Abstract
Human immunodeficiency virus type 1 (HIV-1) assembly takes place at the plasma membrane of cells and is directed by the Pr55Gag polyprotein (Gag). One of the essential steps in the assembly process is the multimerization of Gag. We have developed a novel fluorescence resonance energy transfer (FRET) assay for the detection of protein-protein interactions between Gag molecules. We demonstrate that Gag multimerization takes place primarily on cellular membranes, with the majority of these interactions occurring on the plasma membrane. However, distinct sites of Gag-Gag interaction are also present at punctate intracellular locations. The I domain is a functional assembly domain within the nucleocapsid region of Gag that affects particle density, the subcellular localization of Gag, and the formation of detergent-resistant Gag protein complexes. Results from this study provide evidence that the I domain mediates Gag-Gag interactions. Using Gag-fluorescent protein fusion constructs that were previously shown to define the minimal I domain within HIV-1 Pr55Gag, we show by FRET techniques that protein-protein interactions are greatly diminished when Gag proteins lacking the I domain are expressed. Gag-Tsg101 interactions are also seen in living cells and result in a shift of Tsg101 to the plasma membrane. The results within this study provide direct evidence that the I domain mediates protein-protein interactions between Gag molecules. Furthermore, this study establishes FRET as a powerful tool for the detection of protein-protein interactions involved in retrovirus assembly.
Human immunodeficiency virus type 1 (HIV-1) encodes a structural polyprotein, Pr55Gag (Gag), that directs the retroviral assembly process. Following activation of the HIV protease, Gag is cleaved into the matrix (MA), capsid (CA), and nucleocapsid (NC) subunits. Additional Gag cleavage products include the p6 domain at the C-terminal end, as well as two spacer peptides, SP1 and SP2, located between CA and NC and NC and p6, respectively. Gag is synthesized on cytosolic ribosomes and then makes its way to the plasma membrane, where it acquires a lipid envelope and ultimately buds from the host cell (12). Intracellular assembly intermediate complexes of Gag oligomers have been proposed (21, 22, 27), but the site of formation and the functional role of these intermediate complexes are not yet established.
The uncleaved Gag polyprotein contains distinct functional domains that are essential for the HIV-1 assembly process. The membrane binding (M) domain is located within the MA region of Gag. The M domain is essential for mediating interactions of the Gag polyprotein with the inner leaflet of the plasma membrane, where both myristic acid and electrostatic interactions between basic residues within the N-terminal end of MA and negatively charged membrane phospholipids play a role (14, 46). The interaction (I) domain was originally defined as a region within NC of Rous sarcoma virus (RSV) Gag that is responsible for the production of particles of normal retroviral density (2, 31). The I domain plays a critical role in subcellular localization of Gag and in the formation of intracellular complexes of Gag protein (9, 34, 35). In the context of C-terminal truncations of Gag, the I domain can be localized to the first seven residues of NC (34, 35). The late (L) domain of Gag is located within the p6 region of HIV-1 Gag. The PTAP motif within the L domain of HIV-1 Gag has been shown to interact with Tsg101, and this interaction is essential for the final stages of particle budding (15, 37). Additional sites distinct from the M, I, and L domains have also been implicated in HIV assembly. A dimerization domain within the C-terminal structural domain of CA has been shown to be essential for Gag multimerization in vitro (16, 39, 42). Mutations within the major homology region (MHR), as well as mutations within the C-terminal end of the capsid outside of the multimerization domain itself, result in defects in particle assembly (10, 19, 24, 33, 38). In order to derive a more complete picture of retroviral assembly, we will need to define the specific contribution made by each of these functional domains.
Several distinct assays for Gag-Gag multimerization, including yeast two-hybrid analysis, rescue of nonmyristylated Gag or tagged Gag into particles, identification of Gag complexes in detergent-resistant membranes, velocity sedimentation centrifugation, in vitro capsid assembly, and coimmunoprecipitation and glutathione S-transferase pull-down experiments, have been employed in previous studies (4, 5, 9, 11, 23, 27, 44). However, an assay for the sensitive detection and localization of direct protein-protein interactions between Gag molecules in living cells has not been utilized. In this study, we employed a novel fluorescence resonance energy transfer (FRET) assay for the detection of HIV Gag-Gag interactions. FRET relies on the transfer of energy from a donor fluorophore to an acceptor fluorophore only when the two are within approximately 20 to 60 Å of each other (20). Both wild-type Gag and truncated versions of Gag were fused to cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) for analysis of protein-protein interactions by FRET. Using this approach, we have demonstrated that Gag-Gag multimers are detected at steady state exclusively in membrane fractions. FRET microscopy reveals that although the majority of Gag-Gag interactions occur at the plasma membrane, Gag-Gag interactions are also seen within distinct intracellular membrane compartments. Using Gag-fluorescent protein fusion constructs that were previously used to map the I domain, we now show that the I domain mediates Gag-Gag interactions in living cells. The results from this study establish that FRET is a powerful technique for the analysis of Gag-Gag interactions and indicate that FRET techniques can also be applied to the study of Gag interactions with cellular proteins such as Tsg101.
MATERIALS AND METHODS
Plasmid construction.
This study used Gag protein expression constructs fused to variants of the codon-optimized version of CFP, YFP, or a version of YFP with an alanine-to-lysine mutation at position 206 (A206K) and all variants of the codon-optimized version of enhanced green fluorescent protein (EGFP) (Clontech, Palo Alto, Calif.). The Gag coding sequences for all constructs were derived from the codon-optimized version of HXB2 Gag in expression plasmid pVRC3900 (17). The CFP and YFP expression constructs were created by first replacing the EGFP gene in pEGFP-N3 (Clontech) with the CFP gene from pECFP-N1 or the YFP gene from pEYFP-N1. These new constructs were designated pECFP-N3 and pEYFP-N3, respectively. PCR cloning was then used to amplify the gag gene from the pVRC3900 vector, with a HindIII site at the 5′ ATG and a BamHI site at the 3′ end. The amplified gag gene was then ligated into pEYFP-N3, pECFP-N3, and pEYFP(A206K) by digestion of the BamHI and HindIII sites located within the multiple cloning region. A schematic diagram of the Gag expression constructs employed in this study is shown in Fig. 1A. A farnesylated membrane protein control expression construct, pECFP-f, was constructed by replacing the NheI-BsrGI fragment of pEGFP-f (Clontech) with the NheI-BsrGI fragment from plasmid pECFP-nuc (Clontech). tsg101 was amplified from a HeLa cDNA library (Clontech) and inserted into pCDNA 3.1, and the sequence was verified by automated DNA sequencing. The tsg101 gene was then amplified from this vector by PCR to include a BamHI restriction site at the 3′ end and a HindIII restriction site at the 5′ end and was inserted into the BamHI and HindIII sites of pEYFP-N3.
FIG. 1.
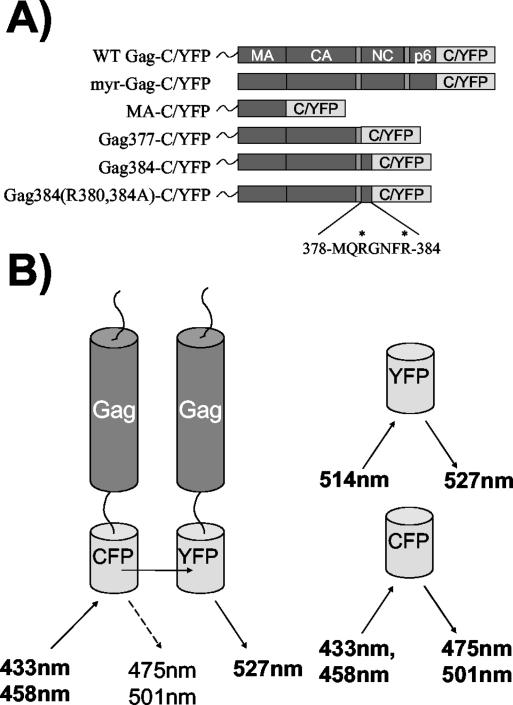
Schematic representation of Gag fusion proteins and Gag-Gag FRET. (A) Diagram of all Gag fusion expression constructs. All six constructs were made as CFP and YFP fusions. (B) Gag-Gag FRET is represented, with the Gag C terminus fused to CFP or YFP with a flexible linker (left). The excitation and emission wavelengths for CFP and YFP are shown to the right. Where two wavelengths are listed, the upper wavelength represents the primary excitation-emission peak and the lower represents the secondary peak.
