the presence of dense material in their interiors. In both of these mutants, the dimeric viral RNA had undergone the stabilization normally associated with maturation, suggesting that this change may depend upon the separation of CA from NC. Alteration of the C-terminal end of CA blocked CA-NC cleavage but also reduced the efficiency of particle formation and, in some cases, severely disrupted the ability of Gag to assemble into regular structures. This observation highlights the critical role of this region of Gag in assembly.
Retrovirus particles are released by budding from the plasma membrane of the host cell. Expression of the major viral structural protein, termed the Gag polyprotein, is sufficient for assembly of retrovirus particles in permissive cells. However, the particles are not infectious at the moment of release: they must undergo an extracellular maturation event before they are competent to infect a new host cell. This conversion to an infectious particle is brought about by the cleavage of viral proteins by the virus-encoded protease (PR). During viral maturation, the Gag polyprotein is always cleaved into at least three cleavage products, termed (from N to C terminus) matrix (MA), capsid (CA), and nucleocapsid (NC) (43).
Maturation causes a wide variety of changes in the particle. Thus, the morphology of immature particles is totally different from that of mature particles: the former are characterized by a darkly staining ring under the viral envelope, enclosing an electron-lucent core; in contrast, the interior of mature particles contains a condensed core. Immature particles are far more stable under mild detergent treatment than are mature particles (17, 20, 29, 41, 51). The conformation of the dimeric genomic RNA is different within immature and mature particles, as demonstrated by differences in the electrophoretic mobility and thermostability of the dimer (12, 42). In gammaretroviruses, the envelope glycoprotein of mature, but not immature, particles is fusogenic (31, 35).
In the murine leukemia viruses (MLVs), Gag is cleaved at three sites during maturation, resulting in the formation of four cleavage products, i.e., MA, p12, CA, and NC (18). Thus, concerted cleavage of each of the Gag proteins in a virion at three sites induces multiple changes in the particle. It would be of great interest to know which cleavage(s) is responsible for each of the changes associated with maturation. As an approach to this question, we have introduced missense mutations into each of the three cleavage sites. In each case, mutations were identified that completely prevented cleavage at the mutated site but had little or no effect on cleavage at the remaining two sites. The properties of these mutants are described below.
MATERIALS AND METHODS
Cells and viruses.
All viruses used in this study were derived from the infectious molecular clone of Moloney MLV we have referred to as pRR88 (12). The plasmid was modified for these experiments by deletion of approximately 5 kb of 5′-flanking cellular DNA. In many experiments, mutant particles were compared with those produced by the D32L mutant, which has a leucine in place of aspartate at the active site of PR and produces completely immature particles (12). In some experiments, a control MLV that produces no detectable virions was used: this was the mutant in which the glycine codon at the N terminus of Gag is replaced with alanine, so that Gag is not modified by myristic acid (34). All experiments were performed with full-length proviral clones. Virus particles were produced by transient transfection of 293T cells using the calcium phosphate method (16).
For analysis of RNA or protein within virions, particles were pelleted from culture fluids through a cushion of 20% sucrose in TNE (100 mM NaCl-10 mM Tris-HCl [pH 7.4]-1 mM EDTA) by centrifugation for 50 min at 25,000 rpm in a Beckman SW28 rotor at 4°C. They were then resuspended in TNE in 1/100 of the original volume of culture fluid.
Site-directed mutagenesis.
Site-directed mutagenesis was performed by overlap extension PCR (37). All mutant clones were verified by sequencing. In all mutations described here, the residue at the P1 position of an MLV Gag cleavage site was changed from the wild-type, hydrophobic amino acid to a charged amino acid. The mutants are designated using “S” for the site that is modified, followed by the one-letter code for the amino acid replacing the wild-type residue. For example, S2D means that the tyrosine at the P1 position of site 2 (i.e., the tyrosine at the C terminus of p12) has been replaced by aspartic acid. In some cases the P2 or P2 and P3 residues, as well as the P1 residue, were changed; in these cases, both or all three of the mutant residues are identified in the designation.
Infectivity assays.
Replication-competent MLV was assayed using the S+L- focus assay (2). Infectivity was also assayed using pLZRS-EGFP, an MLV-based vector that directs the synthesis of green fluorescent protein (GFP) (6) (a gift of Vineet KewalRamani, National Cancer Institute). These assays were performed by cotransfecting 293T cells with the pLZRS-EGFP plasmid and the plasmid containing the MLV genome being analyzed. The supernatant from this transient transfection was used to infect NIH 3T3 cells. Two days later, the cells were trypsinized and cells expressing EGFP were enumerated by flow cytometry analysis using a FACSCalibur instrument (Becton Dickinson). Titers of infectious virus were calculated from the fraction of GFP-negative cells, using the Poisson distribution.
Analysis of viral proteins.
Proteins were characterized by either radioimmunoprecipitation or immunoblotting, using rabbit antisera against p15MA, p30CA, or p10NC (obtained from the AIDS Vaccine Program, National Cancer Institute-Frederick), or the mouse monoclonal antibody F548 directed against p12 (4). In other immunoblotting experiments we used rabbit antisera against PR, reverse transcriptase (RT)-integrase (IN) (19) (a kind gift of Judith Levin, National Institute for Child Health and Human Development), and p15E. Immunoblotting was performed as described previously (3, 36). For radioimmunoprecipitation, transiently transfected 293T cells were starved for 1 h in methionine-cysteine-free minimal essential medium and then labeled for 4 h with 50 μCi each of l-[35S-]methionine and l-[35S-]cysteine (Amersham)/ml in methionine-cysteine-free minimal essential medium containing 10% dialyzed fetal calf serum (Life Technologies). The supernatants were filtered through 0.45-μm filters, and virions in the filtrate were pelleted through 20% sucrose and lysed in 0.8 ml of TNT lysis buffer (200 mM NaCl-20 mM Tris [pH 7.5]-1% Triton X-100 containing phenylmethylsulfonyl fluoride and aprotinin protease inhibitors). Viral proteins were then precipitated out of 0.2 ml of lysate and analyzed by electrophoresis through NuPAGE gels (Novex), which were treated with Amplify (Amersham) before autoradiography.
Detergent sensitivity of virions.
Sensitivity of virions to disruption by nonionic detergents was assessed by resuspending pelleted virions in TN buffer (0.1 M NaCl-0.01 M Tris [pH 7.4]) and adding NP-40 to a concentration of 1%. The suspension was incubated for 1 h at room temperature and then centrifuged for 1 h in an Eppendorf microcentrifuge at 20,800 × g. The supernatant and pellet were analyzed by immunoblotting with antiserum against p30CA.
