Abstract
Protein S-nitrosothiols (PrSNOs) have been implicated in the pathophysiology of neuroinflammatory and neurodegenerative disorders. Although the metabolically instability of PrSNOs is well known, there is little understanding of the factors involved in the cleavage of S-NO linkage in intact cells. To address this issue, we conducted chase experiments in spinal cord slices incubated with S-nitrosoglutathione (GSNO). The results show that removal of GSNO leads to a rapid disappearance of PrSNOs (t1/2 ~ 2h), which is greatly accelerated when glutathione (GSH) levels are raised with the permeable analogue GSH ethyl ester. Moreover, PrSNOs are stable in the presence of the GSH depletor diethyl maleate, indicating that GSH is critical for protein denitrosylation. Inhibition of GSH-dependent enzymes (glutathione S-transferase, glutathione peroxidase and glutaredoxin) and enzymes that could mediate denitrosylation (alcohol dehydrogense-III, thioredoxin and protein disulfide isomerase) do not alter the rate of PrSNO decomposition. These findings and the lack of protein glutathionylation during the chase indicate that most proteins are denitrosylated via rapid transnitrosylation with GSH. The differences in the denitrosylation rate of individual proteins suggest the existence of additional structural factors in this process. This study is relevant to our recent discovery that PrSNOs accumulate in the CNS of patients with multiple sclerosis.
Keywords: nitric oxide, protein nitrosothiol, glutathione, spinal cord, denitrosylation
Introduction
Several neuroinflammatory (e.g. multiple sclerosis (MS)) and neurodegenerative disorders (e.g. Parkinson’s disease, Alzheirmer’s disease) are characterized by nitrosative stress, a condition caused by excessive production of nitric oxide (NO) by nitric oxide synthetases (NOS) (Duncan and Heales, 2005). One of the major consequences of nitrosative stress is the generation of protein (PrSNO) and non-protein nitrosothiols, which are believed to produce significant alterations in tissue function (Stamler, 1994; Broillet, 1999). To understand the consequences of protein S-nitros(yl)ation it is necessary not only to identify the protein targets and how function is altered upon incorporation of the NO moiety but also to know the lifetime of the S-nitrosothiols.
Utilizing rat spinal cord slices, we have recently identified S-nitrosoglutathione (GSNO) as the most likely physiological mediator of protein S-nitros(yl)ation (Romero and Bizzozero, 2006). The generation of PrSNOs via GSNO occurs by S-transnitrosation, a dynamic and reversible process in which the NO group from the glutathione thiol is transferred to a protein thiol (Romero and Bizzozero, 2006). These studies also showed that there is a restricted set of protein thiols (~10% of the total) that can be altered by NO. Using the biotin switch method we demonstrated that the major PrSNOs are those with molecular mass between 30 kDa and 120 kDa. These species are likely the same as to those previously identified in rat cerebellum homogenate incubated with GSNO such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH), creatine kinase, hexokinase, glycogen phosphorylase, NMDA-glutamate receptors, Na+/K+ ATPase α1/α2 subunits, neurofilament heavy chain, α/β-tubulin and β/γ–actin (Jaffrey et al., 2001; Hao et al., 2006).
The physiological or pathological impact of S-nitros(yl)ation depends on the ability of these proteins to retain the NO portion and thus preserve their altered function. Yet, little is presently known about of metabolic stability of PrSNOs and the molecular mechanisms underlying protein denitrosylation in intact cells. Three distinct enzyme systems seem to be responsible for the catabolism of GSNO: (1) alcohol dehydrogenase III (ADH-III) (Jensen et al., 1998), thioredoxin (Trxn) (Nikitovic and Holmgren, 1996) and protein disulfide isomerase (PDI) (Sliskovic et al., 2005). While ADH-III and PDI do not seem to act on PrSNOs, thioredoxin has been shown to catalyze the decomposition of PrSNOs with molecular mass of 23–30 kDa (Sengupta et al., 2007). However, it is still unclear what factors are mediating in the stability of PrSNOs with higher molecular masses, which indeed comprise the majority of the S-nitros(yl)ated species in the spinal cord.
The present study was undertaken to determine the factors involved in the cleavage of S-NO linkage in the nervous tissue. The results clearly show that GSH plays a key role in the stability of NO bound to protein thiols. Evidence is presented supporting the idea that protein denitrosylation in intact cells takes place non-enzymatically by rapid transnitrosation reactions with GSH. There are, however, differences in the denitrosylation rate of individual proteins suggesting the existence of additional structural factors. This study is relevant to CNS disorders characterized by excessive NO production including MS, Parkinson’s disease and stroke. A preliminary account of this work has been published in abstract form (Romero and Bizzozero, 2007).
Materials and Methods
Chemicals
Ascorbic acid, aurothioglucose (ATG), caffeic acid (CA), diethyl maleate (DEM), 5,5′-dithiobis(2-nitrobenzoic) acid (DTNB), L-glutathione (GSH), L-glutathione monoethyl ester (GSH-EE), mefenamic acid (MEF), mercaptosuccinic acid (MSA), methyl methanethiol sulfonate, 4-methylpyrazole (4-MP), phenylarsine oxide (PAO) and streptavidin-agarose were purchased from Sigma-Aldrich (St. Louis, MO). HPDP-biotin was from Pierce (Rockford, IL). N-ethylmaleimide was obtained from ICN biochemicals (Aurora, OH). Hanks’ balanced salt solution (HBSS) was acquired from Gibco-Invitrogen (Carlsbad, CA). Auranofin (AUR) was purchased from BioMol international (Plymouth Meeting, PA). All other chemicals were of the highest purity available. GSNO was prepared by incubating 40 mM GSH with 80 mM NaNO2 in 0.1 N HCl. After incubation in the dark for 2 min, samples were neutralized with 0.2 vol of 0.5 M Tris base and were immediately added to the incubation flasks.
