Abstract
Expression of the alcohol dehydrogenase gene (ADH) of Arabidopsis is known to be induced by environmental stresses and regulated developmentally. We used a negative-selection approach to isolate mutants that were defective in regulating the expression of the ADH gene during seed germination; we then characterized three recessive mutants, aar1–1, aar1–2, and aar2–1, which belong to two complementation groups. In addition to their defects during seed germination, mutations in the AAR1 and AAR2 genes also affected anoxic and hypoxic induction of ADH and other glycolytic genes in mature plants. The aar1 and aar2 mutants were also defective in responding to cold and osmotic stress. The two allelic mutants aar1–1and aar1–2 exhibited different phenotypes under cold and osmotic stresses. Based on our results we propose that these mutants are defective in a late step of the signaling pathways that lead to increased expression of the ADH gene and glycolytic genes.
Exposure to oxygen deprivation (anoxia) and decreased oxygen availability (hypoxia) due to flooding are common plant environmental stresses. After a change from aerobic to anoxic conditions, carbon metabolism of plant cells switches from aerobic respiration to fermentation (Drew, 1990). Biochemical studies have shown that oxygen deficiency in maize roots induces synthesis of approximately 20 anaerobic polypeptides (for review, see Sachs et al., 1980, 1996), several of which have been identified as glycolytic and fermentative enzymes, including ADH and GAPDH (Freeling and Bennett, 1985; Ricard et al., 1989; Russell and Sachs, 1989; Xie and Wu, 1989; Yang et al., 1993). Several cis-acting elements and trans-acting factors involved in anaerobic induction of the ADH genes in maize and Arabidopsis have been identified (Ferl and Laughner, 1989; Dolferus et al., 1994; Kyozuka et al., 1994; Hoeren et al., 1998). Recent reports indicate that Ca2+ may play an important role in the induction of ADH gene expression during anoxia in maize (Subbaiah et al., 1994a, 1994b; Sedbrook et al., 1996).
Most crop plants, including barley, maize, sorghum, and wheat, can tolerate only very transient flooding (Kennedy et al., 1992). In contrast, rice plants can survive much longer under anaerobic conditions. Formation of aerenchyma, characterized by continuous gas spaces in roots and shoots (Drew et al., 1979; Justin and Armstrong, 1987), has been proposed to correlate with tolerance to flooding (Justin and Armstrong, 1987). Campbell and Drew (1983) have shown that lysogenic aerenchyma formation occurs in the root cortex of maize during hypoxia. A recent report indicates that an ethylene signal is required for cell death during aerenchyma formation induced by hypoxia (He et al., 1996). These results, in conjunction with reports that Ca2+ is also involved in the induction of the ADH gene in the early stage of anoxia (Subbaiah et al., 1994a, 1994b; Sedbrook et al., 1996), suggest that multiple signaling events may be involved.
In addition to hypoxia or anoxia, other environmental stresses, including cold and dehydration, also induce the expression of the ADH gene in Arabidopsis (Dolferus et al., 1994). Recent genetic evidence suggests that both an ABA-dependent and an ABA-independent pathway are involved in mediating the response to cold and osmotic stress (Ishitani et al., 1997; Zhu et al., 1997). The molecular relationship, if any, between ADH induction by hypoxia or anoxia and response to cold and osmotic stress is essentially unknown.
We are taking advantage of the amenability of Arabidopsis to genetic manipulation to dissect these complex signaling pathways. It has been shown that mRNA levels for both ADH and GAPC genes increase during anoxia (Chang and Meyerowitz, 1986; Yang et al., 1993). Analysis of transgenic plants containing ADH- or GAPC-promoter::GUS fusion genes show that transcriptional control is involved in anaerobic induction of both genes in Arabidopsis (Yang et al., 1993; Dolferus et al., 1994). We have used a genetic scheme that allowed us to identify regulatory mutants that were defective in the induction of the ADH gene during seed germination. Characterization of some of these mutants revealed that they were also defective in other stress-induced signal transduction pathways that lead to the activation of several glycolytic genes.
MATERIALS AND METHODS
Generation of an ADH-Promoter::GUS Reporter Gene Construct in the Binary Vector pBI101
The genomic clone pAtAdh-7 served as the source of a 2.4-kb HindIII-SmaI fragment, which included approximately 1 kb of sequence upstream from the transcription start site of the ADH gene of Arabidopsis (Chang and Meyerowitz, 1986). The 2.4-kb HindIII-SmaI fragment was subcloned into pBS (plasmid BlueScript, Stratagene), generating pAdh. This plasmid, along with two oligonucleotide primers, was used in PCR to generate an approximately 1-kb ADH-promoter fragment. The upstream primer was complementary to the T7 RNA polymerase-promoter site in the pBS sequences flanking the multilinker region, whereas the downstream primer encompassed the ADH translation initiation codon. The downstream primer was designed to engineer a new HincII site by changing the original 5′-GTTGATAATG-3′ sequence (the translation initiation codon is underlined) to 5′-GTTGACCATG-3′ (the new HincII site is designated by underlining; modified bases are designated in bold). The HincII site served to facilitate subsequent manipulations. The 1-kb PCR product was subcloned into pBS, generating pAdh(Pro). Digesting pAdh(Pro) with XbaI and HincII produced a 1072-bp fragment that was subcloned directionally into the XbaI and SmaI sites of the multilinker region of the binary vector pBI101 (Jefferson et al., 1987). Restriction mapping was used to confirm the presence and the correct orientation of the ADH-promoter insert in the resulting plasmid clone pBI101/Adh-Gus.
