Abstract
Open reading frame 1 of Bamboo mosaic virus (BaMV), a Potexvirus in the alphavirus-like superfamily, encodes a 155-kDa replicase responsible for the formation of the 5′ cap structure and replication of the viral RNA genome. The N-terminal domain of the viral replicase functions as an mRNA capping enzyme, which exhibits both GTP methyltransferase and S-adenosylmethionine (AdoMet)-dependent guanylyltransferase activities. We mutated each of the four conserved amino acids among the capping enzymes of members within alphavirus-like superfamily and a dozen of other residues to gain insight into the structure-function relationship of the viral enzyme. The mutant enzymes were purified and subsequently characterized. H68A, the mutant enzyme bearing a substitution at the conserved histidine residue, has an ∼10-fold increase in GTP methyltransferase activity but completely loses the ability to form the covalent m7GMP-enzyme intermediate. High-pressure liquid chromatography analysis confirmed the production of m7GTP by the GTP methyltransferase activity of H68A. Furthermore, the produced m7GTP sustained the formation of the m7GMP-enzyme intermediate for the wild-type enzyme in the presence of S-adenosylhomocysteine (AdoHcy), suggesting that the previously observed AdoMet-dependent guanylation of the enzyme using GTP results from reactions of GTP methylation and subsequently guanylation of the enzyme using m7GTP. Mutations occurred at the other three conserved residues (D122, R125, and Y213), and H66 resulted in abolition of activities for both GTP methylation and formation of the covalent m7GMP-enzyme intermediate. Mutations of amino acids such as K121, C234, D310, W312, R316, K344, W406, and K409 decreased both activities by various degrees, and the extents of mutational effects follow similar trends. The affinity to AdoMet of the various BaMV capping enzymes, except H68A, was found in good correlations with not only the magnitude of GTP methyltransferase activity but also the capability of forming the m7GMP-enzyme intermediate. Taken together with the AdoHcy dependence of guanylation of the enzyme using m7GTP, a basic working mechanism, with the contents of critical roles played by the binding of AdoMet/AdoHcy, of the BaMV capping enzyme is proposed and discussed.
Bamboo mosaic virus (BaMV), a member of the Potexvirus group, has a positive-strand RNA genome (∼6.4 kb) with a 5′ m7G(5′)ppp(5′)G cap structure and a 3′ poly(A) tail (16). The 4.1-kb open reading frame 1 of BaMV encodes a 155-kDa polypeptide that has been postulated to be involved in the replication of the viral genome and the formation of cap structure at the 5′ ends of viral transcripts. Recently, biochemical studies demonstrated that the N-terminal 442 amino acids of the 155-kDa viral protein possess both enzymatic activities of GTP methyltransferase and S-adenosylmethionine (AdoMet)-dependent guanylyltransferase (13, 14). Residues 514 to 892 contain nucleoside triphosphate-binding and helicase-like motifs, and a recombinant protein containing this region not only has nucleoside triphosphatase activities but also has an RNA 5′-triphosphatase activity that specifically cleaves the γ phosphate off from the 5′ end of nascent RNA (14). The C-terminal 472 amino acids specifically recognize the 3′-untranslated region of BaMV genome (8) and can perform RNA-dependent RNA synthesis (12). According to these observations, formation of the 5′ cap in RNA transcripts of BaMV requires the sequential actions of the methyltransferase to transfer a methyl group from AdoMet to N7 position of GTP, followed by the guanylyltransferase activity to transfer the m7GMP moiety of the m7GTP to the 5′-diphosphate terminus of the viral RNA. Cap formation by BaMV is therefore distinct from that of eukaryotic cellular systems in which the 5′-diphosphate end of RNA is capped first by GTP:mRNA guanylyltransferase, and then the G(5′)ppp(5′)G cap of RNA is methylated by RNA (guanine-N7) methyltransferase (22, 25). The BaMV capping apparatus is also completely different from that of eukaryotic cellular systems with regard to protein primary sequence and genetic organization. The RNA 5′-triphosphatase activity of BaMV is recruited from the central helicase-like domain, and both the methyltransferase and guanylyltransferase activities are executed by a single capping enzyme domain located at the N terminus of the 155-kDa viral protein. In contrast, cellular guanylyltransferase and methyltransferase, from fungi, plants, or metazoans, are encoded by separate polypeptides, and each of the enzymes is structurally conserved during evolution (26).
The order of the cap formation found in BaMV has been demonstrated in other members of the alphavirus-like superfamily, such as Semliki Forest virus (1, 10), Hepatitis E virus (20), Tobacco mosaic virus (21), and Brome mosaic virus (3, 9). Therefore, it is likely that the capping enzymes within the alphavirus-like superfamily have the same catalytic mechanism in spite of the fact that only limited amino acid identities are conserved (24). Elucidating the structure-function relationship of this class of capping enzyme is necessary for completely understanding the viral replication and translation. As a first step toward understanding the capping enzyme domain of BaMV, site-directed mutagenesis was performed, and the roles of several key amino acid residues are discussed here.
MATERIALS AND METHODS
Chemicals.
General chemicals and nucleotides such as GTP, GDP, GMP, m7GTP, m7GDP, S-adenosylhomocysteine (AdoHcy), and guanylylimidodiphosphate (GIDP) were purchased from Sigma. Guanosine-5′-[(α,β)-methyleno]triphosphate (GpCpp), and guanosine-5′-[(β,γ)-methyleno]triphosphate (GppCp)were from Jena Bioscience, whereas AdoMet was from Boehringer Mannheim. Ado[methyl-3H]Met and [α-32P]GTP are products of NEN. [α-32P]m7GTP, synthesized by H68A mutant in the presence of [α-32P]GTP and AdoMet, were purified by high-pressure liquid chromatography (HPLC) by using an anion exchanger (SAX column, 250 by 4.6 mm) from Phenomenex. Elution was performed at a flow rate of 1 ml min−1 with a 26-min linear gradient (40 to 500 mM) of KH2PO4 (pH 5.5), with detection at 254 nm.
Plasmids.