Incubation of rat spinal cord slices
Forty-day old Sprague-Dawley male rats were used throughout. Housing and handling of the animals as well as the euthanasia procedure were in strict accordance with the NIH Guide for the Care and Use of Laboratory Animals, and were approved by the Institutional Animal Care and Use Committee. Animals were killed by decapitation, and spinal cords (SCs) were removed and sliced in sections ~400 μm-thick using surgical grade, carbon steel razor blades. Slices corresponding to ~25 mg of tissue were transferred to flasks containing 3 ml of HBSS supplemented with 10 mM D-glucose, and were incubated for up to 5 h at 37°C under 95% O2/5% CO2. Various drugs were added at the beginning or during the incubation period as indicated in the figure legends. After incubation, tissue sections were collected by low-speed centrifugation and were rinsed twice with 2 ml of ice-cold saline solution. Slices were homogenized by probe sonication in 0.5 ml of HEN buffer (50 mM Hepes pH 7.7, 1 mM EDTA and 0.1 mM neocuproine) to prevent chemical denitrosylation of proteins, and the homogenates were kept at −20°C until use. Protein concentration was assessed with the Bio-Rad DC™ protein assay (Bio-Rad Laboratories; Hercules, CA) using bovine serum albumin as standard.
Determination of protein (PrSHs) and non-protein thiols (NPrSHs)
Non-protein thiols (NPSHs), which are made mostly of GSH and small amount of cysteine and homocysteine, were determined with DTNB. Briefly, an aliquot of the tissue homogenate was mixed with an equal volume of 2% sulfosalicylic acid to precipitate the proteins. After centrifugation at 10,000 g for 15 min, the supernatants were mixed with 0.1 M sodium phosphate buffer pH 7.5 containing 0.3 mM DTNB, 10 mM EDTA and 1% SDS and incubated for 15 min at room temperature. The protein pellets were also dissolved in the same buffer to determine PrSHs. Absorbance was measured at 412 nm using a Hewlett-Packard 8452-A Diode Array Spectrophotometer. The amount of thiol groups was calculated using a molar extinction coefficient of 13,600 cm−1 for the thionitrobenzoate anion (Riddles et al., 1979).
Fluorometric determination of protein nitrosothiols (PrSNOs)
The concentration of PrSNOs in SC samples was assayed with a fluorometric technique (Park and Kostka, 1997). Briefly, aliquots corresponding to 100 μg of protein were precipitated with acetone at −20 °C. Suspensions were centrifuged at 10,000 g for 10 min, and the pellets were washed 4 times with acetone: H2O (4:1, v/v) to ensure the removal of residual GSNO and other low-molecular-weight nitrosothiols. The resulting pellets were dried under nitrogen, dissolved in 190 μl of 60 mM HCl containing 10 μM 2,3–diaminonaphthalene ± 0.2 mM HgCl2, and incubated at room temperature. After 10 min, 10 μl of 2.8 N NaOH were added to stabilize the fluorescent product 2,3-naphthotriazole. Fluorescence intensity was measured at 450 nm in a PerkinElmer LS 65 Luminescence Spectrometer using an excitation wavelength of 363 nm. Emission intensity was converted to PrSNO concentration using a calibration curve generated with increasing amounts of sodium nitrite.
Detection of S-nitros(yl)ated proteins on western blots
S-nitros(yl)ated proteins were identified using the Nitroglo™ nitrosylation detection kit (PerkinElmer Life Sciences, Boston, MA) following the manufacturer’s instructions. In brief, proteins (80 μg), dissolved in HEN buffer containing 2% SDS, were incubated with methyl methanethiosulfonate to block free SH groups. Thiol groups bound to NO were exposed with 3 mM ascorbic acid and titrated with HPDP-biotin in HEN buffer. Biotin-containing proteins were separated by sodium dodecyl sulfate - polyacrylamide gel electrophoresis (SDS-PAGE) and blotted to PVDF membranes. Proteins were then immunostained using a mouse monoclonal anti-biotin antibody (Sigma; 1:1000) and goat anti-mouse IgG conjugated to horseradish peroxidase (Sigma; 1:1000). Blots were developed by enhanced chemioluminescence (ECL). The creatine phosphokinase standard supplied with the Nitroglo™ kit was used as a positive control.
Pull-down of S-nitros(yl)ated proteins
SC proteins, dissolved in HEN buffer containing 0.7% SDS were incubated at room temperature for 2 h with 6 mM N-ethylmaleimide to bock free thiol groups. Excess NEM was removed by acetone precipitation. Proteins were re-dissolved in SDS-containing HEN buffer and incubated with 3 mM ascorbic acid and HPDP-biotin at room temperature for 1 h. HPDP-biotin was removed by acetone precipitation and proteins were diluted to 1 mg/ml in neutralization buffer (20 mM HEPES buffer pH 7.7 containing 100 mM NaCl, 1 mM EDTA, 0.1% SDS and 0.5% Triton X-100). Proteins were then incubated for 1 h at 20° C with 25 μl of streptavidin-agarose previously equilibrated in neutralization buffer. The resin was washed 5 times with neutralization buffer containing 600 mM NaCl, twice with neutralization buffer containing 1 M NaCl and once with neutralization buffer alone. Bound-proteins were eluted from the resin by incubation for 30 min at 37° C with SDS-sample buffer containing 1% 2-mercaptoethanol. Aliquots from the total and bound fractions were separated by SDS-PAGE on 10% polyacrylamide gels and blotted against PVDF membranes. Blots were probed with monoclonal antibodies (1:1000) against β-tubulin (Sigma), β-actin (Abcam Inc., Cambridge, MA), myelin proteolipid protein (PLP) (a gift from Dr. Vijay Kuchroo, HMS) and GAPDH (EnCor Biotechnology, Gainesville, FL) followed by incubation with the appropriate HRP-conjugated secondary antibody. Blots were developed by ECL as described above.
Statistical Analysis
Results were analyzed for statistical significance with Student’s unpaired t test or ANOVA utilizing GraphPad Prism® program (GraphPad Software Inc., San Diego, CA).