Generation of Transgenic Arabidopsis Plants
Root transformation was used to generate transgenic Arabidopsis plants, essentially as described by Valvekens et al. (1988). Rooted transformed plantlets were transferred to Jiffy Mix Plus soil mix (Hummert International, St. Louis, MO). Plants were grown in environmental chambers (Percivall, Boone, IA) at 20°C under a mixture of incandescent and fluorescent lamps (fluence rate, 100 μE m−2s−1) under long-day conditions (16-h light/8-h dark).
The four independent transformants that were obtained were propagated to maturity for subsequent analysis. All results reported here are based on ADH::GUS expression in F4 progeny of the line designated AG2. The F1 progeny of the primary transformant line of AG2 had an approximate 3:1 segregation of kanamycin-resistant/kanamycin-sensitive progeny. Kanamycin-resistant F1 plants were analyzed through the F2 and F3 generations to identify homozygous lines. The presence of a single copy of the ADH::GUS construct in line AG2 was confirmed by genomic Southern analysis.
Chemical Mutagenesis
The AG2 line of the transgenic ADH::GUS was mutagenized. Three 0.5-g aliquots of seeds were placed separately into sterile 50-mL centrifuge tubes and surface-sterilized by soaking in 1.5% (v/v) NaOCl with 0.04% (w/v) sodium dodecyl sulfate for 30 min, followed by five sterile water rinses. Seeds were then soaked in 0.25% (v/v) aqueous ethyl methanesulfonic acid for 10 to 12 h (Somerville and Ogren, 1982) and then rinsed several times with sterile water. The seeds were planted on Jiffy Mix Plus soil mix and grown to maturity. Approximately 4000 plants from each of the three 0.5-g aliquots of mutagenized seeds were grown to maturity. The M2 seeds from each of the resulting populations of seeds were collected, dried, and stored in a sealed container at 4°C.
Allyl Alcohol Selection
Approximately 50,000 seeds from each of the three mutagenized populations were surface-sterilized as described above. The seeds were then soaked in sterile water at room temperature for 6 h. The imbibed seeds were next exposed to 45 mm (aqueous) allyl alcohol for 2 h at room temperature (Jacobs et al., 1988), followed by five rinses with sterile water and treatment overnight with 15 μm GA (aqueous) at 4°C. Each aliquot of seeds was subdivided and transferred into ten 250-mL flasks, each containing liquid germination medium (Valvekens et al., 1988). The flasks were incubated under fluorescent lights in an environmental chamber on a shaker operated at 150 rpm. Green seedlings that appeared after several days were transferred to Petri dishes containing germination medium solidified with 0.8% agar.
Growth Conditions and Stress Treatment
Seeds of line AG2 and aar mutants were surface-sterilized and treated with 15 μm GA at 4°C overnight. Seeds were sown onto plates with Murashige and Skoog medium containing 1% Suc and 0.8% agar and grown at 20°C with 16-h light/8-h dark cycles. After 1 week, seedlings were transferred to fresh Murashige and Skoog plates in a vertical position under the same conditions for an additional 3 weeks. For anoxic treatments, plants were submerged in liquid Murashige and Skoog medium through which nitrogen gas (>99% purity) was bubbled to purge O2. For hypoxic treatments, similar conditions were used except that pure N2 gas was replaced with gas containing 4.5% to 5% O2 and balanced with N2.
We used a modification of the procedures described by Dolferus et al. (1994) for osmotic-stress experiments. Arabidopsis plants were submerged in liquid Murashige and Skoog medium containing 0.6 m mannitol through which air (18% O2) was bubbled for 24 h at 20°C. For cold treatment, plants were submerged in aerated liquid Murashige and Skoog medium at 4°C for 24 h.
GUS and ADH Enzymatic Assays
Histochemical staining and enzyme activity assays for GUS were performed essentially as described by Jefferson et al. (1987). Fluorescence of the 4-methylumbelliferyl product was quantified using a minifluorometer (model TKO-100, Hoefer Scientific Instruments, San Francisco, CA).
ADH enzyme activity assay was performed according to the procedures described by Freeling (1973) and modified by Xie and Wu (1989). The assay uses ethanol as the substrate and measures the production of NADH in a spectrophotometer (model DU 64, Beckman). One unit of ADH enzyme is defined as an increase in A340 of 0.01 per min (Xie and Wu, 1989).
RNA Isolation and RT-PCR Reactions
Total RNA was isolated by an acidic phenol protocol adapted from the procedures described in Chomczynski and Sacchi (1987). Prior to RT-PCR reactions, RNA samples were treated with DNase I twice to deplete contaminating genomic DNA. First-strand cDNA synthesis was performed in a 50-μL reaction mixture containing 2 μg of RNA, 1 μg of random hexamer, 40 μm each of the four deoxynucleotides, 5 μL of 10× buffer, 200 units of Moloney murine leukemia virus RT, and 20 units of ribonuclease inhibitor. The reaction mixture was incubated at 42°C for 60 min and stopped by heating at 65°C for 10 min.
PCR was performed in a DNA Thermal Cycler (Perkin Elmer/Cetus) programmed for 28 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. To find the amounts of cDNA suitable for linear amplification, we used 1, 2, 4, 6, 8, and 10 μL of RT reaction mixture from the hypoxic-treated AG2 line. For all of the genes shown in Table I, there were linear increases of PCR products in reactions with 1, 2, and 4 μL of cDNA (data not shown). Therefore, we used 1 or 2 μL of the RT reaction mixture for subsequent PCR and analyzed the products using agarose-gel electrophoresis. Sequencing the products from the reactions of the AG2 line confirmed the identities of the PCR products for each primer set .