The BaMV capping enzyme domain with a C-terminal hexahistidine tag was produced in Saccharomyces cerevisiae harboring expression vector pYEB3H as described previously (14). Expression vectors for producing mutant capping enzymes were generated from pYEB3H by a PCR-based method (14) in which a pair of 5′-phosphorylated divergent primer with one of the primers containing mutagenic nucleotides at its 5′ end was used in amplification reactions. The desired mutations were confirmed by nucleotide sequencing.
Protein expression and purification.
The expression and purification of the BaMV capping enzyme was described previously (14). Briefly, the viral proteins from the membrane fraction of the recombinant yeast cells, which had been cultured in a galactose-containing medium, were solubilized in Sarkosyl buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM β-2-mercaptoethanol [β-2ME], 0.3% Sarkosyl NL30, and 10% glycerol). The protein was then purified with Ni2+-nitrilotriacetic acid resin and finally dialyzed in Sarkosyl buffer. Protein concentrations were detemined by comparing the intensity of the Coomassie blue-stained protein band following sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) with that of bovine serum albumin by using an imaging densitometer.
Activity assay.
Formation of the covalent m7GMP-enzyme intermediate was used as an indication of guanylyltransferase activity. The standard reaction was carried out at 30°C for 80 min in a 20-μl solution that contained 0.1 μg of the viral enzyme, 0.3 μM [α-32P]GTP (3,000 Ci/mmol), 50 mM Tris (pH 8.0), 5 mM dithiothreitol, 2 mM MgCl2, 10 mM KCl, 1.2% n-octyl-β-d-glucopyranoside, and 100 μM AdoMet, unless otherwise stated. The reaction was stopped by adding SDS to a final concentration of 2%, followed by 3 min of boiling. The reaction mixture was resolved by SDS-10% PAGE, and radiolabeled proteins were visualized by autoradiography or using a phosphorimager. The fraction of radiolabeled protein molecules was quantified as pixels against a standard curve of predetermined amounts of [α-32P]GTP versus their respective pixels determined by using a phosphorimager and by using the assumption that a protein molecule can be labeled only by one molecule of m7GMP. The assay conditions for AdoHcy-dependent guanylation of the enzyme using m7GTP was basically same as that described above, except that AdoMet was replaced by AdoHcy and [α-32P]GTP was replaced by [α-32P]m7GTP. Methyltransferase activity was measured by the monitoring the transfer of [3H]methyl from Ado[methyl-3H]Met to methyl acceptors (15). Reaction was carried out at 30°C for 30 min in a 50-μl solution that contained 0.5 μg of the viral enzyme, 37 mM Tris (pH 8.0), 110 mM NaCl, 3.7 mM β-2ME, 1.2% n-octyl-β-d-glucopyranoside, 0.22% Sarkosyl NL30, 7.4% glycerol, 2.75 μCi of Ado[methyl-3H]Met (70 Ci/mmol), and 10 mM methyl acceptor. SDS (12 μl of 10% solution) and 60 μl of 2 M NH4Cl were added to terminate the reactions, and the mixture were extracted with three 250-μl volumes of phenol. A 100-μl aliquot of the aqueous phase was then mixed with 5 ml of scintillation cocktail solution, and the amount of the radiolabeled acceptor was determined by scintillation counting.
UV-cross-linking of Ado[methyl-3H]Met to the viral enzymes.
The UV cross-linking reaction was adapted from that of Luongo et al. (17) and performed by irradiating a 20-μl solution on ice that contained 0.15 μg of the viral enzyme, 2.75 μCi of S-Ado[methyl-3H]Met (70 Ci/mmol), 25 mM Tris (pH 8.0), 75 mM NaCl, 2.5 mM β-2ME, 0.15% Sarkosyl NL30, 5% glycerol, 2 mM EDTA, and 2 mM dithiothreitol by using a single 15-W germicidal UV lamp held at a distance of 10 cm for 30 min. The resulting products were analyzed by SDS-10% PAGE and visualized by fluorography. [γ-32P]GTP or [α-32P]GTP, instead of S-Ado[methyl-3H]Met, was used in the reaction buffer for GTP-binding assay, and the results were visualized by autoradiography.
RESULTS
Mutation of the capping enzyme domain of BaMV replicase.
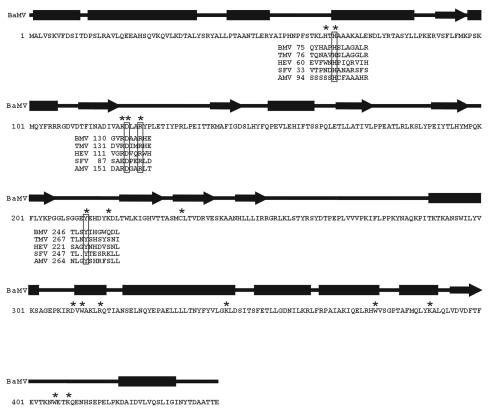
Previous studies demonstrated that the N-terminal 442 amino acids of the BaMV replicase, expressed in S. cerevesiae, exhibited both GTP methyltransferase and AdoMet-dependent guanylyltransferase activities (13, 14). The sequence of capped mRNA formation in BaMV was proposed to be: (i) the methylation of GTP; (ii) formation of a covalent m7GMP-enzyme intermediate; and (iii) transguanylation of m7GMP to the 5′ end of diphosphate RNA from the covalent intermediate (14). In the present study, we used mutational analysis to define amino acid residues critical for the BaMV capping enzyme activities. The residues indicated in Fig. 1 were mutated to alanine. They contain amino acids conserved in the methyltransferase domain among members of alphavirus-like superfamily: H68, D122, R125, and Y213. Lysine residues at positions 121, 218, 344, and 389 were also selected as targets because they are within sequences similar to K×DG and KDLS motifs, which may, respectively, link covalently to GMP during guanyl transfer by mRNA capping enzymes of vaccinia virus (23), Chlorella virus (7), and S. cerevisiae (6) or the mRNA capping enzyme in reovirus (18, 19). In addition, we mutated the sole cysteine residue, C234, in the BaMV capping enzyme, since the palmitoylated cysteine residues in the Semliki Forest virus capping enzyme enhance the binding of the protein to membrane (11). Several tryptophans (W312, W377, and W406) were also mutated in the present study because they may participate in AdoMet binding as in the case of cytosine-DNA methyltransferase (5).