RESULTS
Protein nitrosothiols are metabolically unstable under physiological conditions
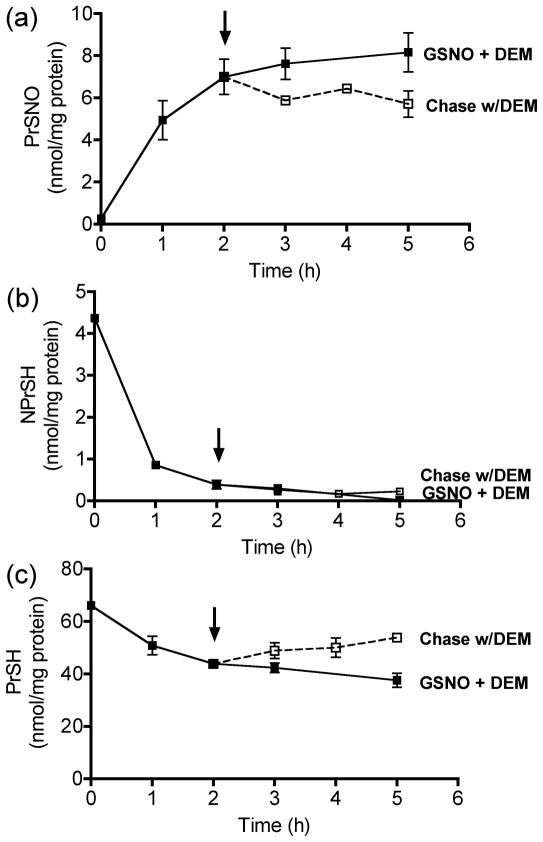
We have recently shown that there is a restricted amount of protein thiols (~10% of total) that is S-nitrosated upon incubation of rat spinal cord slices with GSNO (Romero and Bizzozero, 2006). In the present study we examined the biochemical stability of these PrSNOs utilizing classical chase experiments. To this end, rat SC slices were first incubated with 1 mM GSNO for 2 h, a period that is enough to reach steady-state levels of PrSNOs (5.5 ± 0.2 nmol/mg protein) (Fig. 1a). The medium was then replaced with fresh HBSS, and the slices were incubated for up to 3 h. As measured by fluorescence spectroscopy, PrSNOs levels after the 3h-chase were reduced by 66% (t1/2 ~ 2 h), demonstrating the dynamic nature of protein S-nitrosation. To gain insights into the molecular processes underlying denitrosylation, the concentration of NPrSH (mostly GSH) and PrSH was determined. As depicted in Fig. 1b, the amount of NPrSH steadily declined during the course of the incubation, reaching 40% of the original value after 5 h. This change might be due to decreased de novo synthesis of GSH and/or diminished NADPH-dependent reduction of glutathione disulfide. Interestingly, the levels of PrSH remained unchanged during the same period, indicating that low amounts of GSH are sufficient to keep the reduced status of protein thiols (Fig. 1c). In the presence of GSNO, the concentration of cellular NPrSH decreased at a faster rate than that in controls (Fig. 1b), while PrSH content diminished by 25% at 5 h (Fig. 1c). This indicates that protein thiols become both S-nitrosated and oxidized in the presence of GSNO. After 2 h of incubation with GSNO, there is a small reduction in the average value of PrSH, which seems to correlate with the amount of PrSNO formed. However, due to the small changes and relatively high errors in the assessment of PrSH levels, it is not possible to conclude whether or not denitrosylation is accompanied by an increase of free protein thiols.
Fig. 1.
Changes in the concentration of (a) PrSNO, (b) NPrSH and (c) PrSH after removal of GSNO from the incubation medium. Rat SC slices were incubated for up to 5 h in the absence (control) or presence of 1mM GSNO (+ GSNO). After 2h-incubation with GSNO some sections were rinsed with fresh HBSS solution and the incubation continued for up to 3 h (− GSNO). Slices were finally homogenized in HEN buffer and aliquots of the homogenate were used to determine PrSNO, NPrSH and PrSH as shown in “Materials and Methods”. Values represent the mean ± SEM of 3 separate experiments. Note that protein nitrosothiols are metabolically unstable (t1/2 ~ 2h) under physiological conditions.
Protein denitrosylation was also followed by western blotting after derivatization of protein bound nitrosothiols with biotin. Using this semiquantitative technique, we also found that the vast majority of PrSNOs disappeared after the 3 h-chase period (Fig. 2). However, a few S-nitrosated protein species with molecular mass between 46–69 kDa and 200 kDa were not completely denitrosylated during that period, indicating that denitrosylation of individual proteins does not occur at exactly the same rate.
Fig. 2.

Protein denitrosylation in SC sections. Rat SC slices were incubated with 1 mM GSNO for 2 h. Slices were rinsed in fresh HBSS and the incubation continued for 3 h. After incubation, slices were homogenized in HEN buffer and aliquots of the homogenate were derivatized with HPDP-biotin as described in “Materials and Methods”. PrSNOs were detected by western blotting using anti-biotin antibodies. The molecular weight markers are: myosin (200K), phosphorylase b (97.4K), bovine serum albumin (69K), ovalbumin (46K), carbonic anhydrase (30K), and trypsin inhibitor (21.5K). Note that, with exception of a few species, the majority of PrSNOs disappear after the 3h-chase.
Depletion of intracellular glutathione prevents protein denitrosylation
To determine if GSH is critical for protein denitrosylation, the GSH depletor diethyl maleate (DEM) was included in both the incubation with GSNO and the chase phase. DEM is an α,β-unsaturated dicarboxylic acid that conjugates to GSH irreversibly via a reaction catalyzed by glutathione-S-transferase (GST) (Buchmuller-Rouiller et al., 1995). As shown Fig. 3b, addition of 10 mM DEM reduced NPrSHs to almost undetectable levels within 2 h (the first time point measured) and they remained very low during the chase period. In the absence of NPrSHs, protein denitrosylation was completely arrested, suggesting that GSH plays a key role in the removal of NO from proteins (Fig. 3a and Fig. 4). This idea is further supported by the fact that the steady state level of PrSNOs in absence of GSH (8.2 ± 0.9 nmol/mg protein) (Fig. 3a) is higher than that in the presence of GSH (5.5 ± 0.2 nmol/mg) (Fig. 1a). It is important to note that the patterns of PrSNOs on western blot in the absence (Fig. 2, lane 1) or presence of DEM (Fig.4, lane 1) were identical, indicating that the higher levels of S-nitrosation attained in the absence of GSH is indeed due to an increase in steady-state levels of the nitroso species. The effect of increasing concentrations of DEM (1 μM – 10 mM) on protein denitrosylation and NPrSH concentration was also studied. As depicted in Fig. 5, there is an inverse relationship between NPrSH levels and amount of PrSNOs remaining after the chase giving further support to the notion that cellular concentration of GSH is the key determinant of nitrosothiol stability.