Table I.
Primer sequences for RT-PCR
| Gene | Primera | Product Length |
|---|---|---|
| bp | ||
| ADH | TGTCACACCGATGTTTAC | |
| GGTCGAATCTTTTAGAGTTAA | 525 | |
| PDC1 | GAAGCTGCGGTGGAAGCAAC | |
| TCCTAGAGTTGCACCAACAG | 682 | |
| PDC2 | GGCTCATGAACTTATCGATA | |
| TCTCAGCAAGCACAGCAGAC | 740 | |
| GAPC | GGAGCTGACTACGTTGTTGAGT | |
| CGTTGTCGTACCATGACACCAA | 670 | |
| βATP | GATCATGACATCTCTCGAGG | |
| TGGTAAGGAGCAAGGAGATC | 350 |
The primers shown on top rows represent forward primers used for each gene, whereas those shown on bottom rows correspond to the reverse primers.
We quantified the PCR products by analyzing the digitized images of agarose gels using the ImageQuant software program, version 1.1 (Molecular Dynamics, Sunnyvale, CA). Nucleotide sequences of the primer pairs for amplification of each gene and the sizes of the corresponding products appear in Table I. The accession numbers (in parentheses) for the genes on which the primer sequences were based are as follows: ADH (M12196), GAPC (M64116), PDC1 (U71121), and PDC2 (U71122).
Chemical and Enzyme Suppliers
We purchased synthetic plant hormones (benzylaminopurine, GA3, and kinetin), antibiotics (kanamycin, rifampicin, and vancomycin), and other chemicals from Sigma, and 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid cyclohexylammonium salt from Gold Biotechnology (St. Louis, MO). Restriction and modification enzymes were from New England BioLabs and Promega, and the sequenase and radiochemicals used in DNA sequencing were from Amersham.
RESULTS
Isolation of Mutants Affecting ADH Gene Expression
The genetic scheme we adapted uses the ADH enzyme as a selectable marker. ADH normally functions in alcoholic fermentation, catalyzing the reduction of acetaldehyde to ethanol. However, ADH also uses allyl alcohol (2-propen-1-ol) as a substrate, forming acrolein (2-propenal), which is highly toxic to plant cells. Therefore, in the presence of allyl alcohol, wild-type plants with normal levels of functional ADH enzyme will die, whereas plants that lack ADH will survive (Freeling and Bennett, 1985; Jacob et al., 1988). This property can be exploited to isolate mutants that are defective in regulating the expression of the ADH gene.
Several transgenic Arabidopsis lines that contain one copy of ADH::GUS were constructed. We chose one of these lines, AG2, in which the expression pattern of the ADH::GUS transgene reflected the expression pattern of the endogenous ADH gene (Chang and Meyerowitz, 1986; Dolferus et al., 1994), for ethyl methanesulfonic acid mutagenesis. To screen for mutants affecting ADH gene expression, 150,000 M2 seeds were treated with allyl alcohol (for details, see Methods). A total of 40 seeds germinated and produced green seedlings. These we designated as aar (allyl alcohol resistant) mutants, which we further divided into two classes. The first, with mutations to the endogenous ADH gene, were to have normal ADH::GUS expression. The second, with mutations in genes involved in the signaling pathway(s) leading to the induction of ADH, were to have reduced levels of expression of both the endogenous ADH gene and the GUS reporter gene. To distinguish between these two classes, we stained imbibed seeds of aar mutants for GUS activity. Thirteen of the 40 aar mutants had pronounced GUS staining after 6 h or less of incubation, similar to the parental AG2 plants, indicating that the ADH::GUS gene was expressed. In contrast, the remaining 27 aar plants exhibited no detectable GUS staining even after 24 h of incubation (data not shown). We characterized three of these mutants, aar7, aar10, and aar17, which are described below.
Table II showed that F1 progeny from crosses between line AG2 and each of the three aar mutants were sensitive to allyl alcohol treatment. In addition, allyl alcohol-sensitive and -resistant plants in the F2 progeny from all three crosses showed a 3:1 ratio. These results indicated that each aar mutant contains a monogenic and recessive mutation. Pair-wise crosses among these mutants showed that aar7 and aar17 belonged to the same complementation group, whereas aar10 constituted a second group (Table III). Therefore, aar7 and aar17 were renamed aar1–1 and aar1–2, respectively, whereas aar10 was renamed aar2–1.
Table II.
Segregation test of aar mutants
| Cross | No. of F1 Seeds | aar Seeds in F1a | No. of F2 Seeds | aar Seeds in F2 | WT:aarb | χ2 |
|---|---|---|---|---|---|---|
| AG2 × aar 7 | 42 | 0 | 293 | 73 | 3:1 | 1.65 |
| AG2 × aar 10 | 55 | 0 | 346 | 101 | 3:1 | 2.93 |
| AG2 × aar 17 | 32 | 0 | 414 | 112 | 3:1 | 2.79 |
The aar phenotype corresponds to seeds that can germinate after allyl alcohol treatment.
WT is defined as seeds that fail to germinate after allyl alcohol treatment.
Table III.