FIG. 1.
Primary and predicted secondary structures of the BaMV capping enzyme. The secondary structures were predicted at the PSIPRED Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred/psiform.html). The rectangle and arrow symbolize the α-helix and β-strand, respectively. Amino acid sequences encompassing the conserved residues of several capping enzyme domains of viruses within alphavirus-like superfamily are aligned. BaMV, BMV, TMV, HEV, SFV, and AMV are abbreviations for Bamboo mosaic virus, Brome mosaic virus, Tobacco mosaic virus, Hepatitis E virus, Semliki Forest virus, and Alfalfa mosaic virus, respectively. The residues denoted by asterisk were subjected to mutational analysis in the present study.
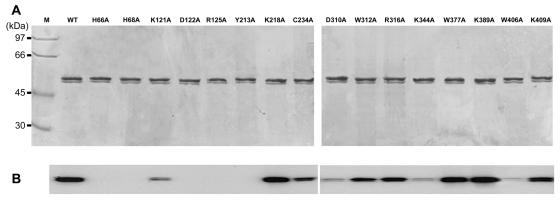
All proteins were purified based on a previously defined protocol, which includes steps of ultracentrifugation, detergent extraction, and metal-affinity chromatography (14) (Fig. 2A). The minor protein band immediately below the major band resulted from the degraded form of the viral enzyme according to Western blotting analysis (data not shown). All mutant proteins, including C234A, retained association with yeast membranes as strongly as the wild-type protein in a manner resistant to sequential extraction with alkaline (Na2CO3, pH 11.8) and high-salt (1 M NaCl) buffers (data not shown). Membrane association thus does not require C234. We also found no indication of palmitoylation of the BaMV capping enzyme when the recombinant yeast cells were grown in medium containing [3H]palmitic acid (data not shown).
FIG. 2.
Protein purification and formation of the covalent m7GMP-enzyme intermediate. (A) The yeast-expressed viral capping enzymes were purified through steps of membrane separation, ionic detergent extraction, and metal affinity chromatography as described in Materials and Methods. Each of the purified enzymes (0.2 μg) was resolved on SDS-PAGE (10% acrylamide) and stained by Coomassie blue. (B) The formation of the m7GMP-enzyme intermediate was assayed by incubating the various enzymes with [α-32P]GTP and AdoMet as described in Materials and Methods.
Mutational effects on the formation of the covalent m7GMP-enzyme intermediate with GTP and AdoMet.
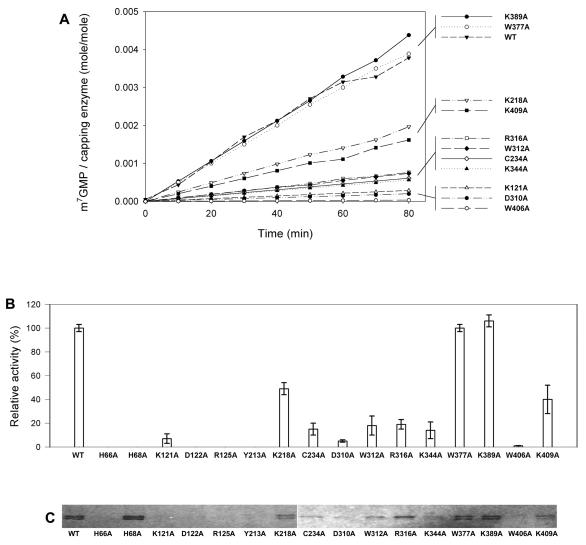
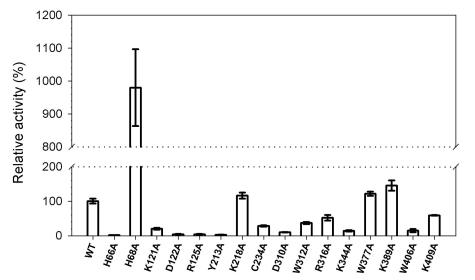
Formation of the covalent intermediate was examined by incubating the enzymes with [α-32P]GTP in the presence of AdoMet. It should be noted that GTP methyltransferase activity might be a prerequisite for the formation of the covalent m7GMP-enzyme intermediate under the reaction conditions. The apparent effects of mutations can be roughly grouped into three categories (Fig. 2B): (i) abolishment of the activity (in proteins with mutations at residue H66, H68, D122, R125, or Y213), (ii) decreases of activity in various extents (in proteins with mutation at K121, C234, D310, W312, R316, K344, W406, or K409), and (iii) minor effects on the activity (mutation at K218, W377, or K389).
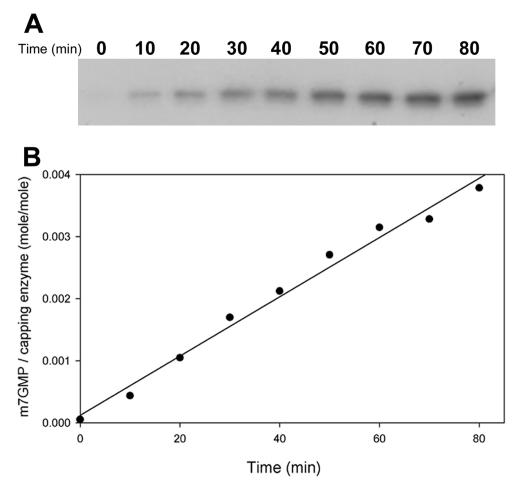
The amounts of radiolabeled protein were quantified over an 80-min period to carefully calculate the formation rate of the covalent m7GMP-protein intermediate. The accumulation of radiolabeled wild-type protein increased linearly throughout the reaction, with a total yield of 0.3% of the input protein in an hour period (Fig. 3). The rates of mutant proteins were determined as was the case in wild type (Fig. 4A), and the relative activities of various BaMV capping enzymes are illustrated in Fig. 4B and also shown in Table 1. Mutation at any of the conserved residues found in alphaviral capping enzymes (H68, D122, R125, or Y213) or at the nonconserved residue H66 completely inactivated the formation of covalent intermediate. Mutation at W406 remained ∼1% activity of the wild type, whereas the activity for those mutations at K218, K409, R316, W312, C234, K344, K121, or D310 decreased from ∼2- to 20-fold. Mutations at W377 or K389 did not have obvious effects.