The forward (sense strand) oligonucleotides used for the above PCR amplifications were GRCAAGCTTGTCGACATGGGCGCCCGCGCCAGC (forward oligonucleotide for all Gag constructs except Myr− Gag-C/YFP), GTCAAGCTTGTCGACATGGCCGCCCGCGCCAGC (Myr− Gag-C/YFP oligonucleotide), GAAGCTTCCATGGCGGTCTCGGA (tsg101 oligonucleotide for amplification and insertion into pcDNA 3.1), and GAAGCTTCCATGGCGGTGTCGGA (tsg101 oligonucleotide for insertion into pEYFP-N3 and pECFP-N3 vectors). Reverse oligonucleotides used for the above PCR amplifications were GGGGATCCTTGTGACGAGGGGTCGCTG (Gag-C/YFP and Myr− Gag-C/YFP), GGATCCGTAGTTCTGGCTCAC (MAC/YFP), CGGGATCCCATGATGGTGGCGCTGTTGGTCAC (Gag377-C/YFP), CGGGATCCGCGGAAGTTGCCGCGCTG (Gag384-C/YFP), CGGGATCCGGCGAAGTTGCCGGCCTGCATCATGATGGTGGC [Gag384(R380,384A)-C/YFP], and GGGATCCGTAGAGGTCACTGAG (Tsg101-C/YFP).
Cells and transfections.
293T cells were used for all fluorometer-based studies, while Mel JuSo cells (human melanoma cell line) (18) were used for all microscopic studies. Mel JuSo cells were provided to us by Markus Thali (University of Vermont). Both cell types were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum and antibiotics at 37°C in 5% CO2. 293T cells were grown in 100-cm2 tissue culture dishes, and Mel JuSo cells were grown in 35-cm2 glass-bottom tissue culture dishes coated with poly-d-lysine (MatTek, Ashland, Mass.). Transfections of all fusion constructs were performed by the calcium phosphate-BBS transfection method (7) or with Lipofectamine 2000 (Invitrogen, Carlsbad, Calif.), with either 10 μg (293T) or 2 μg (Mel JuSo) of total plasmid DNA.
Isolation of cytosolic and membrane fractions and analysis by scanning cuvette fluorometry.
Cells were harvested for analysis at 42 to 48 h posttransfection. Two 10-cm2 dishes of nearly confluent 293T cells were included for each experimental sample. Cells were washed in phosphate-buffered saline, allowed to swell in hypotonic buffer (10 mM Tris-Cl [pH 8.0] plus protease inhibitors) for 15 to 20 min on ice, and broken by Dounce homogenization. The buffer was then adjusted to 0.1 M NaCl, and the nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 10 min. Postnuclear supernatants containing cytosolic and membrane components were then adjusted to 50% iodixanol from a stock solution of 60% iodixanol (Nycomed Pharma, Oslo, Norway). Forty percent and ten percent solutions of iodixanol were layered on top of the 50% iodixanol layer. The preparation was centrifuged in a Beckman SW41 rotor at 41,000 rpm for 2 h at 4°C. The membrane fraction was taken from the 10%-40% iodixanol interface as a 1-ml sample. The cytosolic fraction was taken from the bottom of the tube as a 1-ml sample. Each sample was kept at 4°C and analyzed by fluorometry in a PTI T-format scanning cuvette spectrofluorometer (Photon Technology International, Lawrenceville, N.J.). For FRET analysis, samples were excited at 433 nm, and a resulting emission scan ranging from 450 to 550 nm was obtained. Samples were excited at 514 nm, with a resulting emission scan of 526 to 600 nm, for analysis of YFP emission to compare the relative amount of YFP present. Data were collected from at least three different independent experiments for each expression construct.
Laser confocal fluorescence microscopy.
Mel JuSo cells were grown and transfected in poly-d-lysine-coated glass-bottom cell culture dishes and imaged either as live cells or following fixation in methanol for 10 min. Live cells were maintained in phenol-red-free Dulbecco's modified Eagle medium with 10% fetal bovine serum and antibiotics at 37°C in 5% CO2. Images were obtained with a Zeiss LSM 510 laser scanning confocal microscope (Carl Zeiss Inc., Thornwood, N.Y.) equipped with a Meta multichannel detector, making pixel quantitation possible for a range of wavelengths. Emission scans were obtained by use of Zeiss LSM software. Image reconstruction was performed with Zeiss LSM software and Metamorph image analysis software (Universal Imaging Corporation, Downingtown, Pa.). For some experiments, the 514-nm laser was used to provide high-intensity pulses of light for photobleaching of YFP to specified regions of interest in the cell. For other experiments, the Zeiss LSM software package was used to provide images of CFP or YFP emission by use of the spectral unmixing feature. For this feature, a representative CFP or YFP curve was first generated as a standard, and then image data collected by the multichannel detector were processed to separate the emission spectra of CFP and YFP by use of digital deconvolution algorithms. This provided linear unmixing for resolution of the individual fluorescent protein signals.
Virus-like particle preparation and gradient centrifugation.
Gag fusion constructs were expressed in 293T cells in 10-cm2 dishes as described above. At 60 to 72 h posttransfection, the supernatants were collected, filtered through a 0.45-μm-pore-size filter, and layered on top of 20% sucrose in phosphate-buffered saline. The samples were centrifuged in a Beckman SW28 rotor at 28,000 rpm for 3 h at 4°C. The resultant pellet was resuspended in 1 ml of 1× NTE buffer (100 nM NaCl, 10 mM Tris [pH 8.0], 1 mM EDTA) and layered on top of a 20% to 60% linear sucrose gradient. The gradient and pelleted material were then subjected to centrifugation in a Beckman SW41 rotor at 41,000 rpm for 16 h, after which 20 equal fractions were taken. The bottom 16 samples were analyzed for sucrose density with a refractometer and by scanning fluorometry as described above.
RESULTS
Design of Gag-CFP and Gag-YFP fusion constructs.
In order to utilize FRET as a technique to directly assay specific Gag-Gag interactions, we expressed Gag proteins as both CFP fusion proteins and YFP fusion proteins. Fusion constructs were designed with either CFP or YFP fused to the C terminus of Gag via a small linker region. These fusion constructs retained the myristic acid modification at the N terminus of Gag, allowing proper M domain function. In addition to full-length Gag, a series of truncation mutants fused to both CFP and YFP were designed in order to assess the role of the I domain in mediating Gag-Gag interactions (Fig. 1A). MA-YFP and MA-CFP consist of only the matrix region of Gag fused to YFP or CFP. Gag377 is a truncated version of Gag expressing MA, CA, and SP1 but lacking the entire nucleocapsid region, including the I domain. Gag384 contains the first seven amino acids of the I domain, while Gag384(R380,384A) contains the first seven amino acids of the I domain, with arginine-to-alanine mutations at positions 380 and 384 (Fig. 1A). These two residues have been previously shown to be essential for the function of the I domain in the context of GFP fusion proteins (34). Myristic acid-deficient Gag-CFP and Gag-YFP fusion constructs were also created as membrane binding controls for both FRET microscopy and FRET fluorometry experiments.