Complementation test
| Cross | No. of F1 Seeds Tested | Allyl Alcohol Sensitivitya
|
|
|---|---|---|---|
| Sensitive | Resistant | ||
| aar7 × aar10 | 27 | 27 | 0 |
| aar7 × aar17 | 35 | 0 | 35 |
| aar10 × aar7 | 17 | 17 | 0 |
| aar10 × aar17 | 58 | 58 | 0 |
| aar17 × aar7 | 44 | 0 | 44 |
| aar17 × aar10 | 30 | 30 | 0 |
See footnote in Table II for definition of allyl alcohol sensitivity.
Effects of Mutations in AAR Genes during Seed Germination
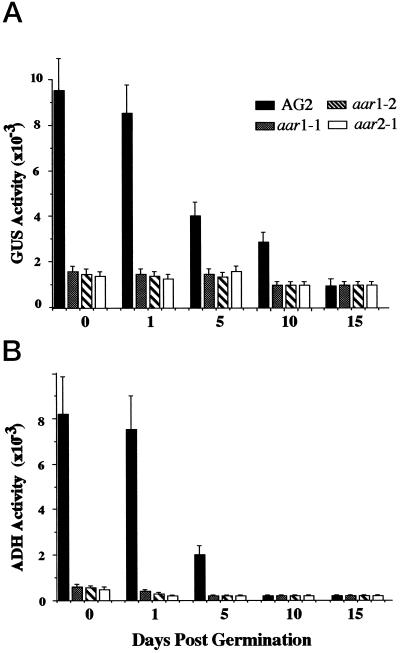
To quantify the effects of mutations on the expression of the ADH::GUS transgene during seed germination, we compared the levels of GUS activity of the wild-type AG2 line and the aar mutants (Fig. 1A). In AG2 GUS activity was highest in imbibing seeds (Fig. 1A, d 0) and 1-d-old germinating seedlings. The activity decreased 50% by 5 d, and had decreased to a steady-state level by 15 d, which was less than 10% of the 1-d level. In contrast, GUS activity was much lower in the imbibing seeds and the germinating seedlings in all three aar mutants. In 1-d-old germinating seedlings, GUS activity in line AG2 was 8- to 10-fold higher than in the aar mutants. In 5-d-old seedlings, GUS activity in line AG2 was 2- to 3-fold higher than in the mutant plants. In contrast, at later developmental stages GUS activity was very similar in line AG2 and in all of the mutants (Fig. 1A, d 15).
Figure 1.
GUS and ADH activities of AG2 and of the aar mutants during seed germination. Seedlings of AG2 and aar1–1, arr1–2, and aar2–1 at different developmental stages were harvested and assayed for GUS and ADH activities as described in Methods. A, GUS activity is expressed as pmol 4-methylumbelliferone min−1 mg−1 protein. B, One unit of ADH enzyme is defined as an increase in A340 of 0.01 per min. The data presented are the averages of three independent treatments. Plants grown at different times were used for replicate treatments. Bar graphs at each time point (from left to right) represent activities for AG2, aar1–1, aar1–2, and aar2–1. Bars indicate sd.
We also measured ADH activity in germinating seedlings of AG2 and aar mutants. Figure 1B shows that the pattern of ADH activity level in AG2 was similar to that of GUS, except that ADH activity decreased more rapidly, reaching an undetectable level in 10-d-old seedlings. However, as in the case for GUS, there was much lower ADH activity in imbibing seeds and germinating seedlings than in any of the three aar mutants. These results showed that mutations in the AAR1 and AAR2 genes affected the expression of both the ADH::GUS transgene and the endogenous ADH gene during germination.
Anoxic and Hypoxic Responses in Mature Plants
To determine whether mutations in the AAR1 and AAR2 genes affected regulation of the ADH::GUS transgene in mature plants in response to anoxia and hypoxia, 4-week-old plants from line AG2 and from the aar1–1, aar1–2, and aar2–1 mutants were subjected to hypoxic or anoxic treatments for 8 h and harvested for the assay of GUS activity. The data showed that after 8 h, there were 10- and 15-fold increases in GUS activity, respectively, in anoxic- and hypoxic-treated AG2 plants (Fig. 2). In contrast, there was no increase in GUS activity in either anoxic- or hypoxic-treated plants in all three aar mutants (Fig. 2).
Figure 2.
Anoxic and hypoxic induction of GUS activity in mature plants. Twenty-day-old AG2 and aar1–1, aar1–2, and aar2–1 plants were subjected to anoxic or hypoxic treatment and assayed for GUS activity as described in Methods. The data presented are the averages of three independent treatments. Bars indicate sd.
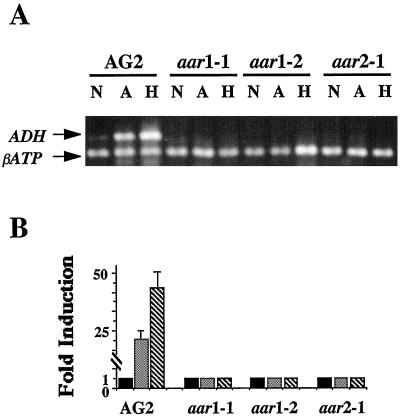
To examine the expression pattern of the endogenous ADH gene, we subjected the 4-week-old AG2 plants and the aar mutants to 8 h of anoxic or hypoxic treatments. Total RNA was isolated and analyzed by RT-PCR. The nuclear gene βATP that encodes the β-subunit of mitochondrial ATP synthase, the expression of which was not affected by either hypoxic or anoxic treatment (T.R. Conley and M.-C. Shih, unpublished data), was used as an internal standard. The results from one set of representative RT-PCR reactions are illustrated in Figure 3. The data show that under normoxic conditions, there was little ADH mRNA in either line AG2 or the three aar mutants (Fig. 3A, lanes N). Levels of ADH mRNA were greatly induced in anoxic- or hypoxic-treated AG2 plants (Fig. 3A, lanes N and H). Quantification of the RT-PCR products indicated that anoxic treatment resulted in a 25-fold increase in ADH mRNA level in AG2, whereas hypoxic treatment resulted in a 50-fold induction (Fig. 3B). In contrast, there was no increase in the ADH mRNA level in either anoxic- or hypoxic-treated plants in any of the three aar mutants. These results demonstrate that mutations in AAR1 and AAR2 genes affected anoxic and hypoxic induction of expression of both the endogenous ADH gene and the ADH::GUS transgene in mature plants.