FIG. 3.
Time course of the formation of the covalent m7GMP-enzyme intermediate of the wild-type enzyme. The activity assays were carried out at 30°C by incubating the wild-type enzyme with [α-32P]GTP and AdoMet for various times as described in Materials and Methods. (A) The reaction products were analyzed by SDS-PAGE (on 10% acrylamide), followed by autoradiography. (B) The amount of [α-32P]GMP linked to enzyme was quantified according to a standard curve of predetermined amounts of [α-32P]GTP versus their respective pixels by phosphorimager. The molar ratios of protein molecule that got radiolabeled are indicated in the ordinate, and the reaction times are indicated in the abscissa.
FIG. 4.
Effects of mutations on the formation of the covalent m7GMP-enzyme intermediate and the AdoMet-binding ability. (A) The reaction rates of guanylation of the enzymes in the presence of [α-32P]GTP and AdoMet were determined from time course studies. The data of H66A, H68A, D122A, R125A, and Y213A are omitted because of their disabilities. (B) The relative activities of various mutants compared to the wild type are illustrated. The data are means of three independent experiments. (C) The binding strength of various BaMV enzymes to AdoMet was assayed by a UV cross-linking reaction as described in Materials and Methods.
TABLE 1.
Mutational effects on GTP methylation, guanylation of the protein, and AdoMet-binding ability
| Wild type or mutant | GTP methylation (%)a | Guanylation (%) in the presence of:
|
Binding of AdoMetd | |
|---|---|---|---|---|
| m7GTP and AdoHcyb | GTP and AdoMetc | |||
| Wild type | 100 | 100 | 100 | ++++ |
| H66A | <1 | ∼0 | ∼0 | − |
| H68A | 980 | ∼0 | ∼0 | +++++ |
| K121A | 20 | 2 | 7 | − |
| D122A | 3 | ∼0 | ∼0 | − |
| R125A | 3 | ∼0 | ∼0 | − |
| Y213A | 2 | ∼0 | ∼0 | − |
| K218A | 120 | 73 | 49 | ++ |
| C234A | 28 | 21 | 15 | + |
| D310A | 10 | 12 | 5 | ± |
| W312A | 37 | 35 | 18 | +± |
| R316A | 51 | 34 | 19 | +++ |
| K344A | 14 | 12 | 14 | + |
| W377A | 120 | NDe | 100 | +++± |
| K389A | 150 | ND | 110 | +++± |
| W406A | 14 | 8 | 1 | ± |
| K409A | 59 | ND | 40 | ++ |
GTP methyltransferase activity (reaction 1) was measured based on the amount of methyl group transferred from S-ado[methyl-3H]Met to GTP. The data are from Fig. 5.
Guanylation of the enzyme when [α-32P]m7GTP and AdoHcy were used as substrates (reaction 2). The data are derived from the respective pixels shown in Fig. 11. Activities under the detectable level are presented as ∼0%.
Guanylation of the enzyme when [α-32P]GTP and AdoMet were used as substrates (reaction 3, which is presumably equivalent to reaction 1 plus reaction 2). The data are from Fig. 4B. Activities under the detectable level are presented as ∼0%.
AdoMet-binding abilities of the enzymes were estimated from the UV cross-linking data shown in Fig. 4C. The symbol “+” and “±” indicate approximate 25 and 10% binding abilities, respectively, of that of the wild-type enzyme, whereas “−” indicates an indiscernible binding ability.
ND, not determined.
The binding activity of mutant proteins to [γ-32P]GTP, [α-32P]GTP, or Ado[methyl-3H]Met were analyzed by UV cross-linking experiments. The results for GTP-binding ability are as yet unavailable, despite extensive efforts. The presumed effects on AdoMet binding appear to correlate roughly with the rates of forming the m7GMP-enzyme intermediate (Fig. 4C and Table 1). For instance, no obvious AdoMet binding could be detected on mutants incapable of forming the covalent intermediate such as H66A, D122A, R125A, and Y213A. An exception to this is the protein H68A, which binds AdoMet in a similar strength as the wild type.
Effects of mutations on methyltransferase activity.
The methyltransferase activities of the various recombinant BaMV proteins were determined by the transfer of 3H-methyl group from Ado[methyl-3H]Met to GTP (Fig. 5). Mutations at H66, D122, R125, or Y213 resulted in barely detectable activities, whereas mutations at K218, K389, or W377 had activities comparable to that of wild type. Mutant proteins with intermediate GTP methyltransferase activities exhibited a trend that correlated well with their abilities on formation of the covalent m7GMP-protein intermediate (K409A > R316A > W312A > C234A > K344A > W406A ≈ D310A) (Fig. 4 and 5).
FIG. 5.
Effects of mutations on the GTP methyltransferase activity. The activity of transferring a methyl group from AdoMet to GTP was assayed as described in Materials and Methods. The activity of the wild type (WT) (5,500 ± 260 cpm) against background (1,900 ± 250 cpm) is referred to as 100% activity. The data are means of three independent experiments.
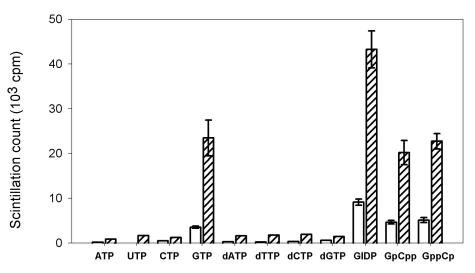
The H68A mutant is interesting in that it had an ∼10-fold increase in GTP methyltransferase activity (Fig. 5), although it was unable to form the covalent m7GMP-protein intermediate (Fig. 2 and 4). The increase of the apparent GTP methyltransferase activity of H68A may be attributed to an increase of the activity itself and/or an accumulation of m7GTP due to an inability of H68A to carry out the next transguanylation reaction. The specificity of methyl acceptors catalyzed by H68A was investigated (Fig. 6). Similar to wild type, H68A preferred to use GTP and its nonhydrolyzable analogues (GIDP, GpCpp, and GppCpp) as methyl acceptors (Fig. 6). Other ribo- and deoxyribonucleotides, including dGTP, were poor substrates. We noted that H68A and the wild-type protein had similar substrate preferences for methylation, with H68A being more active with all of the usable substrates. The greater activities of H68A than the wild type on methylating GIDP, GpCpp, and GppCp suggest that the methyltransferase activity of H68A actually increased.