Fig. 3.
Effect of the GSH depletor diethyl maleate (DEM) on PrSNO stability. Rat SC slices were incubated with 1 mM GSNO plus 10 mM DEM for up to 5 h (GSNO + DEM). After 2h-incubation some sections were rinsed with fresh HBSS solution and the incubation continued for up to 3 h in the presence of 10 mM DEM (chase w/DEM). Slices were finally homogenized in HEN buffer and aliquots of the homogenate were used to determine the levels of (a) PrSNO, (b) NPrSH and (c) PrSH as described in “Materials and Methods”. Values represent the mean ± SEM of 3 separate experiments. Note that protein nitrosothiols are metabolically stable in the absence of GSH.
Fig. 4.
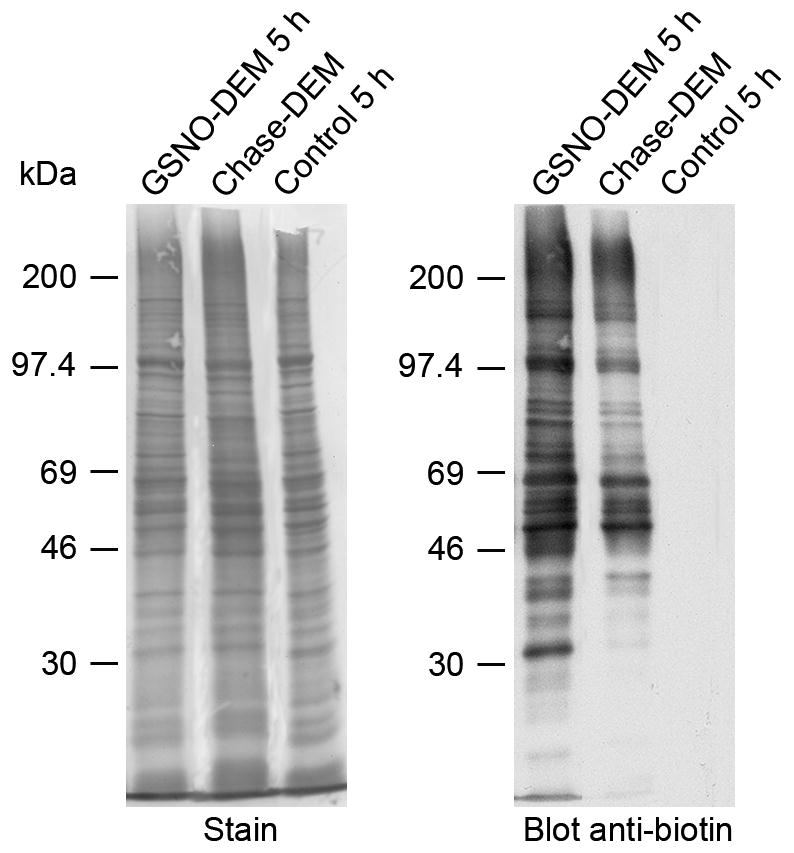
Protein denitrosylation in the absence of GSH. Rat SC slices were incubated with 1 mM GSNO plus 10 mM DEM for 2 h. Sections were then rinsed in fresh HBSS solution and the incubation continued for 3 h in the presence of 10 mM DEM. After incubation, slices were homogenized in HEN buffer and aliquots of the homogenate were derivatized with HPDP-biotin as described in “Materials and Methods”. PrSNOs were detected by western blotting using anti-biotin antibodies. All abbreviations are as in Fig. 2. Note that, with exception of a few species, the majority of PrSNOs are stable in the absence of GSH.
Fig. 5.
Rate of protein denitrosylation at various concentrations of DEM. Rat SC slices were incubated with 1 mM GSNO for 2 h. Sections were then rinsed in fresh HBSS and the incubation continued for 1 h in the presence of DEM (1 μM – 10 mM). After incubation, slices were homogenized in HEN buffer and aliquots of the homogenate were used to determine PrSNO and NPrSH as described in “Materials and Methods”. Values represent the mean ± SEM of 3 separate experiments. Note that there is an inverse relationship between residual [NPrSH] and the amount of PrSNOs after the chase period.
Replenishment of intracellular glutathione increases protein denitrosylation
The participation of GSH in PrSNO decomposition was confirmed using the membrane-permeable glutathione monoethylester (GSH-EE) (Anderson et al., 1985). Addition of 1mM GSH-EE to SC slices pretreated with GSNO/DEM rapidly increased NPrSH levels, reaching 70% and 90% of control values within 1 h and 2 h, respectively (Fig. 6b). Under these conditions PrSNO levels fell at a faster rate (t1/2 < 1h) (Fig 6a) than those in presence of endogenous GSH (t1/2 ~ 2h) (Fig.1a). This difference in denitrosylation rates is likely due to the fact that there is 2.5-times more GSH in the GSH-EE treated SC sections than in untreated slices. Thus, it can be concluded that the denitrosylation rate for the bulk of S-nitroso-proteins under normal concentrations of GSH is very fast. However, incubation with GSH-EE did not cause the total disappearance of PrSNOs, suggesting that some S-nitroso-species (~1.5 nmol/mg protein) are GSH-resistant. Moreover, replenishment of NPrSHs was not enough to bring the PrSH levels back to normal values (Fig. 6c), indicating that that some oxidized thiols are also metabolically stable and/or inaccessible to GSH during this period.
Fig. 6.
Effect of GSH ethyl ester (GSH-EE) on PrSNO stability. Rat SC slices were incubated with 1 mM GSNO plus 10 mM DEM for up to 5 h (GSNO + DEM). After 2h-incubation some sections were rinsed with fresh HBSS solution and the incubation continued for up to 3 h in the presence of 1 mM GSH-EE (chase w/GSH-EE). Slices were finally homogenized in HEN buffer and aliquots of the homogenate were used to determine the levels of (a) PrSNO, (b) NPrSH and (c) PrSH as described in “Materials and Methods”. Values represent the mean ± SEM of 3 separate experiments. Note that protein nitrosothiols are very unstable (t1/2 < 1h) upon replenishment of intracellular GSH.