Figure 3.
RT-PCR analysis of anoxic and hypoxic induction of ADH. Total RNA (1 μg) from anoxic- or hypoxic-treated plants was used in a 100-μL reaction for first-strand cDNA synthesis. One-tenth of the synthesized cDNA from each treatment was then used in subsequent PCR reactions. A, RT-PCR products for ADH were analyzed by agarose-gel electrophoresis. The sizes of PCR products for each gene are 525 bp for ADH and 350 bp for βATP. The notations on top of each lane represent treatment conditions of normoxia (N), anoxia (A), and hypoxia (H). B, Digitized images of the ADH bands in A were quantified and normalized to the βATP band in each lane. The normalized mRNA levels from normoxic-treated plants (solid bars) of AG2 and each aar mutant were used as the reference levels to calculate magnitudes of induction. Checked and striped bars correspond to anoxic- and hypoxic-treated samples, respectively. The data presented are the averages of three independent treatments. Bars indicate sd.
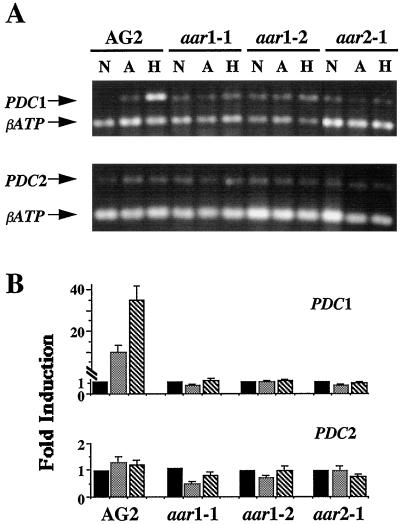
In addition to ADH, PDC was also required to catalyze the conversion of pyruvate to ethanol during alcoholic fermentation. Therefore, we examined whether the expression of PDC1 and PDC2 was affected in the aar1 and aar2 mutants. The data show that there was a low level of PDC1 mRNA present in AG2 plants grown under normoxic conditions (Fig. 4A). However, the PDC1 mRNA level increased by greater than 20- and 40-fold, respectively, in anoxic- and hypoxic-treated AG2 plants (Fig. 4, top panel). In contrast, in the three aar mutants, PDC1 mRNA levels were not induced by either anoxic (lanes A) or hypoxic (lanes H) treatments. Figure 6 also shows that there was a low level of PDC2 mRNA present in AG2 and aar mutant plants grown under normoxic conditions (bottom panels). However, unlike PDC1, levels of PDC2 mRNA were not induced in either anoxic- or hypoxic-treated AG2 plants or in any of the three aar mutants. These results showed that only the expression of PDC1, not that of PDC2, was inducible by anoxic or hypoxic treatment in Arabidopsis, and that mutations in AAR1 and AAR2 genes could affect PDC1 induction.
Figure 4.
RT-PCR analysis of anoxic and hypoxic induction of PDC1 and PDC2. Analysis of RT-PCR products (A) and quantification of levels of induction (B) of PDC1 and PDC2 were performed as described in Figure 3. The symbols used are as described in the Figure 3 legend. The sizes of PCR products are 682 bp for PDC1 and 740 bp for PDC2.
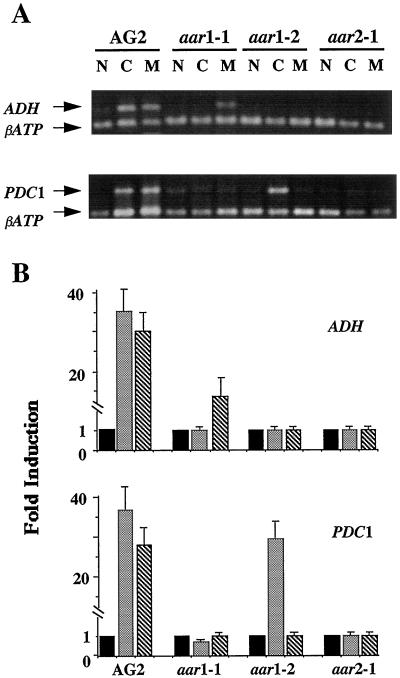
Figure 6.
Effects of cold and mannitol on ADH and PDC1 expression. A, RNA samples from AG2 and aar mutants grown under normal conditions (lanes N), cold treatment (lanes C), and mannitol treatment (lanes M) were subjected to RT-PCR analysis. B, Quantification of cold and mannitol induction of ADH and PDC1 was performed as described for Figure 3B. Solid bars correspond to samples from control experiments, whereas checked and striped bars correspond to samples from cold and mannitol treatments, respectively. The data presented are the averages of three independent treatments. Bars indicate sd.