Analysis of viral RNA.
RNA was extracted from viral pellets as described previously (12). For determination of thermostability of RNA dimers, viral RNA was incubated at a series of temperatures for 10 min using the temperature-gradient setting of a DNA Engine thermocycler (MJ Research) and was then analyzed by nondenaturing Northern analysis as described previously (12, 21). The membrane was probed with a 32P-labeled probe containing sequences from nucleotides (nt) 841 to 1325. The DNA probe was synthesized by PCR and labeled using the random primer method (37).
Copies of MLV genomic RNA and 18S rRNA were enumerated by real-time RT-PCR using the ABI 7700 instrument (Applied Biosystems). After digestion with RNase-free DNase (Promega), the RNA was treated with 2.7 M guanidinium isothiocyanate and precipitated with ethanol in the presence of yeast tRNA as carrier. The precipitate was redissolved in water containing RNasin RNase inhibitor (Promega). RNAs used in the construction of the standard curves were, for viral RNA, a 5-kb transcript of pMXH DNA (9) (a kind gift of Catherine Hibbert) and rRNA purified from 293T cells as described elsewhere (27). These standards were quantitated by their absorbance at 260 nm.
Tenfold serial dilutions of the RNAs were first copied into DNA in 30 μl containing 5 mM MgCl2, 0.5 mM deoxynucleoside triphosphates, 1 mM dithiothreitol, 0.15 μg of random hexamers (Promega), 1× TaqMan buffer A (Applied Biosystems), 20 U of RNase Out (Invitrogen), and 20 U of Superscript II RT (Invitrogen). These reactions were performed for 15 min at 25°C, 40 min at 42°C, and 10 min at 85°C. Twenty microliters of PCR cocktail (1× PCR II buffer [Applied Biosystems], 4.5 mM MgCl2, a 600 nM concentration each of the forward and reverse primers, 100 nM probe, and 1.25 U of AmpliTaq Gold DNA polymerase [Applied Biosystems]) was then added to each tube, and the tubes were heated for 10 min at 95°C and then exposed to 45 cycles of 15 s at 95°C and 1 min at 60°C. For MLV RNA, the primers were 5′TCCAATAAACCCTCTTGCAG (forward primer, nt 44 to 63) and 5′AGGAGACCCTCCCAAGGAAC (reverse primer, complementary to nt 107 to 88) and the probe (modified at its 5′ end with 6-carboxy fluorescein [FAM] and at its 3′ end with 6-carboxy-N,N,N′,N′-tetramethyl-rhodamine [TAMRA]) was 5′TTGCATCCGACTTGTGGTCTCGC (nt 64 to 86). For 18S rRNA, the primers were 5′GCCGCTAGAGGTGAAATTCTTG (forward) and 5′CATTCTTGGCAAATGCTTTCG (reverse), and the probe was 5′-FAM-ACCGGCGCAAGACGGACCAGA-TAMRA.
Total RNA concentrations were determined by Ribogreen fluorescence (Molecular Probes) as described previously (26).
Electron microscopy.
For thin-section electron microscopy, MLV-infected cells were harvested by scraping, centrifuged at 150 × g, and fixed in 2% glutaraldehyde in cacodylate buffer (0.1 M; pH. 7.4). The cell pellet was postfixed in 1% osmium tetroxide in the same buffer. Dehydration and infiltration were carried out in a series of graded ethanols, propylene oxide, and an equal mixture of propylene oxide and epoxy resin. The dehydrated pellet was embedded in Beem capsules with pure resin and cured at 55°C. Thin sections were transferred to copper grids and stained in uranyl acetate and lead citrate. Transmission electron microscopy was performed at 75 kV.
The tannic acid staining procedure was previously described (40). Briefly, the cell pellet was washed three times in cacodylate buffer after osmium postfixation and then incubated in 1% tannic acid in the same buffer for 30 min. The cell pellet was washed three times in the same buffer, then incubated in 1% uranyl acetate for 60 min, and then washed three more times. It was dehydrated, infiltrated, embedded, and sectioned as described above.
Released virus was examined after it was collected on protein G-Sepharose beads (Amersham) as described elsewhere (3). Culture fluids were collected from virus-producing cells every 24 h. Virus particles were harvested from these fluids by pelleting through 20% sucrose, resuspended in TNE, and immunoprecipitated onto protein G-Sepharose beads with goat anti-gp70SU antiserum. The beads with attached virus were dehydrated in a series of graded ethanols. After dehydration with 100% ethanol, the beads were infiltrated with two changes of pure resin and embedded and sectioned as described above.
Electron cryo-microscopy of isolated particles was performed as described previously (48).
RESULTS
Generation of mutants.
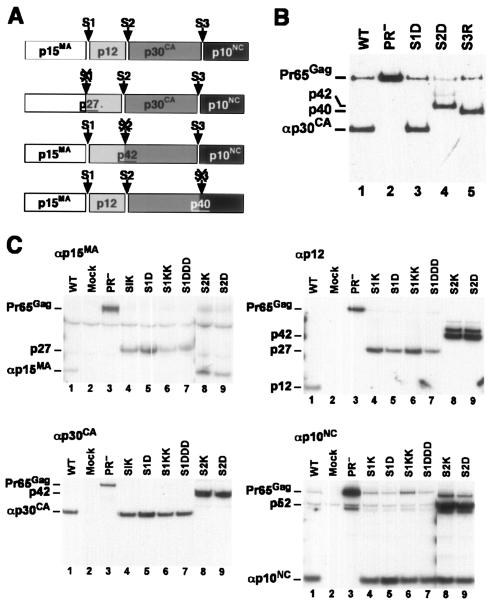
As discussed above, the MLV Gag polyprotein is cleaved at three sites during virus maturation (Fig. 1A). We introduced missense mutations into an infectious MLV clone at each of these sites. In all cases, these mutations replaced hydrophobic residues at the P1 position with charged residues. The wild-type P1 residues were tyrosine (site 1), phenylalanine (site 2), and leucine (site 3).
FIG. 1.