Inhibition of GSH-dependent enzymes has no effect on protein denitrosylation
The above experiments indicate that denitrosylation of the majority of PrSNOs is mediated by GSH either alone or in combination with GSH-dependent enzymes. To discern between these possibilities, we tested the effect various inhibitors of GSH-dependent enzymes on the metabolic decay of PrSNOs (Fig. 7a). These drugs were used at concentrations at least 5-fold higher than those shown to produce complete enzyme inhibition in other cellular systems. The GST inhibitor caffeic acid (Ploemen et al., 1993) did not prevent the denitrosylation of proteins during the chase phase. Addition of mercaptosuccinic acid, one of the most potent Se-glutathione peroxidase (GPx) inhibitors (Chaudiere et al., 1984), did not reduce but rather increased the rate of protein denitrosylation. Glutaredoxin (Grxn) inhibition with 1 mM mefenamic acid (Park and Levine, 1996) had no effect on the half-life of PrSNOs. Together these experiments suggest that free GSH by itself is involved in the removal of NO moiety from proteins.
Fig. 7.
Effect of various enzyme inhibitors of protein denitrosylation. Rat SC slices were incubated with 1 mM GSNO for 2 h. Sections were then rinsed in fresh HBSS solution and the incubation continued for 3 h in the absence or presence of various enzyme inhibitors. After incubation, slices were homogenized in HEN buffer and aliquots of the homogenate were used to determine PrSNOs as shown in “Materials and Methods”. Values represent the mean ± SEM of 3 separate experiments. *Significantly different (p < 0.05) from 2h-GSNO, #Significantly different (p < 0.05) from 3h-chase done in the absence of inhibitors (none). The rate of denitrosylation does not decrease upon inhibition of GST, GPx and Grxn with caffeic acid (CA), mercaptosuccinate (MA) and mefenamic acid (MEF), respectively (panel a). Likewise, the rate of denitrosylation is unaffected by inhibition of ADH-III, Trxn and PDI with 4-methylpyrazole (4-MP), the gold compounds aurothioglucose (ATG) and auranofin (AUR), and phenylarsine oxide (PAO), respectively (panel b).
Inhibition of ADH-III, Trxn reductase and PDI does not affect protein denitosylation
Three GSH-independent enzymes (ADH-III, Trxn and PDI) have been proposed to mediate the denitrosylation of GSNO and could potentially catalyze the removal of NO from S-nitrosoproteins as well. As shown in Fig. 7b, inhibition of ADH-III with 4-methylpyrazole (Delker et al., 1998) had no significant effect on the metabolic decay of PrSNOs. Also, the Trxn reductase inhibitors aurothioglucose and auranofin (Gromer et al., 1998) did not decrease the rate of denitrosylation of SC proteins. Likewise, inhibition of PDI with the dithiol blocking agent phenylarsine oxide (Root et al., 2004) did not prevent the disappearance of PrSNO during the chase period.
Rate of denitrosylation of specific S-nitrosoproteins
To ascertain possible differences in denitrosylation rates between individual S-nitrosoproteins, the levels of 4 distinct S-nitrosoproteins (β-actin, β-tubulin, GAPDH and PLP) were measured before and after the chase period. Quantification of the extent of S-nitrosation of these species was performed using a pull-down/western blot procedure (Jaffrey et al., 2001). To this end, S-nitrosocysteine residues were first converted into biotinylated residues by reaction with HPDP-biotin. Biotin-containing proteins were then isolated with streptavidin-agarose and analyzed by western blotting employing antibodies against each the proteins of interest. As shown in Fig. 8, all four proteins are targets of S-nitrosation, being S-nitroso-GAPDH and S-nitroso-tubulin the most abundant. After the 3 h-chase in the presence of GSH-EE, the amount of all four all S-nitrosoproteins decreased albeit at different rates. While S-nitroso-PLP was almost completely denitrosylated in 3h, the levels of S-nitroso-actin, S-nitroso-GAPDH and S-nitroso-tubulin during the same period diminished by 42%, 41% and 19%, respectively.
Fig. 8.
Rate of denitrosylation of specific S-nitrosoproteins. Rat SC slices were incubated with 1 mM GSNO plus 10 mM DEM for 2 h. Sections were then rinsed in fresh HBSS solution and the incubation continued for 3 h in the presence of 1 mM GSH-EE. After incubation, proteins were derivatized with HPDP-biotin and biotinylated proteins were isolated with streptavidin-agarose as described under “Materials and Methods”. Aliquots from the total (T) and bound (B) fractions were analyzed by western blotting using various monoclonal antibodies. Values are expressed as the percentage of protein that is S-nitrosated and represent the mean ± SEM of 3 separate experiments. *Significantly different (p < 0.05) from 2h-GSNO, # Significantly different (p < 0.05) from untreated controls. Note that the denitrosylation rate varies among the different S-nitrosoproteins. For instance, while PLP is completely denitrosylated in the 3h-chase, a significant amount of S-nitrosotubulin remains intact.
Discussion
In this study we have shown that protein S-denitrosylation in spinal cord sections depends almost exclusively on the intracellular concentration of GSH. This conclusion is based on the following observations: (1) there is an inverse relationship between GSH levels and the amount of PrSNOs remaining after the chase; (2) GSH depletion with DEM prevents PrSNO denitrosylation while minimally affecting the levels of PrSHs; (3) addition of the membrane-permeable GSH-EE accelerates the rate of protein denitrosylation; (4) inhibition of GSH-dependent enzymes (GPx, GST and Grxn) has no effect of protein denitrosylation; and (5) enzymes involved in GSNO decomposition (ADH-III and PDI) and/or protein denitrosylation in other systems (Trxn) do not affect PrSNO stability in the SC cord slices.