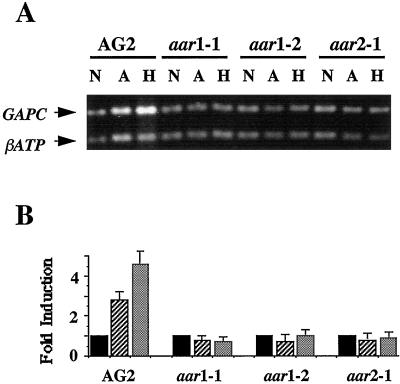
The ADH and PDC genes belong to a class of genes that function exclusively in alcoholic fermentation. The data presented here show that these genes were expressed at extremely low levels under normoxic conditions, but could be greatly induced by anoxic and hypoxic treatments. Russell and Sachs (1989) and Yang et al. (1993) have shown that glycolytic genes whose functions are required for both glycolysis and fermentation were expressed at high levels under normoxia, but could be induced further by anoxia or hypoxia. We chose to examine the expression pattern of the GAPC gene, which encodes cytosolic GAPDH, to determine whether its expression was affected in aar mutants. Figure 5A shows that there was a detectable mRNA level of GAPC in AG2 plants grown under normoxic conditions (lanes N) and that the expression of GAPC was induced by both anoxic and hypoxic treatments (lanes A and H). Quantification of the RT-PCR products indicated that the anoxic treatment resulted in a 3-fold increase and the hypoxic treatment resulted in a 5-fold increase in the GAPC mRNA level in AG2 plants (Fig. 5B). Figure 5 also shows that there were detectable and similar-to-wild-type GAPC mRNA levels in the three aar mutants grown under normoxic conditions. In contrast to the wild-type AG2 line, the expression level of GAPC remained unchanged after the anoxic or hypoxic treatments in all three mutants. Therefore, mutations in AAR1 or AAR2 genes affect anoxic and hypoxic induction of the GAPC gene, as well as induction of ADH and PDC1 genes.
Figure 5.
Effect of aar mutations on anoxic and hypoxic induction of GAPC. A, Levels of GAPC mRNA from normoxic-treated (lanes N), anoxic-treated (lanes A), and hypoxic-treated (lanes H) AG2 plants and aar mutants were analyzed by relative RT-PCR. The sizes of the PCR products are 670 bp for GAPC and 350 bp for βATP. B, Quantification of levels of induction of GAPC by anoxia and hypoxia in AG2 and aar mutants were calculated as described for Figure 3B.
Allelic-Specific Effects of aar1 Mutants under Cold and Osmotic Stress
The expression of the ADH gene is also affected by other environmental conditions, such as cold and osmotic stress, in several plant species (Xie and Wu, 1989; Dolferus et al., 1994). Our RT-PCR analysis showed that in AG2 plants, levels of ADH and PDC1 mRNA were greatly increased by cold or mannitol treatment (Fig. 6). However, the mRNA levels for PDC2 and GAPC were not affected by either treatment (data not shown). Therefore, we examined whether the induction of ADH and PDC1 by cold or mannitol treatment was affected in the aar mutants. In AG2, following 24 h at 4°C, there were 35-fold increases in mRNA levels measured for both ADH and PDC1, whereas exposure to 0.6 m mannitol resulted in approximately 30-fold increases in ADH and PDC1 mRNA levels (Fig. 6, A [lanes C and M] and B). However, the data presented in Figure 6 also show that the three mutants exhibited different allele-specific effects on induction of ADH and PDC1 genes by cold and mannitol treatment. In aar1–1 mannitol treatment resulted in a 15-fold induction of ADH (Fig. 6, A [top panel] and B). In contrast, the induction of ADH by cold treatment (Fig. 6A, lanes C [top panel]) and the induction of PDC1 by cold and mannitol treatments (Fig. 6A, lanes C and M [bottom panel]) were abolished completely in aar1–1. In aar1–2 the induction of PDC1 by mannitol treatment and the induction of ADH by cold and mannitol treatment were abolished, whereas there was a 30-fold induction of the PDC1 gene by mannitol treatment. In aar2–1 the induction of both ADH and PDC1 by cold or mannitol treatment was abolished completely.
DISCUSSION
We have identified and characterized trans-regulatory mutations that affect the expression of the ADH gene in Arabidopsis. The genetic approach used here takes advantage of the fact that ADH catalyzes the conversion of allyl alcohol into a toxic compound (acrolein) that is lethal to wild-type plants. In addition, we have used a transgenic line bearing an ADH::GUS construct as the starting point for mutagenesis. The GUS marker gene allowed us to distinguish between the two classes of mutants that were predicted to be isolated: mutations affecting the promoter or coding region of the endogenous ADH gene, and mutations affecting any step in the signal transduction pathway leading to the activation of the ADH gene. In our screening, about one-third of the putative mutants were potentially members of the first group, whereas the remaining two-thirds potentially belong to the latter group of mutants. Characterization of three of these regulatory mutants has provided us with information for understanding the molecular mechanism of induction of ADH and PDC1 genes by three different environmental stresses. Li et al. (1995) used a similar screening scheme successfully to isolate trans-regulatory mutants of the CAB gene in Arabidopsis.
Expression of ADH in Arabidopsis is known to be induced by several environmental stresses and to be regulated developmentally (Dolferus et al., 1994). The selection scheme we have adapted allowed us to identify mutants that were defective in regulating the expression of the ADH gene in germinating seedlings. Three such mutants, aar1–1, aar1–2, and aar2–1, were also defective in regulating the anoxic and hypoxic induction of the ADH gene in mature plants. These results can be interpreted in two ways. First, the regulatory pathways controlling the expression of ADH during germination and anoxia/hypoxia are identical. Second, two signaling pathways share some common steps and the aar mutants characterized here affect these steps. If the second hypothesis is correct, it is conceivable that some of the other aar mutants identified in these screens may affect ADH expression during germination, but exhibit normal anoxic or hypoxic induction of ADH. Experiments are under way to further characterize the other aar mutants that we have isolated.
