Abstract
Within the vertebrate nervous system, myelination is required for the normal function of neurons by facilitating the rapid conduction of action potentials along axons. Oligodendrocytes are glial cells which myelinate axons in the central nervous system. Disruption of myelination and remyelination failure can cause human diseases such as multiple sclerosis. Despite the importance of myelination, the molecular basis of oligodendrocyte differentiation and myelination are still poorly understood. To understand the molecular mechanisms which regulate oligodendrocyte differentiation and myelination, novel genes were identified using a microarray analysis. The analysis used oligodendrocyte lineage cells isolated from transgenic zebrafish expressing fluorescent proteins in the oligodendrocyte lineage cells. Seven genes not previously known to be involved in oligodendrocyte differentiation were identified, and their expression during oligodendrocyte development was validated.
Keywords: oligodendrocyte, differentiation, myelination, microarray, zebrafish
INTRODUCTION
The insulation of axons in the vertebrate nervous system by myelin is essential for efficient axonal conduction, and is thus required for the normal function of neurons. Oligodendrocytes are glial cells which myelinate axons in the central nervous system (CNS), and are generated from oligodendrocyte progenitor cells (OPCs). In the developing spinal cord, OPCs are generated from the pMN precursor domain of the ventral spinal cord. OPCs express olig2, a basic helix-loop-helix transcription factor (Park et al., 2002b; Zhou and Anderson, 2002), and differentiate into mature oligodendrocytes after migration into the white matter of the spinal cord. The differentiation of oligodendrocytes and the myelination of axons is a highly regulated process controlled by a number of mechanisms, including extrinsic signaling pathways and intrinsic machinery.
In the CNS, myelination disruption and remyelination failure can cause demyelinating human diseases, such as multiple sclerosis (MS). Primary demyelination usually occurs by a direct insult to the oligodendrocytes, and secondary demyelination occurs as a consequence of primary axonal loss (Emery, 2010). Remyelination is the process by which new myelin sheaths are restored to demyelinated axons, enabling them to recover lost function. Although the vertebrate CNS has a very limited regeneration capacity, remyelination generally occurs efficiently. However, despite the efficiency of remyelination in experimental models and in some clinical diseases, remyelination is often inadequate in MS, the most common primary demyelinating disease (Franklin, 2002). In MS, the major cause of primary demyelination is inflammatory damage to myelin and oligodendrocytes, but the reason for the remyelination failure is largely unknown.
To understand the molecular mechanisms which control oligodendrocyte differentiation and myelination, the RNAs expressed by oligodendrocyte lineage cells in culture have been profiled using microarray technology (Dugas et al., 2006; Cahoy et al., 2008; Chen et al., 2009). These studies have revealed distinct classes of RNAs that are differentially expressed during oligodendrocyte differentiation and have led to the identification of new factors such as Gpr17, which encodes a G protein-coupled receptor that controls the timing of oligodendrocyte differentiation (Chen et al., 2009), and myelin gene regulatory factor (Mrf), which encodes a transcription factor necessary for expression of myelin genes (Cahoy et al., 2008). However, screening for genes differentially expressed in oligodendrocyte lineage cells using cultured oligodendrocyte lineage cells has limited potential to isolate genes responsible for oligodendrocyte differentiation and myelination because these processes are also controlled by extracellular ligands and molecules secreted from neurons, such as Jagged, PSA-NCAM, and LINGO-1 (Wang et al., 1998; Charles et al., 2000; Mi et al., 2005). Here, a microarray screening method was used to isolate genes involved in oligodendrocyte differentiation using oligodendrocyte lineage cells sorted from transgenic zebrafish. Oligodendrocyte lineage cells expressing enhanced green fluorescent protein (EGFP) were isolated by fluorescence activated cell sorting (FACS), and the gene expression profiles of undifferentiated OPCs and differentiated oligodendrocytes were compared. Seven genes not previously known to be involved in oligodendrocyte differentiation were identified, and their expression during oligodendrocyte development was validated.