We have previously demonstrated that, due to its chemical stability, extracellular GSNO is more effective than either N2O3 or S-nitrosocysteine to generate PrSNOs in SC slices (Romero and Bizzozero, 2006). In that study we also presented experimental evidence that intact GSNO is able to permeate into cells without the need of putative cell surface transporters like the L-amino acid transporter (Li and Whorton, 2005) or PDI (Ramachandran et al., 2001), and S-transnitrosate proteins directly. We now show that GSH plays a key role in protein denitrosylation process in intact cells. There are two mechanisms by which GSH can remove the NO moiety from PrSNO. One involves a transnitrosation reaction between the glutathione thiol and the protein thiol (Eq. 1).
| (1) |
Alternatively reduced GSH could react with the protein nitrosothiol to form a protein-glutathione mixed disulfide (i.e. glutathionylated protein) (Eq. 2).
| (2) |
While our data do not completely allow us to distinguish between these two processes, transnitrosation reactions (Eq. 1) are generally favored over thiolation reactions (Eq. 2) (Giustarini et al., 2005). It has been suggested that the nitrogen atom in the S-N bond is more positive than the sulfur atom, and thus it is more favorable to nucleophilic attack by the thiolate in GSH (Konorev et al., 2000). In preliminary studies we have found that PrSSG levels in GSNO-treated SC slices (determined with the GSH recycling method (Kleinman et al., 2003), never surpass 1 nmol/mg protein and that there is no difference before and after the chase period (data not shown), suggesting that transnitrosation is the main mechanism underlying protein denitrosylation. This, however, does not rule out that a minor number of proteins are being denitrosylated by glutathionylation as shown in other systems (West et al., 2006). It should be noted, however, that the possibility that protein thiols mediate NO removal from S-nitrosoproteins by forming intra- or intermolecular disulfides (Eq. 3) is unlikely since denitrosylation is abolished when GSH is depleted with DEM, a condition that marginally reduces PrSH.
| (3) |
Utilizing a number of selective inhibitors, we have also excluded the possible role of GSH-dependent (GST, GPx, Grxn) and GSH-independent enzymes (ADH-III, PDI, Trxn) in denitrosylation of the bulk of cellular proteins. Thus, it is fair to conclude that NO is non-enzymatically exchanged between PrSHs and GSH (Eq. 1). In cell-free system, non-enzymatic denitrosation of proteins with GSH is very rapid (t1/2= 2 sec-10 min) (Giustarini et al., 2005). These rates are significantly higher that those obtained in our ex-vivo system. The incomplete removal of GSNO from the cells during the chase with the slow return to a high GSH/GSNO ratio is the most likely reason for that apparent discrepancy. In fact, the rate of total protein denitrosylation in the presence of GSH-EE, a condition that rapidly restores the levels of NPSHs, was significantly faster than in its absence (Fig. 6). In addition, some nitrosothiols may be more stable due to their location inside hydrophobic protein pockets that could be less accessible to GSH. Indeed, we have found that ~ 1.5 nmol of PrSNO/mg protein remains after 3 h of chase period even in the presence of GSH-EE, and that there is a considerable variation in denitrosylation rates between individual proteins (Fig. 8). Another possibility is that NO moiety may be transported between protein thiols before being ultimately transferred to GSH. The idea that most PrSNOs and GSH are in rapid equilibrium with GSNO and PrSHs has several important implications. First, the known effectiveness of GSH at reducing the generation of PrSNOs is probably due to an increase in the rate of denitrosylation rather than to the purported ability of the tripeptide to intercept free NO released from GSNO (He et al., 2007). Second, low GSH concentrations could make cells more susceptible to NO-mediated toxicity as shown in several systems (Rodríguez-Martín et al., 2002; Fass et al., 2004; Che et al., 2005). Third, physiologically significant steady state levels of PrSNOs can only be achieved when the reducing potential of the cell is severely impaired (Beltran et al., 2000). However, this does not rule out that transient S-nitros(yl)ation of proteins may have considerable physiological consequences.
The S-nitros(yl)ation of specific cysteine residues in the four proteins that we have investigated causes significant alterations in their function. For example, S-nitroso-actin polymerizes less efficiently than unmodified actin as a consequence of reduced annealing rate (Dalle-Donne et al., 2000). Likewise, GSNO-induced S-nitrosation and thiolation on tubulin affect its polymerization rate (Landino et al., 2007). The activity of glycolytic enzyme GAPDH is greatly attenuated after nitrosylation of its critical thiol (cys-149) at the active site (Padgett and Whorton, 1995; Mohr et al., 1996). It has been recently shown that GADPH S-nitros(yl)ation also induces its binding to the E3 ubiquitin ligase Siah1 to cause nuclear translocation and to promote apoptosis (Hara et al., 2006). S-nitros(yl)ation of PLP has been linked to decompaction of CNS myelin at the level of the intraperiod line where this protein plays an adhesive role (Bizzozero et al., 2004).
Our interest on protein S-nitros(yl)ation arose from the observation that PrSNOs with molecular masses between 45–200 kDa accumulate in the brain white matter of MS patients (Bizzozero et al., 2005). Both MS and its animal model experimental allergic encephalomyelitis (EAE) are characterized by severe nitrosative stress due to augmented iNOS (NOS-2) expression (Bo et al., 1994; Lin et al., 1993). The occurrence of nitrosothiols in MS was first suggested by Boullerne et al. (1995) who found elevated levels of antibodies to a conjugated S-nitroso-cysteine epitope in sera and cerebrospinal fluid (CSF) from MS patients, a finding later extended to EAE animals (Boullerne et al., 2002). Calabrese et al. (2003) also reported increased concentration of both nitric oxide metabolites and low-molecular-weight nitrosothiols (probably GSNO) in CSF from patients with active MS. Thus, our finding that significant amounts of PrSNOs (0.3 nmol/mg protein) accumulate in the MS white matter is in agreement with these other reports. Levels of GSH are diminished in the white matter of MS patients and this may have contributed to the stability of the PrSNO in the specimens analyzed. Also the S-nitroso species that accumulate in the diseased tissue may be those with relatively long half-lives. Identification of the S-nitros(yl)ated species and knowledge of both the extent of nitrosylation and the metabolic stability are necessary to fully understand the functional consequences of this widespread posttranslational protein modification.
Acknowledgments
This work was supported by PHHS grant NS 47448 from the National Institutes of Health.