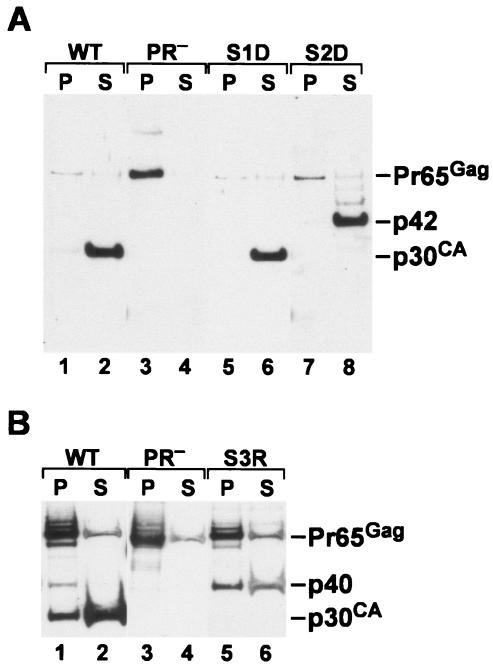
Mutants blocking cleavage at individual sites in MLV Gag. (A) Cleavage scheme of MLV Gag. The top line shows the three cleavage sites and the four Gag cleavage products found in mature MLV particles. The next three lines show the fusion proteins expected if cleavage at each of the three sites were prevented. (B) p30CA-containing proteins in mutant particles. Proteins in wild-type and mutant virions were analyzed by immunoblotting with anti-CA antiserum. Lanes: 1, wild-type MLV; 2, PR− MLV; 3, S1D; 4, S2D; 5, S3R. (C) Protein profiles of site 1 and site 2 mutant particles. Cells producing wild-type or mutant particles were labeled with [35S]methionine-cysteine. Particles were collected and analyzed by immunoprecipitation with monospecific antisera against MLV Gag proteins, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. The four panels show results of immunoprecipitation with anti-p15MA, anti-p12, anti-p30CA, and anti-p10NC. Lanes: 1, wild-type MLV; 2, supernatant from cells transfected with the empty vector pGCcos3neo; 3, PR− MLV; 4, S1K; 5, S1D; 6, S1KK; 7, S1DDD; 8, S2K; 9, S2D.
Cleavage patterns in mutant virions.
The mutations studied here were created with the intention of blocking cleavage at the mutated sites. The respective fusion proteins expected if cleavage failed to occur at sites 1, 2, and 3 are shown in Fig. 1A. To determine whether the cleavages had been successfully inhibited, we transfected the mutant plasmids into 293T cells. Virus particles were collected from the culture fluids of the transfected cells and initially characterized by immunoblotting with antiserum against p30CA. We found (data not shown) that the site 1 and site 2 mutants all released virions at levels indistinguishable from wild type. All site 3 mutants were somewhat defective with respect to virus production; culture fluids of S3R particles (i.e., the mutant in which the C-terminal residue of CA had been replaced with arginine) contained virus at levels 1/10 to 1/4 that of wild type, while all other site 3 mutants tested (S3K, S3KK, S3RR, S3D, S3DD, S3E, and S3EE) produced far lower amounts of virus, roughly 1% of the wild-type level. The defects in particle production presumably represented effects of the changes in Gag upon the assembly process per se and were independent of the interference with cleavage during maturation.
Immunoblotting profiles of virion proteins from selected mutants are shown in Fig. 1B. It can be seen (lane 3) that the site 1 mutant in this experiment (S1D) contained p30CA with the same electrophoretic mobility as in the wild-type control (lane 1). Thus, both of the cleavages that produced p30CA from Pr65 (i.e., the site 2 and site 3 cleavages) occurred normally in these mutant particles. In contrast, mutant S2D contained no p30 but had a 42-kDa protein reacting with this antiserum (lane 4). This result shows not only that cleavage at site 2 was blocked, as anticipated (Fig. 1), but also that cleavage at sites 1 and 3 (generating the N and C termini of p42) was occurring. Similarly, S3R particles contained no p30, but a 40-kDa protein was detected in the immunoblot (lane 5); production of this protein results from inhibition of cleavage at site 3, and requires cleavage at site 2. Similar results were also obtained with the particles produced by all other site 3 mutants (data not shown). Lane 4 of Fig. 1B also shows traces of a larger cleavage intermediate in the S2D particles; this protein could be either p15MA-p12-p30CA or p12-p30CA-p10NC; evidence presented below (Fig. 1C) suggests that it is the latter.
Cleavage profiles in the mutant particles were further examined by both immunoblotting and radioimmunoprecipitation using antisera against individual viral proteins. Several site 1 mutants (i.e., S1K, S1D, S1KK, and S1DDD) were metabolically labeled and tested for the presence of the expected 27-kDa fusion protein by immunoprecipitation with anti-p15MA and anti-p12 sera. As shown in Fig. 1C, all of these mutants contained a 27-kDa protein that was precipitated with both of these sera; the mutant particles also contained p30 and p10, demonstrating that the mutation has no effect on cleavage at site 3. Site 2 mutant particles contained p15MA, p10NC, and the expected 42-kDa p12-CA fusion protein. However, they also contained a larger intermediate that reacted with anti-p10NC antiserum (p52, lanes 8 and 9), suggesting that separation of NC from the p12-CA protein is somewhat inefficient in these particles. We also found that cleavage at site 1 as well as at site 2 is efficient in S3R particles, since they contain p15MA and p12, but no 27-kDa fusion protein (data not shown). We also tested site 1, site 2, and site 3 mutant particles for the cleavages in the Pol and Env proteins. In all cases, we found efficient production of PR, RT, and IN and, as in wild-type particles, partial removal of the R peptide from p15E (data not shown).
In summary, at each of the three cleavage sites, replacement of the hydrophobic residue at the P1 position with a charged residue completely prevented cleavage at that site; there was no detectable effect on cleavages at the remaining two sites, except that site 2 mutants exhibited slightly reduced cleavage at site 3. Immunoblotting and radioimmunoprecipitation data were in complete agreement in these experiments. While the mutations at site 1 and site 2 had no detectable effects on virus assembly and release, all site 3 mutations tested inhibited particle production.
Infectivity of mutant particles.
Particles from each mutant were tested for infectivity, using the S+L- focus assay. This assay is analogous to a plaque assay: only viruses capable of undergoing multiple replication cycles within a few days will register in this assay. As shown in Table 1, we found that the site 1 mutants were able to form foci; some of these mutants had titers approximately 10-fold lower than that of the wild-type control. No focus formation was observed with any site 2 or site 3 mutants.
TABLE 1.