MATERIALS AND METHODS
Animals
Zebrafish embryos used in this study were wild-type AB line, Tg (olig2:egfp) (Shin et al., 2003), Tg(sox10:egfp) (Carney et al., 2006) and raised at 28.5℃ in egg water or embryo medium (15 mM NaCl, 0.5 mM KCl, 1 mM CaCl2, 1 mM MgSO4, 0.15 mM KH2PO4, 0.05 mM NH2PO4, 0.7 mM NaHCO3, EM). Embryos were staged according to days post fertilization (dpf) and morphological criteria (Kimmel et al., 1995).
Trichostatin A treatment
Embryos were dechorionated and incubated in EM with histone-deacetylase inhibitor Trichostatin A (Tsai et al., 2006) at a concentration of 100 ng/ml.
Cell dissociation for flow cytometry
Embryos were raised until appropriate stage and anesthetized with ethyl 3-aminobenzoate methanesulfonic acid on ice. A thousand embryos were chopped into several pieces and transferred into 1.5 ml eppendorf tube. Collected samples were rinsed with calcium-, magnesium-free Ringers solution for 20 min, then washed with Dulbecco's phosphate buffered saline (D-PBS) (GIBCO) three times. Tissue was digested with 100 µl of D-PBS containing 35 µl of Liberase Blendzyme 3, followed by incubation at 29℃ for 15 min with occasional mixing. After incubation, 1 ml of trypsin (GIBCO) was added and incubated at 29℃ for 15 min until complete dissociation. To inactivate trypsin, we added 2 ml of D-PBS containing 10% fetal bovine serum (FBS) and filtered the solution through a cell strainer (BD Falcon) to remove undissociated cell clusters. We collected dissociated cells by centrifugation at 300 g at 4℃ for 5 min and rinsed twice with D-PBS containing 10% FBS. Dissociated cells were collected by centrifugation and resuspended in PBS containing 1 mM EDTA, 25 mM HEPES (pH 7.0) and 1% FBS. Cell suspensions were separated into GFP+ and GFP- cells by BD FACSAria II (Becton Dickinson) performed at room temperature under sterile conditions. Isolated GFP+ and GFP- cells were collected in D-PBS/10% FBS pre-coated tubes with RNase inhibitor.
RNA extraction
Isolated cells were collected by low-speed centrifugation and homogenized in TRIzol solution (Invitrogen) subsequently. Total RNA extraction was performed as described previously (Peterson and Freeman, 2009). Isolated total RNA was cleaned up with RNeasy midi kit (Quiagen) and qualified as a microarray target samples using Bioanalyzer 2100 system (Agilent) and NanoDrop spectrophotometer.
Hybridization of agilent gene chips
1 µg of qualified total RNA was amplified and labeled with Cy3, Cy5 dyes. Hybridization was performed with Zebrafish Agilent Gene Expression Microarray Chips (Agilent), followed by washing step. Hybridized chips were then scanned, normalized and analyzed.
Semi-quantitative and quantitative RT-PCR
cDNA was synthesized using ImProm-II reverse transcription system (Promega). The quantitative real-time PCR was performed using the Lightcycler system (Roche Applied Science) with the Lightcycler-FastStart DNA Master SYBR Green I (Roche Applied Science) according to the manufacturer's instructions. Each reaction was performed in a volume of 20 µl with final concentration of 1×Lightcycler-FastStart DNA Master SYBR Green 1, 3 mM MgCl2, 0.5 µM of each primer and 2 µl of first strand cDNA mixture. Quantitative real-time PCR was carried out for 40 cycles and fluorescence readings were acquired at the end of each amplification cycle at 72℃. Melting curve analysis was performed with continuous fluorescence acquisition from 65 to 95℃ at a temperature transition rate of 0.1℃/s to determine the amplification specificity. All reactions were performed as technical duplicates.