Footnotes
As most investigators in the field of nitric oxide, we use the term S-nitrosation to indicate the addition of an NO+ equivalent (nitrosonium) to a thiolate anion (S−) and the term S-nitrosylation to denote the attachment of an NO radical to a thiyl radical (RS·). Also, as recommended by Bryan et al. (2004), we employ the chimera “S-nitros(yl)ation” when the mechanism RSNO formation is either unknown or includes both pathways.
References
- Anderson ME, Powrie F, Puri RN, Meister A. Glutathione monoethyl ester: preparation, uptake by tissues, and conversion to glutathione. Arch Biochem Biophys. 1985;239:538–548. doi: 10.1016/0003-9861(85)90723-4. [DOI] [PubMed] [Google Scholar]
- Beltran B, Orsi A, Clementi E, Moncada S. Oxidative stress and S-nitrosylation of proteins in cells. Br J Pharmacol. 2000;129:953–960. doi: 10.1038/sj.bjp.0703147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzozero OA, DeJesus G, Howard T. Exposure of rat optic nerves to nitric oxide causes protein S-nitrosation and myelin decompaction. Neurochem Res. 2004;29:1675–1685. doi: 10.1023/b:nere.0000035802.27087.16. [DOI] [PubMed] [Google Scholar]
- Bizzozero OA, DeJesus G, Bixler HA, Pastuszyn A. Evidence of nitrosative damage in the brain white matter of patients with multiple sclerosis. Neurochem Res. 2005;30:139–149. doi: 10.1007/s11064-004-9695-2. [DOI] [PubMed] [Google Scholar]
- Bo L, Dawson T, Wasswlingh S, Mork S, Choi S, Kong PA, Hanley D, Trapp BD. Induction of nitric oxide synthetase in demyelinating regions of MS brains. Ann Neurol. 1994;36:778–786. doi: 10.1002/ana.410360515. [DOI] [PubMed] [Google Scholar]
- Boullerne AI, Petry K, Meynard M, Geffard M. Indirect evidence for nitric oxide involvement in multiple sclerosis by characterization of circulating antibodies directed against conjugated S-nitrosocysteine. J Neuroimmunol. 1995;60:117–124. doi: 10.1016/0165-5728(95)00061-6. [DOI] [PubMed] [Google Scholar]
- Boullerne AI, Rodriguez JJ, Touil T, Brochet B, Schmidt S, Abrous ND, Le Moal M, Pua JR, Jensen MA, Mayo W, Arnason BG, Petry KG. Anti-S-nitrosocysteine antibodies are a predictive marker for demyelination in experimental autoimmune encephalomyelitis: implications for multiple sclerosis. J Neurosci. 2002;22:123–132. doi: 10.1523/JNEUROSCI.22-01-00123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broillet MC. S-nitrosylation of proteins. Cell Mol Life Sci. 1999;55:1036–1042. doi: 10.1007/s000180050354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan NS, Rassaf T, Maloney RE, Rodriguez CM, Saijo F, Rodriguez JR, Feelisch M. Cellular targets and mechanisms of nitros(yl)ation: an insight into their nature and kinetics in vivo. Proc Natl Acad Sci USA. 2004;101:4308–4313. doi: 10.1073/pnas.0306706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmuller-Rouiller Y, Corrandin SB, Smith J, Schneider P, Ransijn A, Jongeneel CV, Mauel J. Role of glutathione in macrophage activation: effect of cellular glutathione depletion on nitrite production and leishmanicidal activity. Cell Immunol. 1995;164:73–80. doi: 10.1006/cimm.1995.1144. [DOI] [PubMed] [Google Scholar]
- Calabrese V, Scapagnini G, Ravagna A, Bella R, Butterfield DA, Calvani M, Pennisi G, Giuffrida-Stella AM. Disruption of thiol homeostasis and nitrosative stress in the cerebrospinal fluid of patients with active multiple sclerosis: evidence for a protective role of acetylcarnitine. Neurochem Res. 2003;28:1321–1328. doi: 10.1023/a:1024984013069. [DOI] [PubMed] [Google Scholar]
- Chaudiere J, Wilhelmsen EC, Tappel AL. Mechanism of selenium-glutathione peroxidase and its inhibition by mercaptocarboxylic acids and other mercaptans. J Biol Chem. 1984;259:1043–1050. [PubMed] [Google Scholar]
- Che T, Pearce LL, Peterson J, Stoyanowsky D, Billiar TR. Glutathione depletion renders rat hepatocytes sensitive to nitric oxide donor-mediated toxicity. Hepathology. 2005;42:598–607. doi: 10.1002/hep.20813. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Milzani A, Giustarini D, Di Simplicio P, Colombo R, Rossi R. S-NO-actin: S-nitrosylation kinetics and the effect on isolated vascular smooth muscle. J Muscle Res Cell Motil. 2000;21:171–81. doi: 10.1023/a:1005671319604. [DOI] [PubMed] [Google Scholar]
- Delker DA, McKnight S, Rosenberg DW. The role of alcohol dehydrogenase in the metabolism of the colon carcinogen methylazoxymethanol. Toxicol Sci. 1998;45:66–71. doi: 10.1006/toxs.1998.2499. [DOI] [PubMed] [Google Scholar]
- Duncan AJ, Heales SJ. Nitric oxide and neurological disorders. Mol Aspects Med. 2005;26:67–96. doi: 10.1016/j.mam.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Fass U, Panickar K, Williams K, Nevels K, Personett D, McKinney M. The role of glutathione in nitric oxide donor toxicity to SN56 cholinergic neuron-like cells. Brain Res. 2004;1005:90–100. doi: 10.1016/j.brainres.2004.01.046. [DOI] [PubMed] [Google Scholar]
- Giustarini D, Milzani A, Aldini G, Carini M, Rossi R, Dalle-Done I. S-nitrosation versus S-glutathionylation of protein sulfhydryl groups by S-nitrosoglutathione. Antioxid Redox Signal. 2005;7:930–939. doi: 10.1089/ars.2005.7.930. [DOI] [PubMed] [Google Scholar]