Infectivity of cleavage-site mutants
| Expt and virus | Infectious units/mla | Relative specific infectivityb |
|---|---|---|
| Expt 1 | ||
| Wild type | 1 × 105 | |
| S2D | <2 × 100 | |
| S2K | <2 × 100 | |
| S3K | <2 × 100 | |
| Expt 2 | ||
| Wild type | 3 × 106 | 1 |
| S1D | 3 × 104 | 0.2 |
| S1K | 3 × 105 | 0.05 |
| S1DDD | 2 × 105 | 0.03 |
| Expt 3 | ||
| Wild type | 9.8 × 105 | |
| S1K | 6.8 × 104 | |
| Expt 4 | ||
| Wild type | 1.9 × 106 | |
| S2D | <1.3 × 103 | |
| S3R | <1.3 × 103 |
Culture fluids were assayed for infectious MLV by the S + L − focus assay (experiments 1 and 2) or by their ability to rescue the pLZRS-EGFP vector into infectious virions (experiments 3 and 4).
In experiment 2, the relative levels of virions in the culture fluids were determined by immunoblotting with anti-CA antiserum, and these results were used to determine the specific infectivities of the mutants, relative to wild-type MLV.
It seemed possible that particles composed of site 2 or site 3 mutant proteins were capable of infecting cells at a low efficiency. Such particles would not be detected by the S+L- focus assay. Therefore, we also tested them for their ability to rescue a GFP-expressing vector genome into infectious particles: since this assay detects individual infectious events, any particle capable of successfully carrying out the infectious process will register in this assay. As shown in Table 1, these mutants were also negative in these tests. We estimate that the specific infectivity of site 2 and site 3 mutant particles was at least 1,000 times lower than that of wild-type particles. In agreement with the S+L- assay, the GFP assay also showed that site 1 mutant particles infected cells roughly 10-fold less efficiently than wild-type particles.
Morphology of mutant particles.
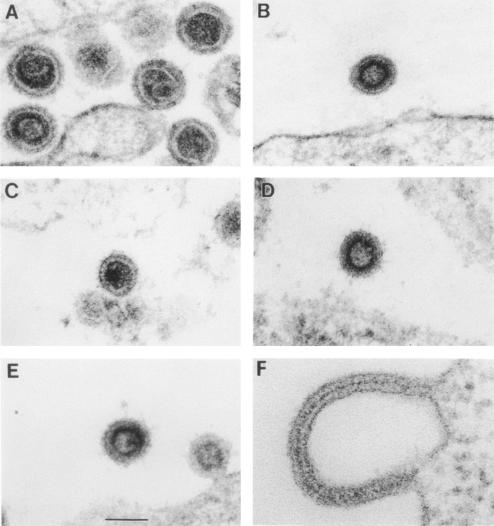
It would be of great interest to determine which of the three cleavage events in MLV maturation is responsible for the striking change in morphology associated with maturation. We therefore performed thin-section electron microscopy on cells producing the cleavage site mutants. As shown in Fig. 2C, particles of the site 1 mutant S1KK were indistinguishable in overall morphology from wild-type controls (Fig. 2A), and (as with wild type) both immature and mature particles could be identified in these cultures. Thus, cleavage between MA and p12 is unnecessary for morphological maturation.
FIG. 2.
Analysis of structure of cell-associated virions by electron microscopy. Cells producing wild-type or mutant particles were fixed and sectioned for electron microscopy. (A) Wild type; (B) PR−; (C) S1KK; (D) S2D; (E) S3R; (F) S3KK. The samples shown in panels A to E were processed by the tannic acid procedure, while that in panel F is a conventional thin-section electron micrograph. Scale bar, 100 nm.
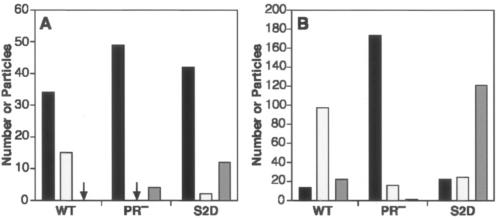
An image from a culture producing S2D mutant particles is shown in Fig. 2D. Very few mature particles were observed in an examination of over 100 site 2 mutant particles; these included S2K as well as S2D particles. A quantitative comparison between wild-type, PR−, and S2D particles in sections of virus-producing cells is shown in Fig. 3A. It can be seen that the majority of particles in both the wild type and S2D appeared immature; however, the remainder in the wild-type culture were typical, mature MLV particles, while those in the S2D-producing cells were aberrant. Most often, they contained a darkly staining interior, like typical mature particles, but this densely staining material was not surrounded by a clear border.
FIG. 3.
Proportions of virions with immature, mature, and aberrant morphologies in sections of virus-producing cells (A) and released particles (B). For the section shown in panel B, particles were collected onto beads by immunoprecipitation and processed as described in Materials and Methods. Black bars, virions with immature morphology (particles with a dark ring surrounding an electron-lucent center); light grey bars, virions with mature morphology (particles with a densely staining interior with a border around it); dark grey bars, virions with aberrant morphology (generally particles with a densely staining interior that is not surrounded by a distinct border). The fact that some PR− particles were scored as mature in panel B is an indication of the level of inaccuracy in these profiles.
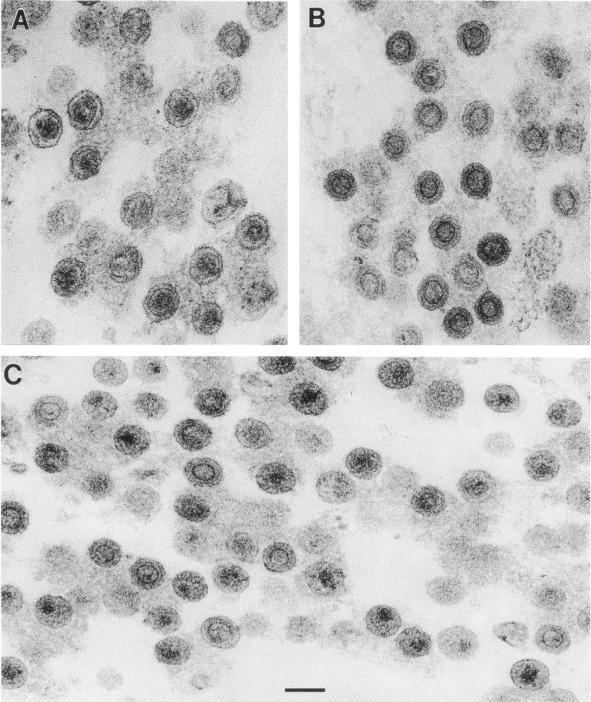
We also examined S2D virions that had been released into the supernatant over a 24-h period. These particles are shown in Fig. 4C and can be compared with wild-type and PR− particles processed in parallel (Fig. 4A and B, respectively). The mutant particles were quite pleomorphic. The majority of the S2D particles lacked the intact, darkly staining ring characteristic of immature particles (Fig. 4B), but they also did not exhibit the distinctly outlined, polygonal core seen in many wild-type particles (Fig. 4A). Many of the mutant particles contained a compact, darkly stained body in the interior; this was roughly ovoid in some particles and fibrillar in others (Fig. 4C). Figure 3B shows the proportions of immature, mature, and aberrant virions observed in these samples; it was evident that the majority of S2D particles were converted, after release, from an immature morphology to an aberrant morphology.