Immunohistochemistry
Embryos were anesthetized until movement had ceased and fixed in 4% paraformaldehyde overnight. Fixed embryos were embedded in 1.5% agar with 5% sucrose blocks and equilibrated by 30% sucrose solution. Frozen blocks were sliced into 10 µm sections on glass slides using cryostat microtome. Sections were rinsed with PBS several times and then blocked in 2% bovine serum albumin with sheep serum. Embryos were processed for immunohistochemistry, following primary antibodies were used: rabbit anti-Sox10 (1 : 250), rabbit anti-MBP (1 : 100), rabbit anti-GFP (1 : 500, Abcam) and mouse anti-HuC/D (16A11 1 : 20, Molecular Probes, Eugene, OR). For fluorescent detection of antibody labeling, we used Alexa Fluor 568-conjugated goat anti-mouse IgG, Alexa Fluor 568-conjugated goat anti-rabbit IgG, and Alexa Fluor 488-conjugated goat anti-rabbit IgG (1 : 500, Molecular Probes). Photos were taken using a confocal lasers canning microscope (LSM 510 Pascal, Carl Zeiss).
RESULTS AND DISCUSSION
Identification of EGFP expression in the oligodendrocyte lineage cells of transgenic zebrafish
To identify the genes involved in oligodendrocyte differentiation and myelination, we isolated OPCs and differentiated oligodendrocytes expressing EGFP from the transgenic zebrafish and compared their transcriptional profiles. Therefore, the embryo stages suitable for collecting OPCs and differentiated oligodendrocytes were first identified. Previously, we have shown that olig2-expressing precursor cells in the pMN precursor domain of the ventral spinal cord first generate motor neurons during neurogenesis and later produce migrating OPCs, which express olig2 continuously to the mature oligodendrocyte stage (Park et al., 2002a; Park and Appel, 2003; Park et al., 2004). As soon as OPCs are specified in the pMN precursor domain, sox10 starts to be expressed specifically in the oligodendrocyte lineage cells. Larvae carrying the Tg(olig2:egfp) and Tg(sox10:egfp) transgenic reporters, which express EGFP under the control of the olig2 and sox10 promoters (Shin et al., 2003; Carney et al., 2006), respectively, were examined for use in the transcriptome analysis.
Expression of EGFP in OPCs and mature oligodendrocytes at several developmental stages in the Tg(olig2:egfp) and Tg(sox10: egfp) larvae was verified by fluorescence microscopy using transverse sections of the spinal cord. By 2 days post-fertilization (dpf), sox10:EGFP+ cells are located in the gray matter of the spinal cord, in which Hu+ neurons are located, indicating that sox10:EGFP+ cells are undifferentiated OPCs (Fig. 1A, arrow-heads). By 5 dpf, sox10:EGFP+ OPCs have migrated to the white matter of the spinal cord and differentiated into mature oligodendrocytes with multiple processes (Fig. 1B, arrows). Labeling of the spinal cord of Tg(olig2:egfp) larvae with anti-Sox10, a marker for OPCs and mature oligodendrocytes, revealed that Sox10+, olig2:EGFP+ cells are located in the gray matter of ventral spinal cord at 2 dpf, indicating that these cells are undifferentiated OPCs (Fig. 1C, arrowheads). Sox10+, olig2:EGFP+ cells migrated to the white matter and differentiated into mature oligodendrocytes possessing multiple process by 5 dpf (Fig. 1D, arrows). These data demonstrated that in the spinal cord of Tg(olig2:egfp) and Tg (sox10:egfp) larvae, EGFP-expressing cells are OPCs at 2 dpf and mature oligodendrocytes at 5 dpf. Thus, OPCs and mature oligodendrocytes can be obtained by sorting EGFP+ cells from Tg (olig2:egfp) and Tg(sox10:egfp) 2 dpf and 5 dpf larvae, respectively.
Fig. 1.