- Gromer S, Arscott LD, Williams CH, Schirmer RH, Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J Biol Chem. 1998;273:20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- Hao G, Derekhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Snyder SH. Nitric oxide-GAPDH-Siah: a novel cell death cascade. Cell Mol Neurobiol. 2006;26:527–538. doi: 10.1007/s10571-006-9011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Wang T, Wang P, Han P, Yin Q, Chen C. A novel mechanism underlying the susceptibility of neuronal cells to nitric oxide: the occurrence and regulation of protein S-nitrosylation is the checkpoint. J Neurochem. 2007;102:1863–1874. doi: 10.1111/j.1471-4159.2007.04651.x. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris H, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Jensen DE, Belka GK, Du Bois GC. S-Nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III. Biochem J. 1998;131:659–668. doi: 10.1042/bj3310659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman WA, Komninou D, Leutzinger Y, Colosimo S, Cox J, Lang CA, Richie JP. Protein glutathiolation in human blood. Biochem Pharmacol. 2003;65:741–746. doi: 10.1016/s0006-2952(02)01560-5. [DOI] [PubMed] [Google Scholar]
- Konorev EA, Kalyanrsman B, Hogg N. Modification of creatine kinase by S-nitrosothiols: S-nitrosylation vs S-thiolation. Free Rad Biol Med. 2000;28:1671–1678. doi: 10.1016/s0891-5849(00)00281-1. [DOI] [PubMed] [Google Scholar]
- Landino LM, Koumas MT, Mason CE, Alston JA. Modification of tubulin cysteines by nitric oxide and nitroxyl donors alters tubulin polymerization activity. Chem Res Toxicol. 2007;20:1693–1700. doi: 10.1021/tx7001492. [DOI] [PubMed] [Google Scholar]
- Li S, Whorton AR. Identification of stereoselective transporters for S-nitroso-L-cysteine: role of LAT1 and LAT2 in biological activity of S-nitrosothiols. J Biol Chem. 2005;280:20102–20110. doi: 10.1074/jbc.M413164200. [DOI] [PubMed] [Google Scholar]
- Lin R, Lin T, Tilton RD, Cross A. Nitric oxide localized to spinal cords of mice with experimental allergic encephalomyelitis: an electron paramagnetic resonance study. J Exp Med. 1993;178:643–648. doi: 10.1084/jem.178.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Stamler JS, Brüne B. Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J Biol Chem. 1996;271:4209–4214. doi: 10.1074/jbc.271.8.4209. [DOI] [PubMed] [Google Scholar]
- Nikitovic D, Holmgren A. S-Nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J Biol Chem. 1996;271:19180–19185. doi: 10.1074/jbc.271.32.19180. [DOI] [PubMed] [Google Scholar]
- Padgett CM, Whorton AR. S-nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am J Physiol. 1995;269:C739–C749. doi: 10.1152/ajpcell.1995.269.3.C739. [DOI] [PubMed] [Google Scholar]
- Park JK, Kostka P. Fluorometric detection of biological S-nitrosothiols. Anal Biochem. 1997;249:61–66. doi: 10.1006/abio.1997.2159. [DOI] [PubMed] [Google Scholar]
- Park JB, Levine M. Purification, cloning and expression of dehydroascorbic acid-reducing activity from human neutrophils: identification as glutaredoxin. Biochem J. 1996;315:931–938. doi: 10.1042/bj3150931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploemen JH, van Ommen B, de Haan A, Schefferlie JG, van Bladeren PJ. In vitro and in vivo reversible and irreversible inhibition of rat glutathione S-transferase isoenzymes by caffeic acid and its 2-S-glutathionyl conjugate. Food Chem Toxicol. 1993;31:475–482. doi: 10.1016/0278-6915(93)90106-9. [DOI] [PubMed] [Google Scholar]
- Ramachandran N, Root P, Jiang XM, Hogg PJ, Mutus B. Mechanism of transfer of NO from extracellular S-nitrosothiols into the cytosol by cell-surface protein disulfide isomerase. Proc Natl Acad Sci USA. 2001;98:9539–9544. doi: 10.1073/pnas.171180998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddles PW, Blakely RL, Zerner B. Ellman’s reagent: 5,5′-dithiobis(2-nitrobenzoic acid)-a reexamination. Anal Biochem. 1979;94:75–81. doi: 10.1016/0003-2697(79)90792-9. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Martín E, Casarejos MJ, Canals S, de Bernardo S, Mena MA. Thiolic antioxidants protect from nitric oxide-induced toxicity in fetal midbrain cultures. Neuropharmacol. 2002;43:877–888. doi: 10.1016/s0028-3908(02)00150-8. [DOI] [PubMed] [Google Scholar]
- Romero JM, Bizzozero OA. Extracellular S-nitrosoglutathione, but not S-nitrosocysteine or N2O3, mediates protein S-nitrosation in rat spinal cord slices. J Neurochem. 2006;99:1299–1310. doi: 10.1111/j.1471-4159.2006.04180.x. [DOI] [PubMed] [Google Scholar]
- Romero JM, Bizzozero OA. Reduced glutathione is required for denitrosylation of CNS proteins. J Neurochem. 2007;102(Suppl 1):P-344. [Google Scholar]
- Root P, Sliskovic I, Mutus B. Platelet cell-surface protein disulphide-isomerase mediated S-nitrosoglutathione consumption. Biochem J. 2004;382:575–580. doi: 10.1042/BJ20040759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta R, Ryter SW, Zuckerbraun BS, Tzeng E, Billiar TR, Stoyanovsky DA. Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry. 2007;46:8472–8483. doi: 10.1021/bi700449x. [DOI] [PubMed] [Google Scholar]
- Sliskovic I, Raturi A, Mutus B. Characterization of the S-denitrosation activity of protein disulfide isomerase. J Biol Chem. 2005;280:8733–8741. doi: 10.1074/jbc.M408080200. [DOI] [PubMed] [Google Scholar]
- Stamler JS. Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- West MB, Hill BG, Xuan Y, Bhatnagar A. Protein glutathionylation by nitric oxide: an intracellular mechanism regulating redox protein modification. FASEB J. 2006;20:E1049–E1060. doi: 10.1096/fj.06-5843fje. [DOI] [PubMed] [Google Scholar]