In addition to the ADH gene, glycolytic and other fermentative genes are induced by anoxia and hypoxia (Yang et al., 1993; Sachs et al., 1996; Rivoal et al., 1997). We have also examined the effects of the aar mutations on the expression of other glycolytic genes. Among these genes, GAPC gene products are required for both aerobic respiration and anaerobic fermentation, whereas ADH and PDC1 gene products are required only for alcoholic fermentation. The expression patterns of these genes reflect their physiological roles. ADH and PDC1 genes are expressed at extremely low levels in plant cells under normal growth conditions and their expression is strongly induced by hypoxia or anoxia. In contrast, the GAPC gene is expressed constitutively at a fairly high level under normal growth conditions, but its expression can be induced further by hypoxia or anoxia. This raises the interesting question of whether similar regulatory elements are involved in the activation of these two different classes of genes. Our data show that mutations in AAR1 and AAR2 genes affected induction of both classes of glycolytic genes by anoxia and hypoxia. These results suggested that similar regulatory elements mediated the induction of these two different classes of genes in response to anoxic and hypoxic stresses.
Cold and osmotic stress also induce the expression of ADH in several plant species (Xie and Wu, 1989; Dolferus et al., 1994). Our results show that both ADH and PDC1 genes could be induced by cold and mannitol treatment in Arabidopsis. Recent genetic and molecular studies suggested that both ABA-dependent and -independent pathways interact and converge to activate the expression of downstream genes (Ishitani et al., 1997, 1998; Zhu et al., 1998). Our results show that the aar2–1 mutant was defective in the induction of ADH and PDC1 by anoxic/hypoxic, cold, and osmotic stresses (Fig. 6). This suggests that the three stress-induced signaling pathways shared some common intermediate steps and that aar2–1 was defective in one of these steps. However, our results also show that the induction of ADH and PDC1 by these three stresses was affected differently in the two allelic mutants, aar1–1 and aar1–2. There, the induction of ADH and PDC1 by the three stresses was abolished, with two exceptions: in aar1–1 there was a 15-fold induction of ADH in response to the mannitol treatment, which was about 50% of the induction level of the wild-type AG2 line (Fig. 6); second, the induction of PDC1 by cold treatment was unaffected in aar1–2.
A report by de Bruxelles et al. (1996) indicated that although induction of the ADH gene by osmotic and cold stresses required some common cis-acting promoter elements, additional, but distinct, regulatory elements were required for each pathway. Their results also showed that regulatory elements different from those involved in cold and osmotic responses were required for induction of ADH by anoxia and hypoxia. This implies that transcription complexes for the ADH gene under cold, osmotic, and low-oxygen stresses may share some common factors, but probably also embody some discrete factors. If this hypothesis is true, one can propose that the AAR1 gene may encode a transcription factor shared by these complexes, and that mutations in aar1–1 and aar1–2 have different effects on the interaction between the AAR1 factor and factors that are specific to each stress condition.
Current information suggests complex patterns of interactions among the signal transduction pathways elicited by cold, osmotic, and low-oxygen stresses in plants (de Bruxelles et al., 1996; Stockinger et al., 1997; Zhu et al., 1997). Ishitani et al. (1997) have isolated mutants that affect cold- and osmotic-stress signaling pathways by a high-throughput screening approach. Some of these mutants are specifically defective in cold- or osmotic-stress-induced signaling pathways (Ishitani et al., 1997; Zhu et al., 1998). It is likely that some of the aar mutants we have isolated are specifically defective in the signaling pathway that corresponds to decreased oxygen. Combining studies of these different classes of mutants should greatly facilitate the elucidation of these signaling pathways.
ACKNOWLEDGMENTS
We thank Drs. Joseph Frankel and Andy Wang for comments on the manuscript. We also thank the Arabidopsis Biological Resource Center at the Ohio State University, Columbus, for providing us with expressed sequence tag clones.
Abbreviations:
- ADH
alcohol dehydrogenase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PDC
pyruvate decarboxylase
- RT
reverse transcriptase
Footnotes
This work was supported by the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (grant nos. 9401454 and 9700603 to M.-C.S.).