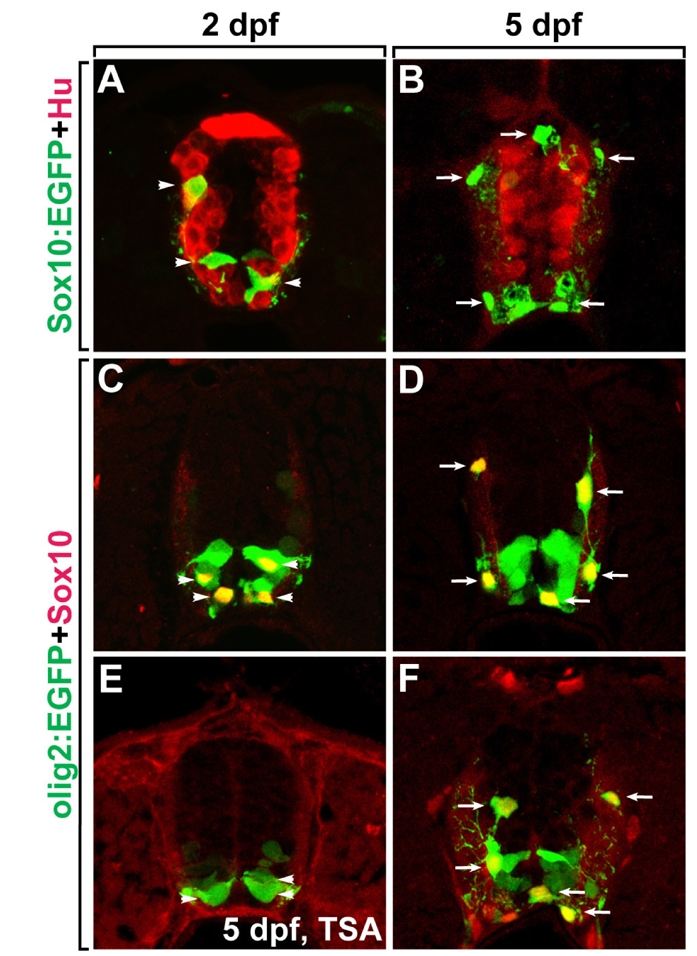
Identification of EGFP expression in oligodendrocyte lineage cells in transgenic zebrafish lines. All panels show transverse sections of the spinal cord, dorsal side at the top of the image. (A, B) Labeling of Tg(sox10:egfp) larvae with anti-Hu at 2 dpf (A) and 5 dpf (B). Arrowheads indicate OPCs in the gray matter and arrows indicate differentiated oligodendrocytes in the white matter of the spinal cord. (C, D) Labeling of Tg(olig2:egfp) larvae with anti-Sox10 at 2 dpf (C) and 5 dpf (D). Arrowheads indicate olig2:EGFP+, Sox10+ OPCs in the gray matter and arrows indicate olig2:EGFP+, Sox10+ differentiated oligodendrocytes in the white matter of the spinal cord. (E, F) Labeling of TSA-treated (E) and untreated (F) Tg(olig2:egfp) larvae with anti-Sox10 at 5 dpf. Arrows indicate olig2:EGFP+, Sox10+ differentiated oligodendrocytes in the white matter of the spinal cord.
To identify the genes specific to oligodendrocytes, a comparison of transcriptional profiles of normal and oligodendrocyte-deficient larvae could be conducted. Previously, oligodendrocyte development was shown to require histone deacetylase (HDAC) activity (Marin-Husstege et al., 2002; Cunliffe and Casaccia-Bonnefil, 2006; Ye et al., 2009), and treatment with the HDAC inhibitor Trichostatin A can block formation of oligodendrocytes in zebrafish (Tsai et al., 2006). Consistent with previous reports, we found that treatment of Tg(olig2:egfp) embryos with TSA from 36 hours post fertilization (hpf) to 5 dpf blocked migration of Sox10+, olig2:EGFP+ oligodendrocytes, which are located in the white matter of the spinal cord in the normal 3 dpf larvae (Fig. 1E and F, arrows indicate mature oligodendrocytes). These data indicated that comparison of transcriptional profiles of EGFP+ cells from TSA-treated and untreated Tg(olig2:egfp) larvae can be used to identify genes expressed specifically in oligodendrocytes.