FIG. 6.
Substrate specificity of the methyltransferase activity. Various nucleotide analogs of GTP were used as acceptors of methyl group under the reaction conditions of methyltransferase as described in Materials and Methods. GIDP, GpCpp, and GppCp are nonhydrolyzable analogues of GTP. The data in the figure are results after subtraction of the backgrounds (1,900 ± 250 cpm). Bars: □, wild-type enzyme; ▨, H68A. The data are means of three independent experiments.
Identification of m7GTP from the reaction products of H68A.
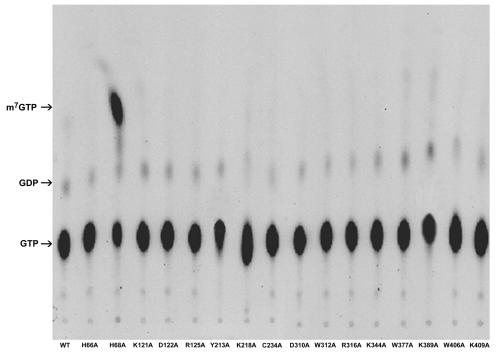
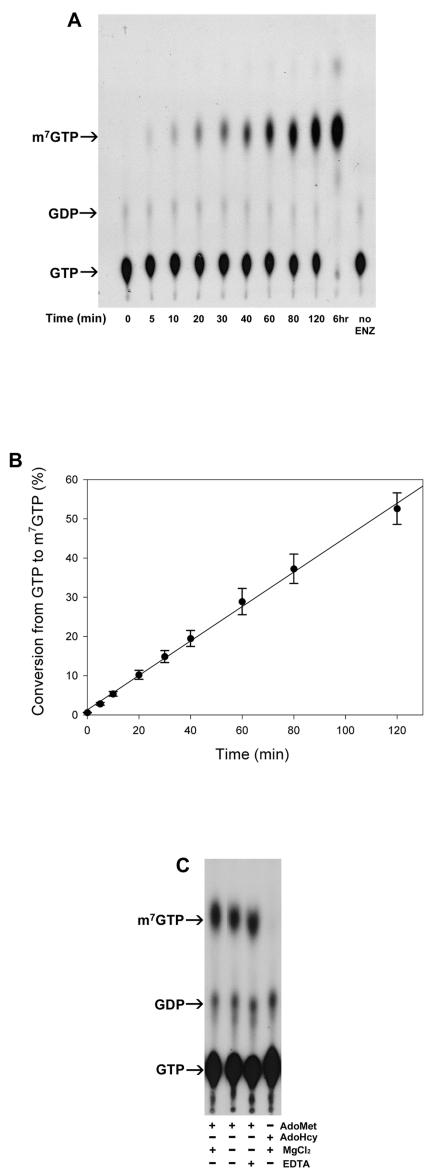
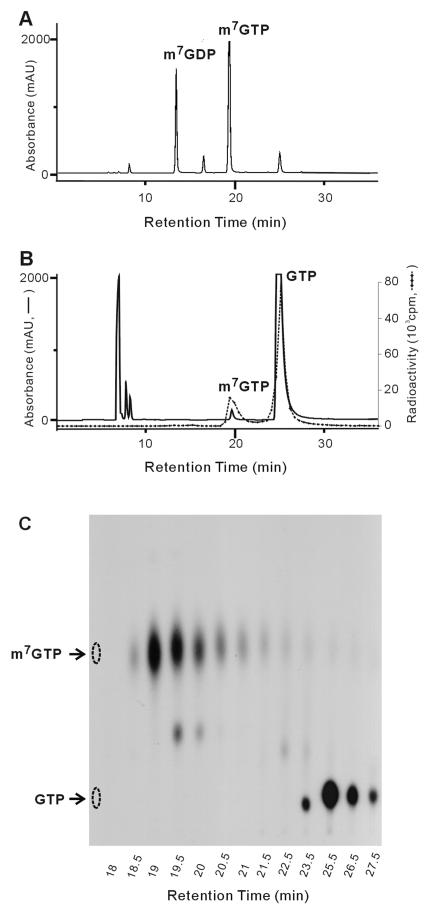
The increase of GTP methytransferaes activity of H68A prompted us to analyze the products of reactions with [α-32P]GTP and AdoMet as substrates. Abundant products of H68A migrated to the same position in a thin-layer chromatograph, as did m7GTP (Fig. 7). The amount of this product increased with the reaction times of up to 120 min (Fig. 8A). The percentage of conversion from [α-32P]GTP to [α-32P]m7GTP was determined by counting the radioactivities of the respective spots with a scintillation counter, and the result shows 50% of GTP being transformed to m7GTP in 2 h (Fig. 8B). The production of m7GTP depended on the presence of AdoMet but was largely unaffected by the presence of 5 mM EDTA and did not require exogenously provided Mg2+ (Fig. 8C). The identity of m7GTP was confirmed by HPLC analysis by using an anion-exchange column (SAX; Phenomenex). The retention time (19 min) was the same as that of the m7GTP standard (Fig. 9), suggesting that H68A did indeed produce m7GTP.
FIG. 7.
Product analysis of the reactions containing various BaMV capping enzymes, [α-32P]GTP, and AdoMet by TLC. Reactions were based on a standard assay for the formation of the covalent m7GMP-enzyme intermediate as described in Materials and Methods. At the ends, proteins were removed by phenol-chloroform extraction, and products in the aqueous phase were spotted onto a polyethyleneimine-cellulose TLC plate, developed with 1.2 M LiCl2, and visualized by autoradiography. The nucleotide standards were visualized by UV (254 nm) illumination, and arrows indicate their migration positions.
FIG. 8.