The basis for Gag-CFP-Gag-YFP FRET is outlined in Fig. 1B. Upon stimulation of the energy donor, CFP, energy is transferred to the acceptor, YFP. YFP then emits light at its characteristic wavelength (peak at 527 nm). We hypothesized that the C-terminal location of these tags, together with the propensity of Gag molecules to align in an N-to-C-terminal radial array (41), would create efficient FRET pairs.
Gag-Gag interactions are observed in membrane fractions but not in cytosolic fractions.
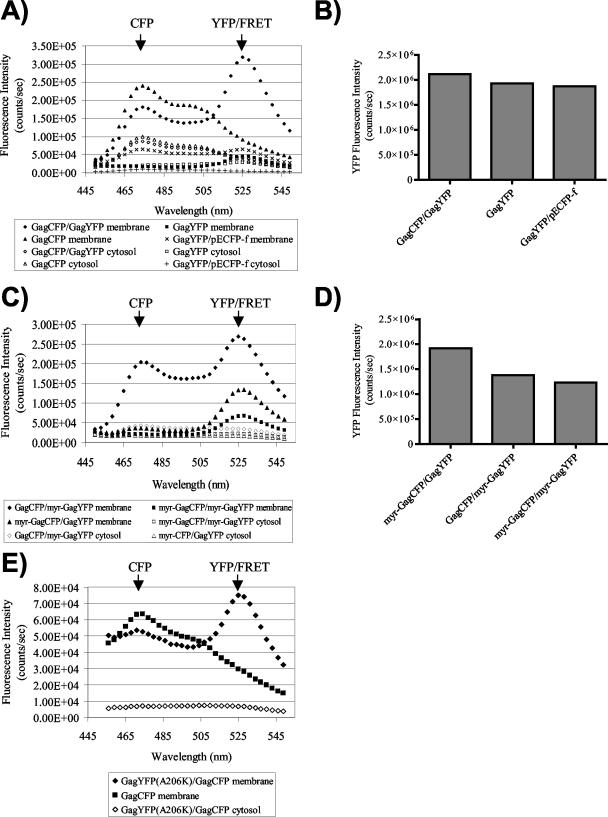
Previous studies have indicated that synthesized Gag proteins are rapidly membrane associated within cells. In order to determine if Gag-Gag interactions can be detected in membrane-free fractions, we employed FRET fluorometry to detect Gag-Gag interactions in both membrane and cytosolic cell preparations. Gag fusion constructs were expressed in 293T cells, followed by isolation of both a membrane fraction (prepared by flotation centrifugation) and a cytosolic fraction. Note that the isolated membrane fraction represents both the plasma membrane and intracellular membranes. The emission spectra of membrane or cytosolic fractions were then obtained in a spectrofluorometer, using an excitation wavelength of 433 nm. When Gag-YFP was coexpressed with Gag-CFP, the membrane fraction exhibited a curve representative of efficient fluorescence energy transfer (Fig. 2A, closed diamonds), while the cytosolic fraction resulted in a CFP emission peak with no YFP emission peak (Fig. 2A, open diamonds). The large CFP emission peak accompanied by the lack of a YFP emission peak for the cytosolic fraction represents non-membrane-bound Gag-CFP and further supports the role of cellular membranes in facilitating Gag-Gag interactions. Gag-YFP expressed alone showed no FRET-like curve for the cytosolic or membrane fractions (Fig. 2A, squares), and Gag-CFP alone also did not reveal a FRET curve (Fig. 2A, triangles). We then examined a control plasma-membrane-bound protein, ECFP-f, for the ability to transfer energy to Gag-YFP. The combination of ECFP-f and Gag-YFP in the membrane fraction resulted in only background levels of YFP emission (Fig. 2A). The lack of FRET between ECFP-f and Gag-YFP was not due to a lower expression level for Gag-YFP in this experiment, as indicated by very similar levels of maximal YFP emission upon excitation at the YFP excitation peak (Fig. 2B). These results establish that the Gag-CFP-Gag-YFP FRET curve in Fig. 2A represents a true Gag-Gag interaction.
FIG. 2.
Analysis of Gag-Gag interactions by FRET fluorometry. Membrane and cytosolic fractions were collected for each sample as described in Materials and Methods. Both fractions were analyzed in a scanning cuvette fluorometer. (A) Gag-CFP-Gag-YFP FRET curve for the membrane fraction (closed diamonds) and the cytosolic fraction (open diamonds). The YFP emission peak at 527 nm is indicated by the arrow. Squares, Gag-YFP alone; triangles, Gag-CFP alone. Gag-YFP coexpressed with pECFP-f is also shown for membrane (×) and cytosolic (+) fractions. (B) Relative levels of cellular YFP expression are shown for the experiment depicted in panel A, as determined by peak YFP output following excitation of cell lysates at 514 nm. (C) Wild-type Gag is able to efficiently interact with Myr− Gag in the membrane fraction (closed diamonds and triangles) but not in the cytosolic fraction (open diamonds and triangles). The rest of the symbols are as for panel A. (D) Relative levels of cellular YFP expression for the experiment shown in panel C, as determined by peak YFP output of cell lysates excited at 514 nm. (E) Gag-YFP(A206K)-Gag-CFP FRET curve for the membrane fraction (closed diamonds) and Gag-CFP curve for the membrane fraction (closed squares). Cytosolic fractions of Gag-YFP(A206K)-Gag-CFP are shown for comparison.
To further evaluate the membrane dependence of Gag-Gag interactions, we compared wild-type Gag-CFP and Gag-YFP interactions to those of myristic acid-deficient (Myr−) constructs. When Myr− Gag-CFP and Myr− Gag-YFP were expressed together, a low level of energy transfer between the pair was observed in the membrane fractions, and none was observed in the cytosolic fractions (Fig. 2C, closed versus open squares). This result is consistent with previous reports indicating that myristylation-deficient Gag associates with cellular membranes in an inefficient manner (21, 36), and it suggests that Gag-Gag multimer formation between myristic acid-deficient Gag proteins occurs with a low efficiency on cellular membranes. Wild-type Gag has previously been shown to rescue Myr− Gag to the plasma membrane and incorporate it into retrovirus-like particles (23). For examination of Gag-Gag interactions with this rescue model, myristic acid-deficient Gag fusion constructs were coexpressed with wild-type Gag fusion constructs, and the cytosolic and membrane fractions were isolated. As shown in Fig. 2C, the cytosolic fractions demonstrated no emission peak at the YFP wavelength (no FRET). Gag-CFP was able to efficiently rescue Myr− Gag-YFP into membrane-bound Gag complexes (Fig. 2C, closed diamonds). Gag-YFP was also able to rescue Myr− Gag-CFP, but with a lower efficiency (Fig. 2C, closed triangles). This difference in the efficiency of rescue was present in three independent experiments and remains unexplained. However, these results indicate that myristylated Gag can rescue nonmyristylated Gag to the membrane fraction, as expected, and demonstrate further that this rescue involves direct Gag-Gag interactions. The YFP emission peak for the Gag-CFP-Myr− Gag-YFP membrane pair was not due to excessive background excitation (bleedthrough) of YFP by the 433-nm laser, as the total levels of YFP expressed in each of the experiments were comparable (Fig. 2D).
As a final control for the influence of the fluorescent protein on Gag-Gag interactions, we obtained a multimerization-incompetent version of YFP (A206K) from the laboratory of David Piston at Vanderbilt University. This mutation disrupts the formation of GFP-GFP homodimers which may form at very high concentrations of GFP (43; data not shown). Figure 2E demonstrates that this mutation did not prevent Gag-CFP-Gag-YFP FRET in membrane fractions, and this observation reproduces the major findings of Fig. 2A. We conclude that the measurement of Gag-CFP-Gag-YFP FRET represents Gag-Gag interactions and is not a consequence of interactions between the fluorescent protein fusion partner.