Microarray-based transcriptome analysis to identify genes involved in oligodendrocyte differentiation
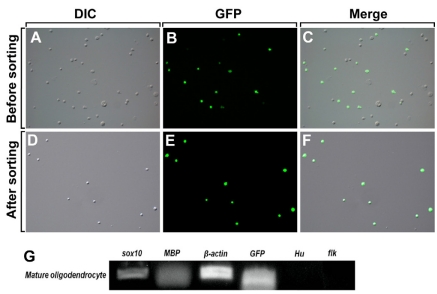
On the basis of the above data, OPCs and mature oligodendrocytes were purified from the dissociated cells of Tg(olig2:egfp) and Tg(sox10:egfp) larvae at 2 dpf and 5 dpf, respectively, using FACS. EGFP+ cells were also purified from TSA-treated and untreated 3 dpf Tg(olig2:egfp) larvae. Prior to sorting of EGFP+ cells, dissociated cells from the Tg(olig2:egfp) and Tg(sox10:egfp) larvae contained 20% EGFP+ cells (Fig. 2A~C). After sorting, an average of 98% of the sorted cells was EGFP+ OPCs or mature oligodendrocytes (Fig. 2D~F). To confirm the purity of the sorted cells, total RNA was isolated from EGFP+ mature oligodendrocytes and semi-quantitative RT-PCR was performed with several PCR primers specific to mature oligodendrocytes, neurons, and blood vessels. Mature oligodendrocyte specific genes were successfully amplified by RT-PCR, but neuron and blood vessel specific genes were not. These data indicate that oligodendrocyte lineage cells were specifically isolated by FACS (Fig. 2G).
Fig. 2.
Sorting of EGFP+ oligodendrocytes from transgenic zebrafish using FACS. Differential interference contrast images (DIC; A, D), fluorescence images (B, E), and combined DIC and fluorescence images of the same samples (C, F). (A~C) Tg(sox10:egfp) larvae were trypsinized to dissociate them into single cells at 5 dpf. (D~F) EGFP+ oligodendrocytes were sorted from Tg(sox10:egfp) larvae by FACS. More than 98% of the sorted cells were EGFP+ oligodendrocytes. (G) Semi-quantitative RT-PCR with RNA purified from the sorted 5 dpf sox10:EGFP+ cells and PCR primers specific to genes expressed in mature oligodendrocytes (sox10, mbp, gfp), neurons (Hu), and blood vessels (flk). β-actin was used as a loading control.
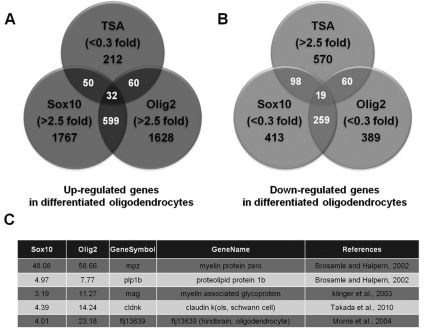
To compare the transcriptional profiles of OPCs and mature oligodendrocytes, we purified RNA from the sorted cells and synthesized dye-labeled cDNAs, which were hybridized to microarray gene chips. A total of 19,956 transcripts were examined with the Agilent zebrafish gene expression microarray. Genes up-regulated by 2.5-fold in mature oligodendrocytes were identified as genes required for oligodendrocyte maturation and genes down-regulated to 0.3-fold or less in mature oligodendrocytes were identified as genes required for the maintenance of OPC status. Fig. 3A and B illustrate diagrammatically the 4348 up-regulated and 1808 down-regulated genes in mature oligodendrocytes compared to OPCs as analyzed by three different algorithms. In addition, a total of 32 up-regulated and 19 down-regulated genes were selected from the intersection of the three algorithms (Fig. 3A, B). To confirm the accuracy of the microarray screen, we searched the expression profile of well-characterized genes required for the oligodendrocyte differentiation, such as mpz, plp1b, mag, cldnk and flj13639, and found that the expression levels of these genes were highly elevated in mature oligodendrocytes (Fig. 3C), indicating that our microarray analysis was successful.