FIG. 4.
Analysis of structure of released virions by electron microscopy. (A) Wild type; (B) PR−; (C) S2D.
It will be recalled that the site 3 mutants all exhibited defects with respect to virion production, with S3R the least severe in this regard. Some S3R particles were clearly mature in appearance. In ∼40% of the S3R particles observed, there was an acentric accumulation of darkly staining material within the otherwise-electron-lucent center (Fig. 2E). (Because the sections were nearly as thick as virions, we are confident that the absence of this feature from the majority of the remaining particles was genuine, rather than an artifact related to the position of the section with respect to the particle.) When cultures expressing other S3 mutants were examined, they were found to contain obvious accumulations of Gag protein at their plasma membranes. However, these accumulations did not exhibit the characteristic radius of curvature required to produce a spherical particle of ∼100 to 120 nm in diameter. Rather, they were extremely pleomorphic. In some fields, a membrane appeared to be surrounded on both sides by a darkly staining accumulation of Gag protein, as shown for the S3KK mutant in Fig. 2F.
Sizes of mutant particles.
To further characterize the mutant particles, we also measured their external diameters. The measurements were made on images obtained by electron cryo-microscopy, so that there was no distortion associated with the fixation, embedding, sectioning, or staining processes used in conventional electron microscopy. We found that both S1KK and S2D particles were slightly smaller than wild-type particles: the mean diameter of wild-type virions was 122.6 ± 14.3 nm (mean ± standard deviation), while that of S1KK virions was 117.0 ± 9.6 nm and that of S2D particles was 111.2 ± 9.9 nm. The differences from wild type were, in both cases, highly significant, with a P value of <10−6 (Student's t test). Unfortunately, the low yield of S3R particles precluded the measurement of significant numbers of S3R virions.
Detergent sensitivity of mutant particles.
One striking difference between immature and mature retrovirus particles is their sensitivity to disruption by mild detergents: the ribonucleoprotein core of detergent-treated immature particles remains intact, while disruption of mature particles by this treatment solubilizes the core proteins. It was of interest to determine whether preventing any of the normal cleavages would produce a detergent-resistant structure within the virion. We therefore exposed mutant particles, as well as wild-type and PR− controls, to 1% NP-40 and then centrifuged the extracts for 1 h at ∼20,000 × g. The retention in particulate structures of p30 (and fusion proteins containing p30) was determined by immunoblotting (Fig. 5). It can be seen that treatment of site 1 mutant particles, like that of wild-type particles, completely solubilized p30. Similarly, p42 was completely solubilized by exposure of site 2 mutant particles to the detergent. However, a significant fraction of the p40 present in S3R particles was found in the pellet in the detergent-treated sample. Thus, separation of CA from NC is apparently required for the complete detergent susceptibility characteristic of mature particles.
FIG. 5.
Detergent sensitivity of mutant particles. Particles were collected from culture fluids, treated with 1% NP-40, centrifuged, and analyzed by immunoblotting with anti-CA antiserum as described in Materials and Methods. (A) Lanes: 1 and 2, wild type; 3 and 4, PR−; 5 and 6, S1D; 7 and 8, S2D. Lanes 1, 3, 5, and 7, pellets; lanes 2, 4, 6, and 8, supernatants. (B) Lanes: 1 and 2, wild type; 3 and 4, PR−; 5 and 6, S3R. Lanes 1, 3, and 5, pellets; lanes 2, 4, and 6, supernatants. In control samples not exposed to NP-40, all of the CA-containing proteins were found exclusively in the pellet fraction in all samples (data not shown).
RNA in mutant particles.
In all normal retrovirus particles, the genomic RNA is in the form of a dimer in which two copies of the RNA are presumably linked together by a limited number of base pairs (43). We previously reported that mature retrovirus particles contain a more stable dimer of genomic RNA than immature particles (11, 12). Therefore, viral maturation involves not only a change in the viral proteins but also a change in the conformation of the viral RNA. We termed this change “maturation” of the dimer. In vitro experiments suggest that maturation of the dimer is due to the action of the NC protein on the nucleic acid (8, 25, 33). In order to determine which of the three cleavages in Gag was required for RNA maturation, we attempted to measure the thermostabilities of dimeric RNAs isolated from site 1, site 2, and site 3 mutant particles.
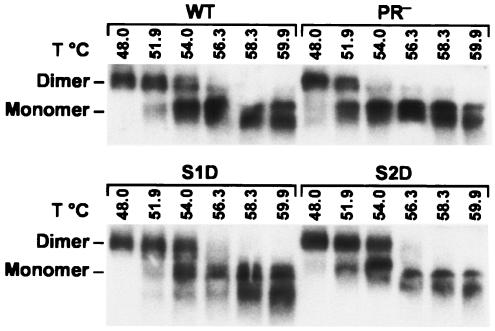
RNA was isolated from virions by proteinase K digestion followed by phenol-chloroform extraction. It was then incubated at various temperatures and analyzed by electrophoresis under nondenaturing conditions, followed by Northern blot analysis. The controls in Fig. 6 show the expected difference between dimers from mature and immature particles: the RNA from mature (wild-type) particles was almost completely dimeric after exposure to 51.9°C and was only partly dissociated into monomers by treatment at 54°C. In contrast, RNA from immature (PR−) particles was substantially dissociated into monomers at 51.9°C and virtually all monomeric after incubation at 54°C. Examination of the remaining two panels in Fig. 6 shows that RNA from a site 1 mutant or a site 2 mutant had the same thermostability as the RNA of the wild-type control. Therefore, neither site 1 nor site 2 cleavage is essential for the maturation of the dimeric RNA.
FIG. 6.
Thermostability of dimeric RNAs extracted from site 1 and site 2 mutant particles. RNAs from the mutant particles and from wild-type and PR− MLV particles were extracted and heated to the indicated temperatures before analysis by nondenaturing Northern analysis as described in Materials and Methods.