Gag-Gag interactions are observed at the plasma membrane by FRET microscopy.
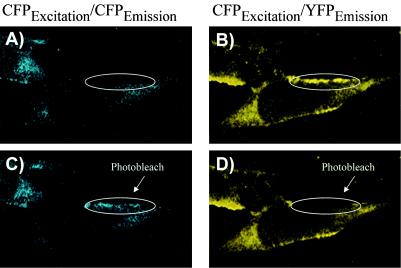
Confocal microscopy is a second method for the detection of Gag-Gag FRET. The advantage to microscopic assays is the ability to localize Gag-Gag interactions within living cells. The laser confocal microscope also allows the photobleaching of specific areas of the cell at 514 nm, the excitation wavelength of YFP. Photobleaching causes a depletion of the acceptor fluorophore (YFP) of the CFP-YFP FRET pair, resulting in an accompanying increase in CFP emission. An enhanced CFP signal following photobleaching thus represents another proof of protein-protein interactions measured by FRET (20). As opposed to the 433- and 475-nm primary excitation and emission wavelengths, the 453- and 501-nm secondary excitation and emission wavelengths for CFP were used in these microscopy experiments due to the available laser source. Gag-CFP and Gag-YFP were coexpressed in Mel JuSo cells and excited at 458 nm with the Zeiss LSM 510 confocal microscope. Mel JuSo cells were chosen for this analysis because they allow levels of both plasma membrane and intracellular particle assembly that can be easily detected at steady state and they have been used in other studies of Gag trafficking (30). Software deconvolution algorithms utilizing characteristic CFP and YFP curves from control cells allowed the specific emission from CFP versus YFP in cells to be resolved; a representative cell is presented in Fig. 3. Plasma membrane fluorescence was prominent for emission at the YFP wavelength (Fig. 3B), while it was quite weak for CFP (Fig. 3A), suggesting efficient energy transfer from CFP to YFP (and little CFP emission). Following photobleaching of YFP with the 514-nm laser, a clear increase in CFP emission was observed within the bleached region due to the loss of the YFP acceptor fluorophore and the release of CFP from the acceptor (Fig. 3C, circled region). Analysis of YFP output following photobleaching revealed a complete loss of the YFP signal (Fig. 3D). This experiment demonstrates that Gag-Gag interactions can be observed within cells by FRET microscopy.
FIG. 3.
Visualization of Gag-Gag FRET by confocal microscopy. Gag-CFP and Gag-YFP were cotransfected in Mel JuSo cells, and images were obtained with a Zeiss LSM 510 confocal microscope equipped with a Meta multichannel detector. Fluorescence excitation was carried out at 458 nm; emission images prepared by linear unmixing techniques for CFP (left panels) and YFP (right panels) are shown. (A) CFP emission image demonstrating weak membrane fluorescence in the indicated region before photobleaching. (B) FRET image indicating efficient plasma membrane Gag-Gag FRET prior to photobleaching of YFP. (C) CFP emission image following photobleaching of YFP in the circled region. Note the increase in CFP emission following YFP photobleaching within the indicated region. (D) FRET image following photobleaching of the indicated area of the cell.
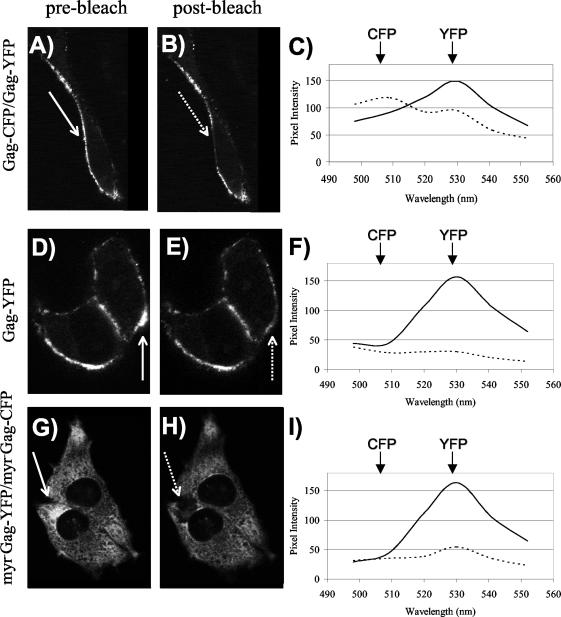
The Meta multichannel detector of the Zeiss LSM 510 confocal microscope allows spectral data to be collected for each point in the imaged cell. We used this feature to derive additional evidence for Gag-Gag interactions by FRET. Mel JuSo cells were transfected with Gag-CFP and Gag-YFP, Gag-YFP alone, or Myr− Gag-CFP and Myr− Gag-YFP. Shown in Fig. 4 are representative cells for each experiment, with the region of interest indicated before and after bleaching with a solid arrow and dashed arrow, respectively (Fig. 4A, B, D, E, G, and H). We then examined the spectral data collected in the region of interest pre- and postbleaching. Similar levels of YFP emission intensity were present in each cell before photobleaching (Fig. 4C, F, and I, solid lines), and a decrease in YFP emission intensity was apparent after photobleaching (Fig. 4C, F, and I, dotted lines). Following photobleaching, cells expressing Gag-CFP and Gag-YFP exhibited an increase in CFP emission intensity (Fig. 4C, dotted line). This increase in CFP emission reflects FRET between Gag molecules in the specified region of interest prior to bleaching. When Gag-YFP was expressed in the absence of Gag-CFP, the same decrease in YFP intensity was observed, with no accompanying increase in CFP intensity (Fig. 4F). Similarly, when Myr− Gag-CFP and Myr− Gag-YFP were coexpressed and photobleaching was performed in the cytoplasm of the cell, no increase in CFP emission intensity was observed (Fig. 4I). These studies provide further evidence of FRET between Gag molecules and indicate that Gag-Gag interactions can be effectively localized within the cell by this technique.
FIG. 4.
Analysis of Gag-Gag interactions by FRET microscopy and spectral analysis. (A) Gag-CFP and Gag-YFP were cotransfected in Mel JuSo cells, and images were obtained with a Zeiss LSM 510-Meta confocal microscope. The image represents YFP excitation-emission before photobleaching. The arrow indicates the selected plasma membrane region to be bleached. (B) The same cell as that shown in panel A is depicted following photobleaching at 514 nm. (C) Emission scans were obtained from the selected region of interest, with excitation at 458 nm (CFP excitation), before (solid line) and after (dashed line) photobleaching of YFP. Peak emission wavelengths for CFP (secondary peak) and YFP are indicated. (D) Gag-YFP distribution before bleaching. (E) Gag-YFP after photobleaching of indicated region. (F) Spectra obtained before (solid line) and after (dashed line) photobleaching of cells from panels D and E. (G) Cotransfection of Myr− Gag-YFP and Myr− Gag-CFP before photobleaching. (H) The same cell as that shown in panel G is depicted after photobleaching of indicated region of the cytoplasm. (I) Spectral analysis of the cell shown in panels G and H.
Gag-Gag FRET is present in VLPs.