Production of m7GTP by H68A. (A) Reactions containing H68A, [α-32P]GTP, and AdoMet were carried out for various times under standard assay conditions for the formation of the covalent m7GMP-enzyme intermediate. The reaction products were analyzed by TLC. (B) The fraction of conversion was determined by comparing the amounts of [α-32P]m7GTP produced with that of residual [α-32P]GTP. (C) AdoMet, AdoHcy, MgCl2, and EDTA were included at different combinations, as indicated in the reaction mixture. The final concentrations of AdoMet and AdoHcy were 100 μM, whereas Mg2+ and EDTA, where present, were at 5 mM. Arrows indicate the migration positions of standards m7GTP, GDP, and GTP on the TLC plate.
FIG. 9.
Separation of the reaction products of H68A by HPLC. The reaction of H68A with [α-32P]GTP and AdoMet and the operation conditions of HPLC were as described in Materials and Methods. (A) Elution profile of a mixture of m7GTP and m7GDP standards. (B) Elution profile of the reaction products of H68A. To obtain enough m7GTP for UV detection, the reaction products for HPLC analysis were a mixture of reactions with 0.3 μM [α-32P]GTP and 0.1 mM AdoMet and with 1 mM GTP and 1 mM AdoMet. The solid line follows the absorption of 254 nm, whereas the dotted line denotes radioactivity. (C) The eluants after HPLC separation were examined by TLC. Arrows on the left side of the TLC plate indicate the migration positions of m7GTP and GTP standards.
Mutational effects on the formation of the covalent m7GMP-enzyme intermediate with m7GTP and AdoHcy.
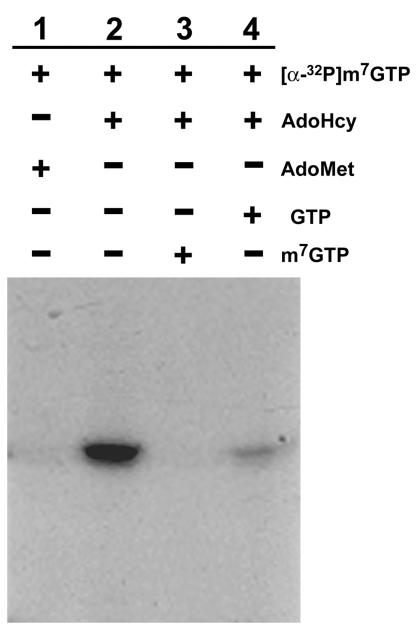
In the proposed model of mRNA capped pathway in BaMV, m7GTP is first formed by the GTP methyltransferase activity, and then the m7GMP portion of m7GTP is transferred by guanylyltransferase activity to the 5′ end of diphosphate RNA via a covalent intermediate of enzyme and m7GMP. According to this model, the BaMV capping enzyme could use m7GTP directly for forming the cap structure. This hypothesis was tested by incubating the wild-type enzyme with [α-32P]m7GTP purified from the reaction products of H68A by HPLC as described above. The covalent intermediate was observed in a reaction that required AdoHcy and used m7GTP as the substrate (Fig. 10, lane 2). AdoMet could not replace AdoHcy in this particular protein guanylation reaction (Fig. 10, lane 1). m7GTP and GTP decreased the reaction (Fig. 10, lanes 3 and 4) presumably by competing for access to the active site. The requirement of AdoHcy for the formation of the covalent intermediate when m7GTP was used as the substrate raised a question as to whether AdoMet and GTP could be formed during the reaction by AdoHcy accepting a methyl group from m7GTP. We did not observe such a reverse reaction by thin-layer chromatography (TLC) analysis of the reaction mixture of the wild-type capping enzyme, [α-32P]m7GTP, and AdoHcy (data not shown). Therefore, the AdoHcy-dependent guanylation of the enzyme using m7GTP might represent the first half reaction of m7GTP guanylyltransferase of the BaMV capping enzyme following the catalysis of m7GTP formation. Since the formation of the covalent m7GMP-enzyme intermediate could be assayed directly with m7GTP as the substrate, mutant enzymes that had impaired activity of forming the covalent intermediate using GTP and AdoMet were incubated with [α-32P]m7GTP in the presence of AdoHcy to determine whether the impairments resulted from defects of GTP methylation or transguanylation of m7GMP from m7GTP to the enzyme. The proteins with mutation at H66, H68, D122, R125, or Y213 were unable to form the covalent complex in the presence of m7GTP and AdoHcy (data not shown). Mutations of other tested residues reduced the activity of forming the covalent intermediate, and the adversely affected degree was in the order of K121 > W406 > K344, D310 > C234 > W312, R316 > K218 (Fig. 11 and Table 1), which was roughly comparable to the adverse effects on GTP methyltransferase activity.
FIG. 10.
Reaction conditions for the formation of the covalent m7GMP-enzyme intermediate when m7GTP was used as the substrate. The reactions, catalyzed by the wild-type enzyme, were carried out at 30°C for 80 min under conditions similar to those for the standard assay for the formation of the m7GMP-enzyme intermediate, except that [α-32P]GTP was replaced by [α-32P]m7GTP, and AdoMet, AdoHcy, GTP, and m7GTP (each 100 μM) were included in the reaction mixture at different combinations as indicated.
FIG. 11.
Effects of mutations on the formation of the covalent m7GMP-enzyme intermediate. The reactions containing various BaMV capping enzymes, [α-32P]m7GTP, and AdoHcy were carried out as described in Materials and Methods. [α-32P]m7GTP was purified by HPLC from the reaction mixture of H68A, [α-32P]GTP, and AdoMet.
DISCUSSION
Eukaryotic mRNA is capped at the 5′ end by an m7GMP moiety. This cap structure plays an important role in translation and mRNA stability. Crystal structures of the mRNA capping enzyme of chlorella virus have provided insights into the molecular details of the capping process of mRNA and clearly demonstrated that the lysine of K×DG motif links to GMP during transguanylation (7). The knowledge gained from chlorella virus capping enzyme may not be helpful in understanding the capping mechanism in viruses of alphavirus-like superfamily owing to the absence of both sequence similarity and the K×DG motif and to the presence of a GTP methyltransferase activity, a finding characteristic of the capping enzymes in the alphavirus-like superfamily. As an initial characterization of the functions of critical amino acids of the BaMV capping enzyme, 16 residues were individually substituted with alanine in the present study. The mutational effects on enzymatic properties are summarized in Table 1.