When a similar analysis was attempted on RNA from S3R mutant particles, the signal on the nondenaturing Northern blots proved too weak for analysis of dimeric stability. To determine whether these particles were deficient in viral RNA, we therefore measured the level of viral RNA in these particles using real-time RT-PCR. To estimate the background due to release of viral RNA from cells not producing virions, we also collected culture fluid from cells transfected with the G2A mutation; this mutant Gag protein is not myristylated and, as far as is known, does not assemble into virions (34, 38). The results of these tests are shown in Table 2. When the amounts of viral RNA/ml of S3R culture fluid were adjusted for the low levels of particles per milliliter (i.e., the amount of pelletable p40/ml, compared to the amount of pelletable p30/ml in wild-type culture fluid), the results indicated that these particles packaged viral RNA ∼2.5-fold less efficiently than wild-type particles. This RNA is apparently associated with virions, since the level of viral RNA in culture fluid from cells expressing unmyristylated Gag is far lower than that in the S3R culture fluid.
TABLE 2.
RNA packaged in S3R particlesa
| Virus | Particles/ml | vRNA/ml | vRNA/particle | rRNA/ml | rRNA/particle |
|---|---|---|---|---|---|
| Wild type | 1 | 1 | 1 | 1 | 1 |
| S3R | 0.2 | 0.08 | 0.4 | 0.4 | 1.9 |
| Myr−b | ND | 0.000015 | 0.08 |
Copy numbers of MLV genomic RNA (vRNA) and 18S rRNA were determined in RNAs extracted from viral pellets as described in Materials and Methods. These values are expressed relative to wild-type values. The relative levels of virions in the pellets were measured by immunoblotting with anti-CA antiserum and are expressed relative to the wild-type values. Dividing copy numbers by amounts of virus yielded relative copies of the RNA per particle. ND, not detectable by immunoblotting.
Expressing unmyristylated Gag.
We also assessed the genomic RNA content of S3R particles by Northern blotting. The genomic RNA in the particles was generally detectable but was significantly more degraded than that extracted from wild-type particles (data not shown). In some experiments, the viral RNA from S3R particles was distributed in a smear over a broad molecular weight range, while that extracted in parallel from wild-type particles was almost entirely in a discrete ∼8-kb band as expected.
When total RNA in virion preparations was measured by Ribogreen analysis, we found that S3R particles contained (per unit of Gag protein) approximately the same amount of RNA as wild-type virions (data not shown). Thus, it appears that some other RNA species compensates for the deficiency in genomic RNA in S3R particles. We recently analyzed the RNA content of MLV particles that failed to package genomic RNA efficiently because of mutations in the NC domain. We found that the principal RNA species in these virions was rRNA (27). We therefore determined the rRNA content of S3R particles. As shown in Table 2, these particles appeared to contain somewhat more rRNA per particle than wild-type particles.
DISCUSSION
Maturation converts an assembled retrovirus particle into an infectious particle. Maturation occurs as a result of concerted cleavages at a small number of sites within the Gag, Pol, and (in some retroviruses) Env protein molecules of the virion, and it is accompanied by a wide variety of changes in the particle. Our goal in these experiments was to gain some insight into the contribution that each of the three cleavages in MLV Gag makes to the physical changes in the particle and in its acquisition of infectivity.
One of the most striking changes associated with retroviral maturation is the conversion from the immature morphology, with a densely staining ring surrounding a relatively electron-lucent interior, to the mature form, in which the ring is absent and the center of the particle is filled with densely staining material. We found that site 1, site 2, and site 3 mutants all lacked the ring and contained densely staining material in their interiors, in contrast to fully immature PR− particles. Thus, no single cleavage event is absolutely required for the loss of the ring or deposition of this material. Perhaps the interior core in mature particles is composed of RNA associated with NC, and cleavage anywhere in Gag is sufficient for the release of RNA, together with RNA-bound proteins, from the membrane-associated MA domain.
Our results with individual mutants can be briefly summarized as follows. First, mutants at site 1 (preventing cleavage between p15MA and p12) are released at essentially the same level as wild-type MLV. S1KK particles (in which the leucine and tyrosine residues at the C terminus of MA have been replaced with lysines) are slightly smaller than wild type. (Obviously, this is an effect of the change in Gag sequence on the assembly process itself and tells us nothing about the significance of site 1 cleavage.) Particles in which this cleavage was blocked were still infectious (Table 1), with a specific infectivity only ∼10-fold lower than that of wild-type control particles. Thus, the cleavage event between MA and p12 appears to be merely “fine-tuning,” improving the efficiency of replication by roughly an order of magnitude. Perhaps this is not surprising, since other studies have shown that a substantial portion of p12 is tolerant to alanine substitution (52), and under some conditions virtually the entire MA domain of HIV-1 can be deleted without loss of infectivity (32).
In contrast, site 2 mutants displayed no detectable infectivity: their specific infectivity was at least 1,000-fold lower than that of wild-type particles. Like site 1 particles, they were slightly smaller than wild-type controls. Further, they were frequently different in appearance from either mature or immature particles: unlike wild-type particles, they did not possess a distinct outline or “capsid shell” around the core, but unlike PR− particles they often contained a compact body within the interior of the virion (Fig. 2 to 4). We also observed that if cleavage at site 2 were blocked, then the cleavage between CA and NC was somewhat inefficient (for example, note the accumulation of a larger intermediate, as well as p42, in S2D particles [Fig. 1B]). This observation suggests that cleavage at site 1 and/or site 3 is partially dependent upon cleavage at site 2. Indeed, in normal MLV maturation, cleavage at site 2 precedes that at sites 1 and 3 (28, 49, 50).
At site 3, replacement of the C-terminal leucine in CA with arginine allowed the formation, at low efficiency, of virions roughly similar to mature MLV particles (Fig. 2E). However, many of these particles were abnormal in appearance, with a small accumulation of electron-dense material acentrically placed within the “core” of the particle. All of the other changes that we made at the C terminus of CA reduced the ability of Gag to assemble into released virions by a factor of approximately 100. The amount of genomic RNA per amount of Gag protein was somewhat lower in the S3R particles than in wild type, and it also appeared to be more susceptible to degradation than that in controls. The deficit in genomic RNA in these particles may be partially compensated for by an increase in rRNA content.