Gag expression alone is sufficient for the production of virus-like particles (VLPs), and Gag-GFP expression results in the release of VLPs that resemble authentic immature HIV virions (34, 35). Since the Gag molecules making up these particles are closely associated, we expected to see a strong FRET signal from purified VLPs incorporating Gag-CFP and Gag-YFP. Gag-CFP and Gag-YFP were expressed in 293T cells, and supernatants were harvested, pelleted through sucrose, and then subjected to equilibrium density centrifugation followed by spectrofluorometry. Efficient Gag-Gag FRET was observed for fractions corresponding to those of typical retroviral density when excited at 433 nm (Fig. 5A, gray bars). In contrast, VLPs consisting of Gag-YFP alone produced a low YFP emission signal, representing direct Gag-YFP excitation from the 433-nm wavelength as opposed to energy transfer from the Gag-CFP molecule (Fig. 5A, black bars). We then compared the total amounts of Gag-YFP in the YFP-alone particles and in the CFP-YFP particles (Fig. 5B). When the fractions were excited at 514 nm, Gag-YFP particles showed an intense fluorescence signal at 527 nm, while the particles consisting of both Gag-YFP and Gag-CFP displayed a fourfold lower fluorescence signal. The higher levels of YFP from the Gag-YFP-alone particles proves that the major signal seen in Fig. 5A for the Gag-YFP and Gag-CFP particles is not due to the direct excitation of YFP at 433 nm, but is rather due to energy transfer. Although the detection of Gag-Gag interactions within VLPs is hardly surprising, these data provide further validation of the Gag-Gag FRET technique and should facilitate future studies of protein-protein interactions occurring within VLPs.
FIG. 5.
Analysis of Gag-Gag interactions in VLPs by FRET fluorometry. VLPs were produced by cotransfection of Gag fusion proteins in 293T cells. At 48 h posttransfection, the supernatants were pooled and pelleted through 20% sucrose. The resuspended pellets were layered on top of a 20% to 60% linear sucrose gradient and centrifuged to equilibrium. Sixteen equal fractions were collected and analyzed by scanning cuvette fluorometry. (A) The fluorescence emission intensity at 527 nm is shown for both Gag-YFP and Gag-CFP particles upon excitation at 433 nm (CFP-YFP FRET). (B) Fluorescence intensity of the same particles as for panel A following excitation at 514 nm (YFP peak).
Gag-Gag interactions are observed at intracellular locations.
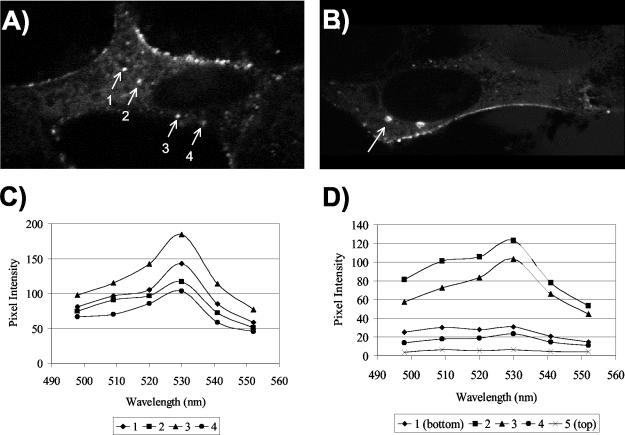
The majority of Gag-GFP in a steady-state analysis is present at the plasma membrane (34, 35). However, Gag intermediate complexes may form within the cell on membranes distinct from the plasma membrane (21, 29). Recent work implicating the vacuolar protein sorting pathway in Gag trafficking suggests that Gag may target endosomal membranes (15, 32). For assessment of the presence of intracellular Gag-Gag FRET, Mel JuSo cells were cotransfected with Gag-CFP and Gag-YFP and fixed in methanol at 24 to 30 h posttransfection. Intracellular foci of fluorescence were apparent that were clearly distinct from the plasma membrane. Four regions of interest corresponding to punctate intracellular foci were isolated and analyzed for FRET by confocal microscopy (Fig. 6A). The fluorescence emission of each region was assessed after excitation at 458 nm (CFP excitation). A distinct (YFP) peak at 527 nm was demonstrated for each of these sites (Fig. 6C), representing Gag-CFP-Gag-YFP FRET. In order to verify that these sites were not simply punctate plasma membrane regions on the top or bottom of the cell, we performed optical sectioning through the z axis of a representative cell. As shown in Fig. 6B, one punctate, intracellular region of interest was isolated and analyzed for FRET in five optical slices. Efficient energy transfer between Gag molecules, shown by a peak at 527 nm, was seen only for the middle two slices of the cell, while no signal was seen for the top or bottom of the cell (Fig. 6D). Taken together, these data provide evidence for intracellular sites of Gag-Gag multimerization. Further studies are required to define the kinetics of Gag-Gag interactions at these sites and their importance in retroviral assembly.
FIG. 6.
Gag-Gag interactions are observed by FRET at intracellular locations. Gag-CFP and Gag-YFP were coexpressed in Mel JuSo cells and imaged by confocal microscopy. (A) Four separate intracellular punctate regions were analyzed for Gag-Gag FRET. The image shown is for YFP excitation (514 nm) and emission. (B) One intracellular region (arrow) was selected from a cell expressing Gag-YFP and Gag-CFP and analyzed for three-dimensional localization within the cell. Image acquisition was optimized for YFP. (C) Emission spectral scans for each region shown in panel A following data collection with CFP excitation (458-nm laser) and multichannel spectral scanning of the regions of interest. (D) Spectral data from z-section optical slices taken starting from the bottom of the cell (section 1) and proceeding to the top of the cell shown in panel B. Data were collected following excitation with a 458-nm laser and multichannel spectral scanning. Note that sections 2 and 3 demonstrate FRET (YFP peak emission) and are internal optical slices.
The I domain mediates Gag-Gag interactions.
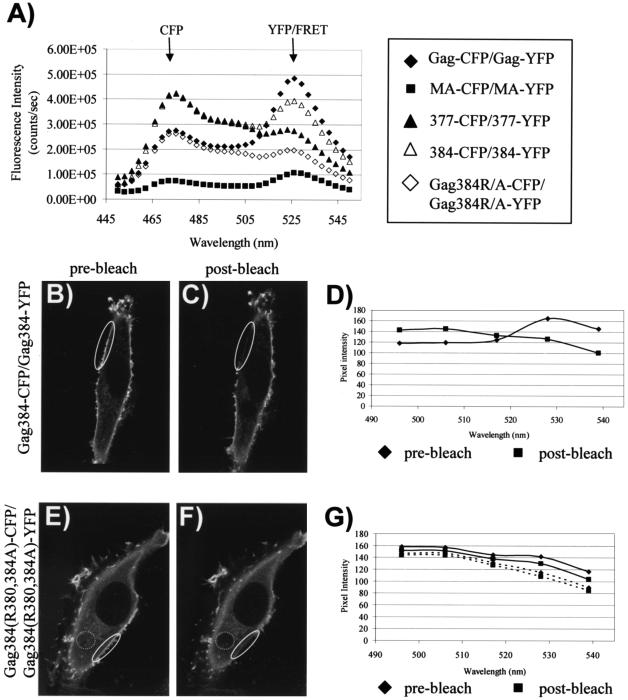
The results described above establish that FRET can be used to measure direct Gag-Gag interactions both within the cell and in VLPs. We next sought to apply this technique to define the role of the I domain. The minimal sequence of the I domain that is necessary for the formation of detergent-resistant Gag complexes and particles of normal density has been mapped to the first seven residues of the nucleocapsid region (34). Truncated molecules incorporating the first 132 (MA), 377, and 384 residues of Gag were fused to CFP and YFP (represented schematically in Fig. 1A). Substitutions of alanine for two arginine residues within the first seven residues of NC were introduced into the Gag384-CFP/YFP constructs to create Gag384(R380,384A)-CFP/YFP. The YFP versions of each Gag construct were then transfected individually or together with the corresponding CFP construct. After cells were harvested and nuclei were removed, whole-cell lysates were analyzed for the amount of YFP fluorescence emission following excitation at the YFP wavelength. We next obtained emission curves corresponding to excitation at 433 nm for each YFP-alone construct and each CFP-YFP matched pair. These curves were normalized by the total amount of YFP present so that the YFP intensities of all samples were directly comparable. We then subtracted the emission curve corresponding to the YFP-alone signal from the curve obtained with the CFP-YFP pair (Gag-CFP-Gag-YFP curve − Gag-YFP-alone curve, MA-CFP-MA-YFP curve − MA-YFP-alone curve, etc.). The resulting emission data, representing the true FRET curve with the direct YFP background stimulation subtracted, are presented in Fig. 7A. In this figure, the relative amount of peak fluorescence at 527 nm indicates the relative amount of Gag-Gag interactions occurring with each paired construct.