Fig. 3.
Comparison of the transcriptional profiles obtained from three different algorithms. (A, B) Venn diagrams show the numbers of genes detected by three different algorithms, including the numbers of overlapping genes. (A, B) The intersection of the three databases contains 32 genes that were up-regulated in differentiated oligodendrocytes (A) and 19 genes that were down-regulated in differentiated oligodendrocytes (B). (C) Microarray profiles were validated by examining the expression profiles of genes known to be specifically expressed in differentiated oligodendrocytes, including mpz, plp1b, mag, cldnk and flj13639.
Validation of microarray data by quantitative RT-PCR (qRT-PCR)
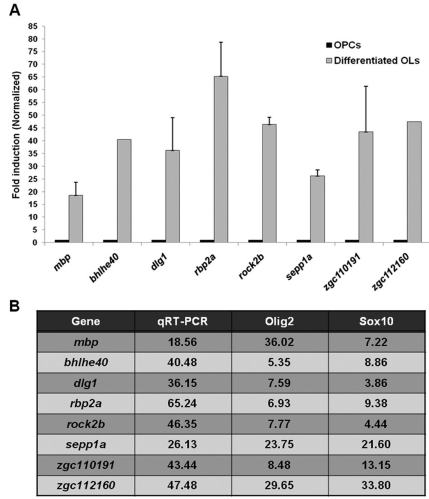
To validate the microarray data, we performed a qRT-PCR analysis of selected genes which exhibited highly elevated expression levels in the differentiated oligodendrocytes. We selected seven genes not previously known to be involved in oligodendrocyte differentiation. The expression levels for bhlhe40 (basic helix-loop-helix family, member e40, NM_212679), dlg1 (discs, large (Drosophila) homolog 1, NM_199526), rbp2a (retinol binding protein 2a, cellular, NM_153004), rock2b (rho-associated, coiled-coil containing protein kinase 2b, NM_001093747), sepp1a (selenoprotein P, plasma, 1a, NM_178297), zgc:110190 (NM_001020570) and zgc:112160 (NM_001017724) were evaluated by qRT-PCR using RNA purified from the sorted OPCs and mature oligodendrocytes. Myelin basic protein (mbp) was used as a positive control, and the expression levels of the selected genes are presented as the relative induction in differentiated oligodendrocytes compared to OPCs. Consistent with the microarray profiles, qRT-PCR demonstrated that the expression of these seven genes was highly elevated in differentiated oligodendrocytes (Fig. 4), suggesting that all seven genes may be involved in oligodendrocyte differentiation.
Fig. 4.
Validation of microarray data by qRT-PCR. Selected target genes were evaluated by qRT-PCR using the RNA purified from the sorted OPCs and differentiated oligodendrocytes. (A) qRT-PCR results were normalized to β-actin, and data are presented as relative induction of differentiated oligodendrocytes compared with OPCs. Myelin basic protein (mbp) was used as a positive control. (B) Fold changes determined by qRT-PCR and microarray analysis are presented.
Basic helix-loop-helix family member e40 (bHLHB2, also known as Dec1/Eip1/Sharp2/Stra13/Clast5) is a novel cAMP-inducible bHLH protein family member. Stra13 is required for the proliferation and differentiation of nerve cells. Stra13 controls growth arrest, and represses the expression of cell proliferation associated genes through an HDAC1-independent mechanism (Sun and Taneja, 2000). Dec1 is expressed in the nuclei of endothelial, glial and neuronal cells in brain tissue (Preusser et al., 2005). Since the bHLHB2 expression level is elevated in differentiated oligodendrocytes, we hypothesize that bHLHB2 plays a role in oligodendrocyte differentiation by regulating the OPC cell cycle. Discs-large homolog 1 (Dlg1) is a multi-domain scaffolding protein that is involved in protein trafficking in polarized cells and membrane addition in Drosophila (Lee et al., 2003; Gorczyca et al., 2007). Dlg1 is expressed in myelin-forming Schwann cells, and enriched in Schwann cell paranodes where myelin outfoldings preferentially arise. Interestingly, dlg1 interacts with kinesin 13B (kif13B) and Sec8, which play a central role in oligodendrocyte membrane formation by regulating the recruitment of vesicles that transport myelin proteins, suggesting that dlg1 function may be required for oligodendrocyte differentiation (Bolis et al., 2009). Retinol binding protein 2a (Rbp2a/CRBP) is a cytoplasmic retinol-specific binding protein and is expressed in the lateral ganglionic eminences (LGE), where a second wave of OPCs originate in the telencephalon (Kessaris et al., 2008). Retinol is naturally converted into retinoic acid which regulates the timing of oligodendrocyte differentiation in vivo (Barres et al., 1994). Therefore, we hypothesize that Rbp2a may play a role in oligodendrocyte development together with retinol.