Particles in which cleavage was blocked at site 1 or 2 still contained mature dimeric RNA, with the same thermostability as that extracted from mature MLV particles (Fig. 6). The fact that inhibition of cleavage at site 1 or site 2 still permits the stabilization of genomic RNA dimers accompanying viral maturation strongly suggests that the separation of CA and NC is the event leading to stabilization of the viral RNA. (We cannot exclude the alternate possibility that other PR-mediated cleavages, such as those in the Pol region, are responsible for the stabilization.) It would seem reasonable to imagine that when NC is released from CA during maturation, it can coat the genome and then, by virtue of its nucleic acid chaperone activity, catalyze the conformational rearrangement of the dimeric RNA. (In avian retroviral particles, chemical cross-linking of NC to RNA in mature particles is several orders of magnitude more efficient than cross-linking of Gag to RNA in immature particles [41].) However, studies with recombinant HIV-1 proteins have shown that the Gag polyprotein possesses nucleic acid chaperone activity very similar to that of its cleavage product, NC (7). Indeed, both HIV-1 NC and the HIV-1 Gag polyprotein can stabilize dimeric linkages in transcripts containing retroviral sequences in vitro (7, 8). Therefore, the requirement that Gag be cleaved before the genomic RNA within the virion attains its most stable conformation is probably due to a change in the access of the viral proteins to the RNA accompanying maturation, rather than a difference in the chaperone activities of Gag and NC. The present results suggest that it is cleavage between CA and NC that affords the NC domain increased access to the RNA. It is also possible, of course, that Gag and NC differ quantitatively or qualitatively with respect to their chaperone activity in ways that our in vitro assays (7) failed to detect. Our results here would then imply that the difference in activity depends upon cleavage of NC from CA. A prior study with HIV-1 also concluded that cleavage of Gag at the N terminus of NC is critical for maturation of the genomic RNA dimers (39).
It seems likely that the p10NC moiety of the p40 fusion protein is bound to RNA within the site 3 mutant particles; perhaps this is the acentrically located mass visible in electron micrographs of these particles (Fig. 2E). The fusion protein is apparently less effective than wild-type NC in protecting the genomic RNA against nucleolytic degradation, either within the particle or during extraction. The attachment of this protein to RNA may also explain the fact that a substantial fraction of p40 remains pelletable after disruption of the particle with NP-40 (Fig. 5).
We also found that the ratio of genomic RNA to Gag protein in S3R particles was somewhat lower than in wild-type particles (Table 2). This observation implies that genomic RNA encapsidation is reduced by the S3R mutation, and it might suggest that the region of Gag responsible for the encapsidation extends a short distance into the CA domain. However, it is also possible that each S3R particle contains the same amount of viral RNA as a wild-type particle but is, on average, somewhat larger and contains two to three times as much Gag protein as a normal particle. We also cannot completely exclude the possibility that some of the RNA detected in viral pellets comes from cellular debris, not from virions.
It is interesting to compare our results with those of prior studies on HIV-1 maturation. In HIV-1, the first cleavage event is not at the N terminus of CA but at the N terminus of NC (30). CA is directly linked to MA in HIV-1 Gag. Interfering with the cleavage between MA and CA prevents the formation of the normal conical capsid shell, resulting instead in particles with dense, acentric cores (15). One explanation for this phenotype could be that the retention of CA at the membrane, through its linkage to MA, prevents its condensation into the normal mature core. Alternatively, the phenotype might result from the lack of the normal CA N terminus. Structural studies on the HIV-1 CA protein show that there is a rearrangement of the N-terminal region of CA following the release of the N terminus during maturation (13, 14, 24, 44) and that this rearrangement (in which the conserved proline at the N terminus forms a buried salt bridge with a conserved aspartate or glutamate residue in CA) leads to new CA-CA interactions (46). Virions in which the proline residue at the N terminus is replaced by leucine, an alteration which does not interfere with cleavage (10), have a morphology similar to those in which the cleavage is blocked (15); this is also true if the aspartate at position 51 in CA, the other partner in the buried salt bridge, is changed to alanine (45, 46). These observations strongly support the idea that the morphogenesis of the conical capsid shell in normal HIV-1 maturation depends upon formation of the salt bridge between the N-terminal proline and the internal aspartate residues. This salt bridge is not, however, required for the deposition of dense material in the interior of the particle.
The most dramatic effect of the S2D mutation upon the morphology of MLV particles was the elimination of the border that surrounds the core of a normal mature particle. It is important to remember that in MLV there is a protein between MA and CA and that cleavage at site 1 still occurs in site 2 mutant particles (Fig. 1). Therefore, the CA-containing fusion protein (p42) is not tethered to the membrane in these particles and should be free to “collapse” into the interior of the particle. The absence of the outer boundary of the core in S2D particles is thus not due to retention of CA at the membrane; rather, these results are consistent with the hypothesis that release of the normal N terminus of CA from Gag is essential for formation of this border, in close analogy with those described above in HIV-1. While the densely staining bodies seen within many S2D particles (Fig. 4C) may be NC-RNA complexes, cleavage of NC from the p42 fusion protein is somewhat inefficient in these particles (Fig. 1); thus, another possibility is that these structures are composed of complexes of the p12-CA-NC fusion protein with RNA.
Our observation that even small alterations at the C terminus of MLV CA can drastically disrupt the normal process of particle assembly (Fig. 2F) is similar to results with HIV-1; it is clear that this region plays a critical role in determining both the efficiency of assembly and budding and the radius of curvature of the resulting particle (15, 22, 23). The effects of our mutations on assembly are obviously due to the changes we introduced into the Gag protein, not to the interference with site 3 cleavage. In the case of HIV-1, it has been suggested that a crucial element in this region is an α-helical structure spanning the CA-spacer cleavage site (1). In MLV, the C-terminal region of MLV CA is important for assembly (47) and contains an extraordinary run of charged residues. Some deletions in this region produce results very similar to those we observed with point mutations at the extreme C terminus, with a dramatic loss of the ability of Gag to assemble into particles with the normal radius of curvature. The properties of the deletions in this region supported the hypothesis that it contains an α-helix (5).
In conclusion, we have found that no individual cleavage event is required for the loss of immature morphology of PR− particles. However, cleavage between p12 and CA is essential for formation of the normal core structure found in mature MLV particles. Cleavage between CA and NC is probably required for the stabilization of genomic RNA dimers associated with MLV maturation. Cleavage between MA and p12 plays no critical role in the conversion of MLV particles to an infectious form.
Acknowledgments
We thank David Thomas and Wei-Shau Hu for the use of a real-time PCR instrument and for their generous help with these experiments; Vineet KewalRamani for the use of the FACS analyzer; Wei-Shau Hu for communicating results prior to publication; Catherine Hibbert for the MLV RNA standard; and Michelle Gignac and Kelly Stanard for their skilled assistance with electron microscopy.