FIG. 7.
The I domain mediates Gag-Gag interactions. (A) Gag fusion constructs were expressed in 293T cells and harvested by Dounce homogenization. Whole-cell lysates were analyzed by fluorometry. The intensities of curves were normalized based on the amount of YFP expressed. The YFP-alone emission curve for each pair of constructs was subtracted from the curve of the CFP-YFP combination (i.e., MA-CFP-MA-YFP curve − MA-YFP-alone curve), resulting in an emission peak at 527 nm representing FRET with the background subtracted for each construct. (B) Gag384-CFP and Gag384-YFP were cotransfected in Mel JuSo cells, and images were obtained with a Zeiss LSM 510-Meta confocal microscope. The image represents YFP excitation-emission before photobleaching. (C) The same cell as that shown in panel B following photobleaching of the indicated regions at 514 nm. (D) Emission spectra were obtained from the selected regions of interest before (diamonds) and after (squares) photobleaching. The excitation wavelength for the scan was 458 nm. (E) Gag384(R380,384A)-CFP cotransfected with Gag384(R380,384A)-YFP. The image represents YFP excitation-emission. (F) The same cell as that shown in panel E following photobleaching at 514 nm. (G) Spectral data obtained from the indicated regions of panels E and F before bleaching (diamonds) and after bleaching (squares).
By this analysis, full-length Gag-CFP and Gag-YFP demonstrate a curve that is representative of FRET (Fig. 7A, filled diamonds). Gag384-YFP and Gag384-CFP also exhibit a curve representative of FRET, indicating that the N-terminal I domain is sufficient to confer protein-protein interactions between Gag molecules (Fig. 7A, open triangles). The intensity of this peak is less than that exhibited with a complete NC region, in agreement with previous studies showing that the I domain is redundant and additive within NC (34, 35). Substitution of alanine for arginine residues 380 and 384 resulted in a loss of FRET, indicating that these residues in the context of the minimal I domain are critical for Gag-Gag interactions (Fig. 7A, open diamonds). Similarly, coexpression of Gag377-CFP and Gag377-YFP, constructs lacking the I domain, resulted in a very low-level FRET signal (Fig. 7A, closed triangles). Finally, MA-CFP and MA-YFP expression showed no evidence of FRET (Fig. 7A, closed squares). We conclude from this analysis that the I domain mediates Gag-Gag interactions within cells and that Gag proteins lacking the I domain [MA, Gag377, and Gag384(R380,384A)] are predominantly monomeric.
It has been previously demonstrated that the I domain enhances the plasma membrane localization of Gag. Gag384-GFP, bearing the minimal I domain, was noted to be more prominently plasma membrane localized than Gag384(R380,384A), which lacks a functional I domain (34). However, constructs lacking the I domain still demonstrated a low degree of plasma membrane fluorescence. We therefore performed experiments to test whether the plasma-membrane-associated Gag in constructs lacking the I domain was multimeric or monomeric. To do this, we expressed Gag384-CFP plus Gag384-YFP or Gag384(R380,384A)-CFP plus Gag384(R380,384A)-YFP in Mel JuSo cells and derived spectral emission curves from the plasma membrane following excitation of CFP (458 nm). Gag384 displayed a distribution pattern similar to that of full-length Gag, but with a higher amount of diffuse cytoplasmic signal (Fig. 7B). A plasma membrane region (Fig. 7B and C, ovals) was subjected to YFP photobleaching, and emission spectra were obtained pre- and postbleaching. Prior to bleaching, a YFP (FRET) emission peak was noted (Fig. 7D, diamonds). After photobleaching, a decrease in YFP emission and an increase in CFP emission was noted, indicating the release of CFP from the FRET pair (Fig. 7D, squares). For the I-domain-deficient constructs, the same procedure was followed. We selected a cell for this analysis in which some degree of plasma membrane fluorescence was apparent by YFP excitation and emission imaging (Fig. 7E). After photobleaching, this signal was no longer apparent (Fig. 7F, solid oval). However, no significant difference in the emission scan for this region was apparent following photobleaching (Fig. 7G, diamonds versus squares). Note that the images demonstrating photobleaching (Fig. 7B, C, E, and F) were obtained with stimulation of YFP (514 nm) to document YFP photobleaching, while the spectral curves (Fig. 7D and G) were obtained with CFP excitation (458 nm) to detect FRET. As further validation of the monomeric nature of the I-domain-deficient construct, we selected a cytoplasmic region for the same analysis (Fig. 7E and F, dotted circles). Emission curves (Fig. 7G, dotted lines) again revealed minimal differences pre- and postbleaching from the cytoplasm, consistent with the presence of monomeric Gag (lack of CFP-YFP FRET). These data provide evidence that Gag384-Gag384 multimers are present on the plasma membrane, while the Gag384(R380,384A) on the plasma membrane and in the cytoplasm is predominantly monomeric. Together with the results of spectrofluorometry shown in Fig. 7A, these results provide direct evidence that the I domain is required for Gag-Gag interactions within cells.
Interactions between Gag and Tsg101 are evident by FRET.
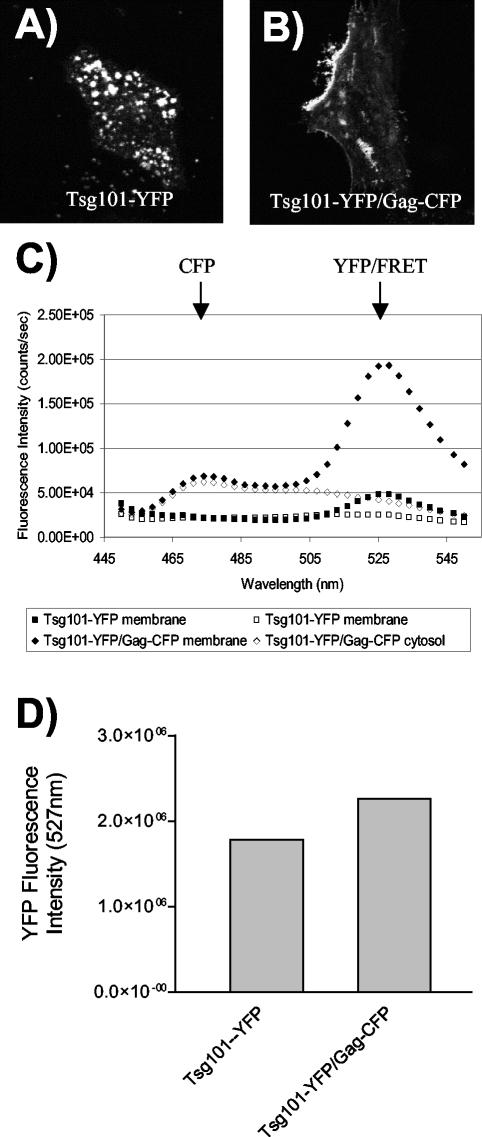
Tsg101 has been identified as a host cell factor that binds the PTAP motif within the p6 region of HIV-1 Gag (13, 15, 37). The functional significance of this interaction has been established, as downmodulation of Tsg101 resulted in a defect in the late-stage budding of immature particles and a dominant-negative truncated Tsg101 construct disrupted particle budding (8, 15). Although the Gag-Tsg101 interaction has been well documented by yeast two-hybrid analysis and by in vitro binding studies, the interaction has not been directly measured and localized within living cells. To do this, we created a Tsg101-YFP fusion protein expression construct and coexpressed this construct with Gag-CFP in Mel JuSo cells. Tsg101-YFP alone exhibited a cytoplasmic and vesicular distribution pattern (Fig. 8A). However, when Tsg101-YFP was coexpressed with Gag-CFP, the Tsg101-YFP signal was redistributed to the plasma membrane (Fig. 8B). In order to test for direct Gag-Tsg101 interactions within cells, we next performed spectrofluorometric analysis of cytoplasmic and membrane fractions. Cells expressing Tsg101-YFP alone or both Gag-CFP and Tsg101-YFP were separated into membrane and cytosolic fractions by flotation and then were analyzed for emission curves upon excitation at 433 nm. When coexpressed, Gag-CFP and Tsg101-YFP demonstrated a curve representative of FRET for the membrane fraction (Fig. 8C, closed diamonds), while Tsg101-YFP alone exhibited a low background level of excitation (Fig. 8C, closed squares). The cytosolic fractions showed no appreciable peak corresponding to the YFP emission wavelength, indicating that there was no Gag-Tsg101 interaction in the cytoplasm of cells (Fig. 8C, open diamonds). Similar levels of YFP were present in both samples, as shown by excitation of the samples at 514 nm and measurement of the emission intensity at 527 nm (Fig. 8D). These data further establish that Gag-Tsg101 interactions occur in living cells and suggest that the interaction occurs in a membrane compartment. This compartment is predominantly the plasma membrane in this steady-state analysis, as demonstrated by imaging (Fig. 8B). These data establish FRET as a useful tool for the detection of both Gag protein homomeric interactions and Gag heteromeric interactions with cellular proteins.