LITERATURE CITED
- Campbell R, Drew MC. Electron microscopy of gas space (aerenchyma) formation in adventitious roots of Zea mays L. subjected to oxygen shortage. Planta. 1983;157:350–357. doi: 10.1007/BF00397407. [DOI] [PubMed] [Google Scholar]
- Chang C, Meyerowitz E. Molecular cloning and DNA sequence of the Arabidopsis thaliana alcohol dehydrogenase gene. Proc Natl Acad Sci USA. 1986;83:1408–1412. doi: 10.1073/pnas.83.5.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–160. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- de Bruxelles GL, Peacock WJ, Dennis ES, Dolferus R. Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 1996;111:381–391. doi: 10.1104/pp.111.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, Jacobs M, Peacock W, Dennis E. Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol. 1994;105:1075–1087. doi: 10.1104/pp.105.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew MC. Sensing soil oxygen. Plant Cell Environ. 1990;13:681–693. [Google Scholar]
- Drew MC, Jackson MB, Giffard S. Ethylene-promoted adventitious rooting and development of cortical air spaces (aerenchyma) may be adaptive response to flooding in Zea mays L. Planta. 1979;153:217–224. doi: 10.1007/BF00384595. [DOI] [PubMed] [Google Scholar]
- Ferl R, Laughner B. In vivo detection of the regulatory factor binding sites of Arabidopsis thaliana Adh. Plant Mol Biol. 1989;12:357–366. doi: 10.1007/BF00017576. [DOI] [PubMed] [Google Scholar]
- Freeling MM. Simultaneous induction by anaerobiosis or 2,4-D of multiple enzymes specified by two unlinked genes: differential ADH1-ADH2 expression in maize. Mol Gen Genet. 1973;127:215–227. doi: 10.1007/BF00333761. [DOI] [PubMed] [Google Scholar]
- Freeling MM, Bennett DC. Maize Adh1. Annu Rev Genet. 1985;19:297–323. doi: 10.1146/annurev.ge.19.120185.001501. [DOI] [PubMed] [Google Scholar]
- He C-J, Page WM, Drew MC. Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiol. 1996;112:463–472. doi: 10.1104/pp.112.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeren FU, Dolferus R, Wu Y, Peacock WJ, Dennis ES. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) low oxygen. Genetics. 1998;149:479–490. doi: 10.1093/genetics/149.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK. Genetic analysis of osomotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell. 1998;10:1151–1162. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M, Dolferus R, Van Den Bossche D. Isolation and biochemical analysis of ethyl methanesulfonate-induced alcohol dehydrogenase null mutants of Arabidopsis thaliana (L.) Heynh. Biochem Genet. 1988;26:105–122. doi: 10.1007/BF00555492. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin SHF, Armstrong W. The anatomical characteristics of roots and plant response to soil flooding. New Phytol. 1987;106:465–495. [Google Scholar]
- Kennedy RA, Rumpho M, Fox TC. Anaerobic metabolism in plants. Plant Physiol. 1992;100:1–6. doi: 10.1104/pp.100.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Stam P. Genetic analysis. In: Koncz C, Chua N-H, Schell J, editors. Methods in Arabidopsis Research. Singapore: World Scientific; 1992. pp. 83–99. [Google Scholar]
- Kyozuka J, Olive M, Peacock W, Dennis E, Shimamoto K. Promoter elements required for developmental expression of the maize Adh1 gene in transgenic rice. Plant Cell. 1994;6:799–810. doi: 10.1105/tpc.6.6.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-M, Culligan K, Dixon RA, Chory J. CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell. 1995;7:1599–1610. doi: 10.1105/tpc.7.10.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard B, Rival J, Pradet A. Rice alcohol cytosolic glyceraldehyde-3-phosphate dehydrogenase contains two subunits differentially regulated by anaerobiosis. Plant Mol Biol. 1989;12:131–139. doi: 10.1007/BF00020498. [DOI] [PubMed] [Google Scholar]
- Rivoal J, Thind S, Pradet A, Ricard B. Differential induction of pyruvate decarboxylase subunits and transcripts in anoxic rice seedlings. Plant Physiol. 1997;114:1021–1029. doi: 10.1104/pp.114.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DA, Sachs MM. Differential expression and sequence analysis of the maize glyceraldehyde-3-phosphate dehydrogenase gene family. Plant Cell. 1989;1:793–803. doi: 10.1105/tpc.1.8.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs MM, Freeling M, Okimoto R. The anaerobic proteins of maize. Cell. 1980;20:761–767. doi: 10.1016/0092-8674(80)90322-0. [DOI] [PubMed] [Google Scholar]
- Sachs MM, Subbaiah CC, Sabb IN. Anaerobic gene expression and flooding tolerance in maize. J Exp Bot. 1996;47:1–15. [Google Scholar]
- Sedbrook JC, Kronebusch PJ, Borisy GG, Trewavas AJ, Masson PH. Transgenic aequorin reveals organ-specific cytosolic Ca2+ responses to anoxia in Arabidopsis thaliana seedlings. Plant Physiol. 1996;111:243–257. doi: 10.1104/pp.111.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL (1982) Isolation of photorespiration mutants in Arabidopsis thaliana. In M Edelman, RB Hallick, N-H Chua, eds, Methods in Chloroplast Molecular Biology. Elsevier Biomedical Press, Amsterdam, The Netherlands, pp 129–138
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain containing transcriptional factor that binds to the C-repeat/DRE, a cis-acting DNA regu-latory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Bush DS, Sachs MM. Elevation of cytosolic calcium precedes anoxic gene expression in maize suspension-cultured cells. Plant Cell. 1994a;6:1747–1762. doi: 10.1105/tpc.6.12.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah CC, Zhang J, Sachs MM. Involvement of intracellular calcium in anaerobic gene expression and survival of maize seedlings. Plant Physiol. 1994b;105:369–376. doi: 10.1104/pp.105.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvenkens D, Van Montagu M, van Lijsebttens M. Agrobacterium tumefaciens-mediated transformation of Arabi-dopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Wu R. Rice alcohol dehydrogenase genes: anaerobic induction, organ specific expression and characterization of cDNA clones. Plant Mol Biol. 1989;13:53–68. doi: 10.1007/BF00027335. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kwon HB, Peng H-P, Shih M-C. Stress responses and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiol. 1993;101:209–216. doi: 10.1104/pp.101.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K, Hasegawa PM, Bressan RA. Molecular aspects of osmotic stress in plants. Crit Rev Plant Sci. 1997;16:253–277. [Google Scholar]
- Zhu J-K, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell. 1998;10:1181–1192. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]