Rho-GTPase family proteins play important roles in coordinating the remodeling of the actin cytoskeleton during myelination. A key effecter of Rho-GTPase is Rho-kinase (ROCK), a serine/threonine kinase that regulates cell migration, proliferation and survival (Mueller et al., 2005). ROCK is expressed in Schwann cells at the onset of myelination, and pharmacological inhibition of ROCK activity causes Schwann cells form aberrant short myelin segments. However, after ROCK inhibition is removed, new myelin segments are formed normally (Melendez-Vasquez et al., 2004), indicating that ROCK plays an important role together with Rho-GTPase in the progression of the Schwann cell membrane encasement of the axon during myelination. Rho-associated coiled-coil containing protein kinase 2b (Rock2b) is a ROCK family protein and our microarray data showed that Rock2b expression was highly elevated in myelinating oligodendrocytes, suggesting the possibility that Rock2b function is involved in myelin formation in CNS oligodendrocytes. Selenoprotein P, plasma, 1a (Sepp1a) is a selenium binding protein. Selenium is an essential micronutrient necessary for normal growth and function of the brain, and selenium deficiency causes a CNS demyelinating disease. Selenium is required for the normal morphological development of oligodendrocytes and their survival in vitro (Eccleston and Silberberg, 1984), and is also required for the normal up-regulation of myelin genes in differentiating oligodendrocytes (Gu et al., 1997). Therefore, significant up-regulation of Sepp1a in mature oligodendrocytes indicates the possibility that Sepp1a is involved in selenium signaling in oligodendrocyte differentiation. We also examined the previously unannotated genes zgc:110191 and zgc:112160. These two novel factors are expressed at elevated levels in differentiated oligodendrocytes.
ACKNOWLEDGEMENTS
This work was supported by the Korea Research Foundation Grant funded by the Korean Government (KRF-2008-313-C00764).
References
- 1.Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- 2.Bolis A, Coviello S, Visigalli I, Taveggia C, Bachi A, Chishti AH, Hanada T, Quattrini A, Previtali SC, Biffi A, Bolino A. Dlg1, Sec8, and Mtmr2 regulate membrane homeostasis in Schwann cell myelination. J Neurosci. 2009;29:8858–8870. doi: 10.1523/JNEUROSCI.1423-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carney TJ, Dutton KA, Greenhill E, Delfino-Machin M, Dufourcq P, Blader P, Kelsh RN. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–4630. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- 5.Charles P, Hernandez MP, Stankoff B, Aigrot MS, Colin C, Rougon G, Zalc B, Lubetzki C. Negative regulation of central nervous system myelination by poly-sialylated-neural cell adhesion molecule. Proc Natl Acad Sci USA. 2000;97:7585–7590. doi: 10.1073/pnas.100076197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Wu H, Wang S, Koito H, Li J, Ye F, Hoang J, Escobar SS, Gow A, Arnett HA, Trapp BD, Karandikar NJ, Hsieh J, Lu QR. The oligodendrocyte-specific G protein-coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nat Neurosci. 2009;12:1398–1406. doi: 10.1038/nn.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eccleston PA, Silberberg DH. The differentiation of oligodendrocytes in a serum-free hormone-supplemented medium. Brain Res. 1984;318:1–9. doi: 10.1016/0165-3806(84)90056-7. [DOI] [PubMed] [Google Scholar]
- 10.Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330:779–782. doi: 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- 11.Franklin RJ. Why does remyelination fail in multiple sclerosis? Nat Rev Neurosci. 2002;3:705–714. doi: 10.1038/nrn917. [DOI] [PubMed] [Google Scholar]