Based on the biochemical effects the mutants were classified into four groups. The first group includes mutations at H66, D122, R125, and Y213, which apparently affected activities of both the GTP methyltransferase and the formation of the covalent m7GMP-enzyme intermediate. D122, R125, and Y213 define residues conserved among alphavirus-like methyltransferases (24). The critical roles of the corresponding residues (or some of them) in the enzymes of Semliki Forest virus (2), Sindbis virus (27), and Brome mosaic virus (3, 4) have been demonstrated recently. Although H66 is not a conserved residue, it is as essential as D122, R125, and Y213 for the catalytic function of the BaMV capping enzyme. The second group is characterized by H68, in which the GTP methyltransferase activity increased by almost an order of magnitude, but the transguanylation activity was abolished. The production of m7GTP by H68A was confirmed by HPLC analysis, and the purified m7GTP was able to label the wild-type BaMV capping enzyme in the presence of AdoHcy. The activity of H68A strongly supports the idea that the activity of AdoMet-dependent guanylyltransferase results from sequential activities of GTP methyltransferase and m7GTP guanylyltransferase. The unique properties of H68A also suggest that the conserved H68 residue may form a phosphoamide bond to m7GMP during the catalytic pathway of transguanylation. Nonetheless, structural evidence such as peptide mapping is needed to test this hypothesis in the future. Amino acids of the third group (K121, C234, D310, W312, R316, K344, W406, and K409) are important, but not essential, for both activities of the GTP methyltransferase and the formation of the m7GMP-enzyme intermediate. The residual activities upon mutation are in a range of from ∼2 to 60%. In general, the extents of mutational effects on the two activities among these amino acids follow similar trends. Lastly, mutations at K218, W377, and K389 caused insignificant changes in enzymatic activities.
The results summarized above not only confirm the essential roles of the conserved residues but also illustrate the critical roles of the neighboring amino acids such as H66 and K121. Besides, the present study also identifies several important residues, such as C234, D310, W312, R316, K344, and W406. The amino acid segments encompassing H66 to H68, K121 to R125, Y213, and D310 to R316 may form part of the active site. The results also indicate the great importance of KDLA (amino acids 121 to 124) and KLDS (amino acids 344 to 347) motifs; nonetheless, their specific activities remained to be determined before a comparison to the KDLS motif of reovirus mRNA capping enzyme is possible.
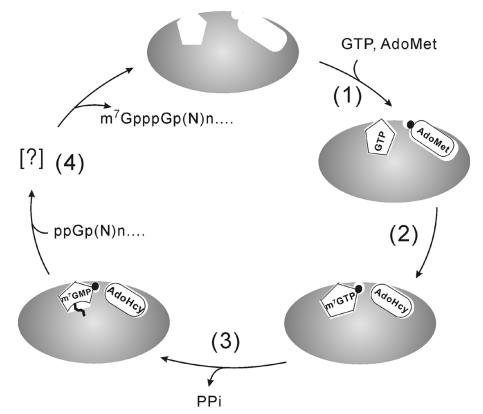
It was surprising to find that AdoHcy was needed for transfer of m7GMP from m7GTP to the enzyme. We hypothesize that AdoHcy induces a conformational change in the protein essential for either better binding to m7GTP or the catalysis of m7GMP transfer. AdoMet is likely to have the same role, raising the question of why it was unable to replace Adottcy. We propose that AdoMet and m7GTP could not bind simultaneously to the enzyme due to a steric hindrance between the methyl groups of the two substrates. This explanation is based on a reasonable assumption that the binding sites of AdoMet/AdoHcy and GTP/m7GTP are close to each other. In support of this model, comparisons of the enzymatic properties of the various mutants of the BaMV capping enzymes, except H68A, show that the affinity to AdoMet correlates not only with the GTP methyltransferase activity but also with the ability to form the m7GMP-enzyme adduct with either GTP-AdoMet or m7GTP-AdoHcy as substrates. This observation seems to disprove a second possible explanation of two separate sites for AdoMet and AdoHcy. In conclusion, a working model for formation of capped RNA in BaMV is proposed (Fig. 12) in which the binding pockets of AdoMet/AdoHcy and GTP/m7GTP are adjacent or even overlapped with some critical amino acids participating in the binding of the two substrates simultaneously. Binding of AdoMet may cause a conformational change in the protein that may enhance the binding of GTP or bring the two substrates to closer proximity, facilitating the transfer of the methyl moiety from AdoMet to GTP. Subsequently, the m7GMP moiety of m7GTP is transiently transferred to an unidentified active-site residue, whereas AdoHcy remains in the active site, keeping the enzyme in the right conformation. In the last step, the m7GMP is in turn transferred from the active-site residue to the 5′-diphosphate end of nascent RNA. According to this model, a mutation that greatly impairs the binding ability to AdoMet would also impair its ability to bind AdoHcy and consequently reduce the activities of not only GTP methyltransferase but also m7GTP guanylyltransferase. Nonetheless, biophysical and/or structural studies are necessary to validate and further elaborate on this model.
FIG. 12.
Schematic representation of the formation of capped RNA in BaMV. The proposed mechanism includes (step 1) binding of GTP and AdoMet, (step 2) transfer of methyl group from AdoMet to GTP, (step 3) formation of a phosphoamide bond between m7GMP and an active-site residue, and (step 4) transfer of m7GMP to the 5′-diphosphate end of viral RNAs.
Acknowledgments
This study was supported by grants NSC 90-2313-B-005-088 and NSC 91-2313-B-005-084 from the National Science Council of the Republic of China.