Mark Yeager is the recipient of a Burroughs Wellcome Clinical Scientist Award in Translational Research.
REFERENCES
- 1.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassin, R. H., N. Tuttle, and P. J. Fischinger. 1971. Rapid cell culture assay for murine leukaemia virus. Nature 229:564-566. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, S., M. Oshima, J. Mirro, K. Nagashima, and A. Rein. 2002. Reversal by dithiothreitol treatment of the block in murine leukemia virus maturation induced by disulfide cross-linking. J. Virol. 76:10050-10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chesebro, B., W. Britt, L. Evans, K. Wehrly, J. Nishio, and M. Cloyd. 1983. Characterization of monoclonal antibodies reactive with murine leukemia viruses: use in analysis of strains of Friend MCF and Friend ecotropic murine leukemia virus. Virology 127:134-148. [DOI] [PubMed] [Google Scholar]
- 5.Cheslock, S. R., D. T. Poon, W. Fu, T. D. Rhodes, L. E. Henderson, K. Nagashima, C. F. McGrath, and W. S. Hu. 2003. Charged assembly helix motif in murine leukemia virus capsid: an important region for virus assembly and particle size determination. J. Virol. 77:7058-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dardalhon, V., N. Noraz, K. Pollok, C. Rebouissou, M. Boyer, A. Q. Bakker, H. Spits, and N. Taylor. 1999. Green fluorescent protein as a selectable marker of fibronectin-facilitated retroviral gene transfer in primary human T lymphocytes. Hum. Gene Ther. 10:5-14. [DOI] [PubMed] [Google Scholar]
- 7.Feng, Y. X., S. Campbell, D. Harvin, B. Ehresmann, C. Ehresmann, and A. Rein. 1999. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 73:4251-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, Y. X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng, Y. X., D. L. Hatfield, A. Rein, and J. G. Levin. 1989. Translational readthrough of the murine leukemia virus gag gene amber codon does not require virus-induced alteration of tRNA. J. Virol. 63:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schroder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294-307. [DOI] [PubMed] [Google Scholar]
- 11.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 14.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 15.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, M., L. Jelinek, R. S. Jones, J. Stegeman-Olsen, and E. Barklis. 1993. Assembly and composition of intracellular particles formed by Moloney murine leukemia virus. J. Virol. 67:5163-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson, L. E., R. Sowder, T. D. Copeland, G. Smythers, and S. Oroszlan. 1984. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J. Virol. 52:492-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, S. C., D. L. Court, M. Zweig, and J. G. Levin. 1986. Murine leukemia virus pol gene products: analysis with antisera generated against reverse transcriptase and endonuclease fusion proteins expressed in Escherichia coli. J. Virol. 60:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan, A. H., P. Krogstad, D. J. Kempf, D. W. Norbeck, and R. Swanstrom. 1994. Human immunodeficiency virus type 1 virions composed of unprocessed Gag and Gag-Pol precursors are capable of reverse transcribing viral genomic RNA. Antimicrob. Agents Chemother. 38:2929-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khandjian, E. W., and C. Meric. 1986. A procedure for Northern blot analysis of native RNA. Anal. Biochem. 159:227-232. [DOI] [PubMed] [Google Scholar]
- 22.Krausslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang, C., J. Hu, J. B. Whitney, L. Kleiman, and M. A. Wainberg. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the Gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 25.Muriaux, D., H. De Rocquigny, B. P. Roques, and J. Paoletti. 1996. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 271:33686-33692. [DOI] [PubMed] [Google Scholar]
- 26.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muriaux, D., J. Mirro, K. Nagashima, D. Harvin, and A. Rein. 2002. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 76:11405-11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naso, R. B., W. L. Karshin, Y. H. Wu, and R. B. Arlinghaus. 1979. Characterization of 40,000- and 25,000-dalton intermediate precursors to Rauscher murine leukemia virus gag gene products. J. Virol. 32:187-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, J., and C. D. Morrow. 1993. Mutations in the protease gene of human immunodeficiency virus type 1 affect release and stability of virus particles. Virology 194:843-850. [DOI] [PubMed] [Google Scholar]
- 30.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Gottlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic acid chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 34.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rein, A., D. E. Ott, J. Mirro, L. O. Arthur, W. Rice, and L. E. Henderson. 1996. Inactivation of murine leukemia virus by compounds that react with the zinc finger in the viral nucleocapsid protein. J. Virol. 70:4966-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 38.Schultz, A. M., and A. Rein. 1989. Unmyristylated Moloney murine leukemia virus Pr65gag is excluded from virus assembly and maturation events. J. Virol. 63:2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shehu-Xhilaga, M., H. G. Kraeusslich, S. Pettit, R. Swanstrom, J. Y. Lee, J. A. Marshall, S. M. Crowe, and J. Mak. 2001. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 75:9156-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simionescu, N., and M. Simionescu. 1976. Galloylglucoses of low molecular weight as mordant in electron microscopy. I. Procedure, and evidence for mordanting effect. J. Cell Biol. 70:608-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, L., G. Schatz, and V. M. Vogt. 1990. Properties of avian retrovirus particles defective in viral protease. J. Virol. 64:5076-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoltzfus, C. M., and P. N. Snyder. 1975. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J. Virol. 16:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 44.Tang, C., Y. Ndassa, and M. F. Summers. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537-543. [DOI] [PubMed] [Google Scholar]
- 45.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, M. Q., and S. P. Goff. 2003. Defects in virion production caused by mutations affecting the C-terminal portion of the Moloney murine leukemia virus capsid protein. J. Virol. 77:3339-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshinaka, Y., and R. B. Luftig. 1977. Characterization of Rauscher leukemia virus (RLV) P40-42, an intermediate cleavage product of the group specific antigen (Gag) precursor polypeptide, P65-70. Biochem. Biophys. Res. Commun. 79:319-325. [DOI] [PubMed] [Google Scholar]
- 50.Yoshinaka, Y., and R. B. Luftig. 1977. Murine leukemia virus morphogenesis: cleavage of P70 in vitro can be accompanied by a shift from a concentrically coiled internal strand (“immature”) to a collapsed (“mature”) form of the virus core. Proc. Natl. Acad. Sci. USA 74:3446-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshinaka, Y., and R. B. Luftig. 1977. Properties of a P70 proteolytic factor of murine leukemia viruses. Cell 12:709-719. [DOI] [PubMed] [Google Scholar]
- 52.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]