FIG. 8.
Gag-Tsg101 interactions are detected by FRET. (A) Tsg101-YFP was expressed in Mel JuSo cells and imaged by confocal microscopy; the image shows YFP excitation-emission. (B) Tsg101-YFP coexpressed with Gag-CFP; the image shows YFP excitation-emission. (C) Gag-CFP and Tsg101-YFP were expressed in 293T cells, and the membrane and cytosolic fractions were isolated and analyzed by scanning cuvette fluorometry. Gag-CFP-Tsg101-YFP coexpression resulted in a FRET curve for the membrane fraction but not for the cytosolic fraction (closed and open diamonds, respectively). Tsg101-YFP alone displayed no FRET curve (closed and open squares). (D) Similar expression levels and membrane binding percentages were observed for Tsg101-YFP alone and in the presence of Gag-CFP from the experiment shown in panel C.
DISCUSSION
The pathway taken by Gag en route to the plasma membrane is poorly understood. Recent studies indicated that Gag interacts directly with Tsg101, a ubiquitin enzyme 2 variant family protein that is involved in vesicular sorting pathways in the cell, suggesting that Gag may utilize components of this pathway to reach the plasma membrane (13, 15, 37). Gag molecules must form multimers during retroviral assembly, and several investigators have suggested the existence of assembly intermediate complexes of Gag multimers within the cell (21, 22, 29). However, the site of initial Gag-Gag interactions remains unknown. We have now developed a tool to address some of the key questions regarding Gag multimerization and trafficking based upon CFP-YFP FRET.
Using fluorometry-based assays, we found Gag-Gag interactions to occur almost exclusively on cellular membranes (Fig. 2). In our studies, the plasma membrane was the predominant site of Gag-Gag interactions, as measured at steady state. However, intracellular sites of Gag-Gag interactions are clearly present in cells (Fig. 6). When these data are combined with the subcellular fractionation and fluorometry results, we can conclude that these intracellular sites of Gag-Gag interaction are intracellular membranes. We suggest that these sites represent endosomes and that they may be late endosomes or multivesicular bodies similar to the late endosome assembly sites identified in human macrophages (32). Gag may form multimeric intermediates on late endosomes, and these intermediate complexes could then traffic to the plasma membrane. Alternatively, Gag multimers appearing in intracellular membrane compartments may represent a degradative pathway for misfolded or mistargeted Gag complexes. Our steady-state data do not allow us to distinguish between these possibilities. Further studies combining live cell imaging over time, colocalization with endosomal markers, and Gag-Gag FRET should help to clarify this issue.
The domains mediating Gag-Gag multimerization have previously been mapped by use of multiple techniques. Yeast two-hybrid analysis identified a large region consisting of the C-terminal third of CA and most of NC as the region mediating retroviral Gag-Gag interactions (11). The incorporation of Gag-beta-galactosidase fusion proteins into VLPs was similarly mapped to the C terminus of CA and to NC (40). Velocity sedimentation analysis suggested a role for MA, CA, and p2 in the formation of oligomers (26). Using a combination of in vitro Gag-Gag interaction experiments and particle rescue, Burniston and colleagues demonstrated that the primary determinant of Gag-Gag interactions resides in NC but also showed that the capsid dimer interface and MA can mediate multimerization in the absence of NC (4). Moreover, their work established a strong correlation between Gag-Gag interactions and the binding of Gag to RNA. A structural role for RNA in retroviral particle assembly has been supported by other data, including the ability of RNA to catalyze in vitro capsid assembly (5, 6) and the incorporation of cellular RNA into virions in the absence of packageable virion RNA (28). We believe that the domains mediating Gag-Gag multimerization and RNA interactions are inseparable from the determinant of particle density and detergent-resistant complex formation known as the I domain.
The I domain was first described by John Wills and coworkers as the domain conferring normal density upon the truncated RSV Gag molecule Bg-Bs (2). Bg-Bs itself consists of RSV MA, p2, p10, and the N-terminal half of CA and is released from cells in the form of light-density particles. The fusion of Bg-Bs with two separate fragments of HIV NC resulted in the restoration of particles with a normal density. Subsequent work established the importance of basic residues within NC to the function of the RSV I domain (3). Our laboratory has performed mapping studies of the HIV-1 I domain with Gag-GFP fusion proteins (34, 35). Those studies demonstrated that a small N-terminal segment of NC is sufficient to mediate I domain function and that two basic residues in this segment are critical to the functioning of this minimal I domain. Recently, we reported that the I domain is required for the formation of Gag complexes that can “float” in centrifugation experiments but are distinct from lipid rafts (9). A uniting hypothesis to explain the function of the I domain is that it is required for Gag-Gag multimerization in cells and that it enhances multimerization through interactions with viral or cellular RNA molecules. However, we have until now had little direct evidence that the I domain mediates Gag-Gag interactions. The present work provides substantial additional evidence. It is important to note that the same amino acid substitutions (R380,384A) that abolished I domain function in previous work (34) eliminated Gag-Gag interactions (Fig. 7), demonstrating a precise correlation between I domain function and Gag-Gag interactions measured by FRET.
We demonstrate here that the I domain is required for Gag-Gag multimerization in cells. This result may seem to be in conflict with the facts that CA readily forms dimers in vitro and that the MA structure has been solved as a trimer. The data presented here do not support the ability of these regions of Gag to form multimers in vivo in the absence of the I domain. In particular, we found no evidence for the existence of MA-MA multimers in cells by using a sensitive spectrofluorometry technique. We note, however, that the I domain may act as a nucleic acid tether, bringing other regions of Gag together that are involved in direct protein-protein interactions. Thus, the dimerization domain within CA and other regions N terminal to the I domain may be the site(s) of direct protein-protein contact, while the I domain may act indirectly to facilitate multimerization. It has been demonstrated that leucine zipper domains can substitute for the I domain in particle assembly (1, 45). Although this finding supports the role of the I domain in Gag-Gag multimerization, it does not prove that the I domain itself is the site of protein-protein contact between higher order Gag oligomers. The fact that both dimeric and trimeric leucine zippers functioned well in this context (1) may indicate that while the I domain brings Gag monomers together, protein-protein interactions within other domains allow the larger multimeric complex to form. We plan further experiments to explore this idea, using cell-based FRET assays as the readout. It should also be noted that the potential influence of the CFP or YFP tag on the mapping studies will have to be addressed by using alternative techniques to identify intracellular Gag-Gag multimers in the absence of a fusion protein.