REFERENCES
- 1.Ahola, T., and L. Kääriäinen. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 92:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahola, T., P. Laakkonen, H. Vihinen, and L. Kääriäinen. 1997. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 71:392-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahola, T., and P. Ahlquist. 1999. Putative RNA capping activities encoded by Brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J. Virol. 73:10061-10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahola, T., J. A. den Boon, and P. Ahlquist. 2000. Helicase and capping enzyme active-site mutations in Brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 74:8803-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, X., S. Kumar, J. Posfai, J. W. Pflugrath, and R. J. Roberts. 1993. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell 74:299-307. [DOI] [PubMed] [Google Scholar]
- 6.Fresco, L. D., and S. Buratowski. 1994. Active site of the mRNA-capping enzyme guanylyltransferase from Saccharomyces cerevisiae: similarity to the nucleotidyl attachment motif of DNA and RNA ligase. Proc. Natl. Acad. Sci. USA 91:6624-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Håkansson, K., A. J. Doherty, S. Shuman, and D. B. Wigley. 1997. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzyme. Cell 89:545-553. [DOI] [PubMed] [Google Scholar]
- 8.Huang, C.-Y., Y.-L. Huang, M. Meng, Y.-H. Hsu, and C.-H. Tsai. 2001. Sequences at the 3′ untranslated region of Bamboo mosaic potexvirus RNA interact with the viral RNA-dependent RNA polymerase. J. Virol. 75:2818-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong, F., K. Sivakumaran, and C. Kao. 1999. The N-terminal half of the Brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology 259:200-210. [DOI] [PubMed] [Google Scholar]
- 10.Laakkonen, P., M. Hyvönen, J. Peränen, and L. Kääriäinen. 1994. Expression of Semliki Forest virus nsP1-specific methyltransferase in insect cells and in Escherichia coli. J. Virol. 68:7418-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laakkonen, P., T. Ahola, and L. Kääriäinen. 1996. The effects of palmitoylation on membrane association of Semliki Forest virus RNA capping enzyme. J. Biol. Chem. 271:28567-28571. [DOI] [PubMed] [Google Scholar]
- 12.Li, Y.-I., Y.-E. Cheng, Y.-L. Huang, C.-H. Tsai, Y.-H. Hsu, and M. Meng. 1998. Identification and characterization of the Escherichia coli-expressed RNA-dependent RNA polymerase of bamboo mosaic virus. J. Virol. 72:10093-10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, Y.-I., Y.-J. Chen, Y.-H. Hsu, and M. Meng. 2001. Characterization of the AdoMet-dependent guanylyltransferase activity that is associated with the N terminus of bamboo mosaic virus replicase. J. Virol. 75:782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, Y.-I., T.-W. Shih, Y.-H. Hsu, Y.-T. Han, Y.-L. Huang, and M. Meng. 2001. The helicase-like domain of plant Potexvirus replicase participates in formation of RNA 5′ cap structure by exhibiting RNA 5′-triphosphatase activity. J. Virol. 75:12114-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao, H. J., and V. Stollar. 1997. Methyltransferase activity of the insect orbivirus JKT-7400: photoaffinity labeling of a minor virion protein, VP4, with S-adenosylmethionine. Virology 235:235-240. [DOI] [PubMed] [Google Scholar]
- 16.Lin, N.-S., B.-Y. Lin, N.-W. Lo, C.-C. Hu, T.-Y. Chow, and Y.-H. Hsu. 1994. Nucleotide sequence of the genomic RNA of Bamboo mosaic potexvirus. J. Gen. Virol. 75:2513-2518. [DOI] [PubMed] [Google Scholar]
- 17.Luongo, C. L., C. M. Contreras, D. L. Farsetta, and M. L. Nibert. 1998. Binding site for S-adenosyl-l-methionine in a central region of mammalian reovirus λ2 protein: evidence for activities in mRNA cap methylation. J. Biol. Chem. 273:23773-23780. [DOI] [PubMed] [Google Scholar]
- 18.Luongo, C. L., K. M. Reinisch, S. C. Harrison, and M. L. Nibert. 2000. Identification of the guanylyltransferase region and active site in reovirus mRNA capping enzyme λ2. J. Biol. Chem. 275:2804-2810. [DOI] [PubMed] [Google Scholar]
- 19.Luongo, C. L. 2002. Mutational analysis of a mammalian reovirus mRNA capping enzyme. Biochem. Biophys. Res. Commun. 291:932-938. [DOI] [PubMed] [Google Scholar]
- 20.Magden, J., N. Takeda, T. Li, P. Auvinen, T. Ahola, T. Miyamura, A. Merits, and L. Kääriäinen. 2001. Virus-specific mRNA capping enzyme encoded by Hepatitis E virus. J. Virol. 75:6249-6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merits, A., R. Kettunen, K. Makinen, A. Lampio, P. Auvinen, L. Kääriäinen, and T. Ahola. 1999. Virus-specific capping of Tobacco mosaic virus RNA: methylation of GTP prior to formation of covalent complex p126-m7GMP. FEBS Lett. 455:45-48. [DOI] [PubMed] [Google Scholar]
- 22.Mizumoto, K., and Y. Kaziro. 1987. Messenger RNA capping enzymes from eukaryotic cells. Prog. Nucleic Acids Res. Mol. Biol. 34:1-28. [DOI] [PubMed] [Google Scholar]
- 23.Niles, E. G., and L. Christen. 1993. Identification of the vaccinia virus mRNA guanyltransferase active site lysine. J. Biol. Chem. 268:24986-24989. [PubMed] [Google Scholar]
- 24.Rozanov, M. N., E. V. Koonin, and A. E. Gorbalenya. 1992. Conservation of the putative methyltransferase domain: a hallmark of the “Sindbis-like” supergroup of positive-strand RNA viruses. J. Gen. Virol. 73:2129-2134. [DOI] [PubMed] [Google Scholar]
- 25.Shuman, S. 1995. Capping enzyme in eukaryotic mRNA synthesis. Prog. Nucleic Acids Res. Mol. Biol. 50:101-129. [DOI] [PubMed] [Google Scholar]
- 26.Shuman, S. 2002. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 3:619-625. [DOI] [PubMed] [Google Scholar]
- 27.Wang, H. L., J. O'Rear, and V. Stollar. 1996. Mutagenesis of the Sindbis virus nsP1 protein: effects on methyltransferase activity and viral infectivity. Virology 217:527-531. [DOI] [PubMed] [Google Scholar]