- 12.Gorczyca D, Ashley J, Speese S, Gherbesi N, Thomas U, Gundelfinger E, Gramates LS, Budnik V. Postsynaptic membrane addition depends on the Discs-Large-interacting t-SNARE Gtaxin. J Neurosci. 2007;27:1033–1044. doi: 10.1523/JNEUROSCI.3160-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu J, Royland JE, Wiggins RC, Konat GW. Selenium is required for normal upregulation of myelin genes in differentiating oligodendrocytes. J Neurosci Res. 1997;47:626–635. doi: 10.1002/(sici)1097-4547(19970315)47:6<626::aid-jnr8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 14.Kessaris N, Pringle N, Richardson WD. Specification of CNS glia from neural stem cells in the embryonic neuroepithelium. Philos Trans R Soc Lond B Biol Sci. 2008;363:71–85. doi: 10.1098/rstb.2006.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 16.Lee OK, Frese KK, James JS, Chadda D, Chen ZH, Javier RT, Cho KO. Discs-Large and Strabismus are functionally linked to plasma membrane formation. Nat Cell Biol. 2003;5:987–993. doi: 10.1038/ncb1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melendez-Vasquez CV, Einheber S, Salzer JL. Rho kinase regulates schwann cell myelination and formation of associated axonal domains. J Neurosci. 2004;24:3953–3963. doi: 10.1523/JNEUROSCI.4920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, Chang J, Thill G, Levesque M, Zhang M, Hession C, Sah D, Trapp B, He Z, Jung V, McCoy JM, Pepinsky RB. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- 20.Mueller BK, Mack H, Teusch N. Rho kinase, a promising drug target for neurological disorders. Nat Rev Drug Discov. 2005;4:387–398. doi: 10.1038/nrd1719. [DOI] [PubMed] [Google Scholar]
- 21.Park H, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Developmental Biology. 2002a;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- 22.Park H, Shin J, Appel B. Spatial and temporal regulation of ventral spinal cord precursor specification by Hedgehog signaling. Development. 2004;131:5959–5969. doi: 10.1242/dev.01456. [DOI] [PubMed] [Google Scholar]
- 23.Park HC, Appel B. Delta-Notch signaling regulates oligodendrocyte specification. Development. 2003;130:3747–3755. doi: 10.1242/dev.00576. [DOI] [PubMed] [Google Scholar]
- 24.Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002b;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- 25.Peterson SM, Freeman JL. RNA isolation from embryonic zebrafish and cDNA synthesis for gene expression analysis. J Vis Exp. 2009;pii:1470. doi: 10.3791/1470. doi: 10.3791/1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preusser M, Birner P, Ambros IM, Ambros PF, Budka H, Harris AL, Hainfellner JA. DEC1 expression in 1p-aberrant oligodendroglial neoplasms. Histol Histopathol. 2005;20:1173–1177. doi: 10.14670/HH-20.1173. [DOI] [PubMed] [Google Scholar]
- 27.Shin J, Park HC, Topczewska JM, Mawdsley DJ, Appel B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods Cell Sci. 2003;25:7–14. doi: 10.1023/B:MICS.0000006847.09037.3a. [DOI] [PubMed] [Google Scholar]
- 28.Sun H, Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci USA. 2000;97:4058–4063. doi: 10.1073/pnas.070526297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai HH, Macklin WB, Miller RH. Netrin-1 is required for the normal development of spinal cord oligodendrocytes. J Neurosci. 2006;26:1913–1922. doi: 10.1523/JNEUROSCI.3571-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 31.Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]