We note that studies utilizing Gag-fluorescent protein fusions have some limitations. The precise folding time for GFP remains uncertain, but if delayed, it may significantly influence the observed intracellular location of fluorescence and of FRET. One result of this may be that our assay could miss cytosolic Gag-Gag interactions that may occur early after translation while revealing strongly the membrane-associated Gag population by virtue of delayed folding and delayed fluorescence. New methods allowing the performance of rapid pulse-chase analysis in living cells will be required to address this possibility. It is also possible that the C-terminally fluorescent proteins utilized here may themselves alter the behavior of the protein. While we do not think that this is the case for full-length Gag, we cannot be certain that some of the smaller constructs (MA-CFP/YFP, for example) act entirely as they would in the absence of the fluorescent tag. To address this point, studies are under way in our laboratory to directly compare the behavior of Gag truncation constructs with the corresponding fluorescent protein fusion constructs.
The FRET technique as applied to Gag-Gag interactions can also be applied to the interactions of Gag with cellular proteins. We have shown energy transfer between Gag and Tsg101 in fluorometry experiments (Fig. 8). Although the interaction between Gag and Tsg101 has already been established through other means, the results from these studies provide confirmation of direct protein-protein interactions between Gag and Tsg101 in a cell-based system and also demonstrate that the predominant interaction site is the plasma membrane. It is interesting that we observed a marked redistribution of Tsg101 in the presence of Gag, a finding that is consistent with recent studies by Martin-Serrano and colleagues (25). One caveat regarding this result is that we used an expressed fusion protein, rather than endogenous Tsg101, to detect this interaction. Alternative methods are needed to prove that the same effect occurs with endogenous Tsg101. Studies are currently under way to identify potential intracellular sites of Gag-Tsg101 interactions and to define the kinetics of the interaction in living cells.
In summary, we have developed a novel FRET-based assay for the monitoring of Gag-Gag interactions and of the interactions of Gag with cellular proteins. This technique has provided direct evidence that the I domain mediates Gag-Gag interactions as well as additional evidence for sites of intracellular Gag-Gag interactions. We believe that this assay will facilitate the identification of intracellular Gag protein intermediate complexes and will be a useful tool for dissecting the trafficking pathway by which Gag reaches the plasma membrane.
Acknowledgments
This work was directly supported by grant R01 AI40338. P.S. is also supported by grants AI52007, AI055441, and AI47985 and an Elizabeth Glaser Pediatric AIDS Foundation basic research grant. A.D. was supported by grant T32 CA009385.
We thank the Vanderbilt Cell Imaging Core Facility (supported by NIH grants CA68485, DK20593, and DK58404), David Piston and Sam Wells for help with FRET microscopy, and Charles Cobb for instruction and access to the PTI spectrofluorometer. We thank Mark Rizzo and the Piston lab for the donation of the YFP A206K construct. We thank Anne Kenworthy for helpful suggestions, Gary Nabel (VRC, NIH) for the gift of the codon-optimized Gag expression construct, Markus Thali (University of Vermont) for Mel JuSo cells and suggestions, and Erik Prentice for advice and assistance with confocal imaging.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, R. P., T. D. Nelle, and J. W. Wills. 1993. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 67:6487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. A., and H. Okayama. 1988. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques 6:632-638. [PubMed] [Google Scholar]
- 8.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding, L., A. Derdowski, J. J. Wang, and P. Spearman. 2003. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J. Virol. 77:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franke, E. K., H. E. Yuan, K. L. Bossolt, S. P. Goff, and J. Luban. 1994. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J. Virol. 68:5300-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 13.Freed, E. O. 2003. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 11:56-59. [DOI] [PubMed] [Google Scholar]
- 14.Garnier, L., J. B. Bowzard, and J. W. Wills. 1998. Recent advances and remaining problems in HIV assembly. AIDS 12(Suppl. A):S5-S16. [PubMed] [Google Scholar]
- 15.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 16.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 17.Huang, Y., W. P. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75:4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. P., M. Demmer-Dieckmann, T. Meo, M. R. Hadam, and G. Riethmuller. 1981. Surface antigens of human melanoma cells defined by monoclonal antibodies. I. Biochemical characterization of two antigens found on cell lines and fresh tumors of diverse tissue origin. Eur. J. Immunol. 11:825-831. [DOI] [PubMed] [Google Scholar]
- 19.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73:3079-3086. [DOI] [PubMed] [Google Scholar]
- 20.Kenworthy, A. K. 2001. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24:289-296. [DOI] [PubMed] [Google Scholar]
- 21.Lee, Y. M., B. Liu, and X. F. Yu. 1999. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J. Virol. 73:5654-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lingappa, J. R., R. L. Hill, M. L. Wong, and R. S. Hegde. 1997. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136:567-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luban, J., K. B. Alin, K. L. Bossolt, T. Humaran, and S. P. Goff. 1992. Genetic assay for multimerization of retroviral Gag polyproteins. J. Virol. 66:5157-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa, Y., D. J. Hockley, M. V. Nermut, and I. M. Jones. 2000. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morikawa, Y., W. H. Zhang, D. J. Hockley, M. V. Nermut, and I. M. Jones. 1998. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J. Virol. 72:7659-7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muriaux, D., J. Mirro, K. Nagashima, D. Harvin, and A. Rein. 2002. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 76:11405-11413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nermut, M. V., W. H. Zhang, G. Francis, F. Ciampor, Y. Morikawa, and I. M. Jones. 2003. Time course of Gag protein assembly in HIV-1-infected cells: a study by immunoelectron microscopy. Virology 305:219-227. [DOI] [PubMed] [Google Scholar]
- 30.Nydegger, S., M. Foti, A. Derdowski, P. Spearman, and M. Thali. 2003. HIV-1 egress is gated through late endosomal membranes. Traffic 4:902-910. [DOI] [PubMed] [Google Scholar]
- 31.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raposo, G., M. Moore, D. Innes, R. Leijendekker, A. Leigh-Brown, P. Benaroch, and H. Geuze. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718-729. [DOI] [PubMed] [Google Scholar]
- 33.Reicin, A. S., A. Ohagen, L. Yin, S. Hoglund, and S. P. Goff. 1996. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 70:8645-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55(Gag). J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Poblotzki, A., R. Wagner, M. Niedrig, G. Wanner, H. Wolf, and S. Modrow. 1993. Identification of a region in the Pr55gag-polyprotein essential for HIV-1 particle formation. Virology 193:981-985. [DOI] [PubMed] [Google Scholar]
- 39.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, C. T., H. Y. Lai, and C. C. Yang. 1999. Sequence requirements for incorporation of human immunodeficiency virus gag-beta-galactosidase fusion proteins into virus-like particles. J. Med. Virol. 59:180-188. [PubMed] [Google Scholar]
- 41.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H. G. Krausslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]
- 43.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913-916. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, W. H., D. J. Hockley, M. V. Nermut, Y. Morikawa, and I. M. Jones. 1996. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J. Gen. Virol. 77:743-751. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, W., and M. D. Resh. 1996. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J. Virol. 70:8540-8548. [DOI] [PMC free article] [PubMed] [Google Scholar]